Abstract
Molecular dynamics simulations using a combined quantum mechanical/molecular mechanical potential are used to determine the two-dimensional free energy profiles for the mechanism of RNA transphosphorylation in solution and catalyzed by the hairpin ribozyme. A mechanism is explored whereby the reaction proceeds without explicit chemical participation by conserved nucleobases in the active site. The ribozyme lowers the overall free energy barrier by up to 16 kcal/mol, accounting for the majority of the observed rate enhancement. The barrier reduction in this mechanism is achieved mainly by the electrostatic environment provided by the ribozyme without recruitment of active site nucleobases as acid or base catalysts. The results establish a baseline mechanism that invokes only the solvation and specific hydrogen-bonding interactions present in the ribozyme active site and provide a departure point for the exploration of alternate mechanisms where nucleobases play an active chemical role.
Keywords: ribozyme catalysis, hairpin ribozyme, electrostatic interactions
INTRODUCTION
An intriguing question in the understanding of the catalytic mechanism of RNA enzymes, or ribozymes, concerns the ability of RNA to accelerate a variety of chemical transformations without the aid of the diverse set of functional groups available to protein enzymes (Takagi et al. 2004). A number of factors have been implicated to be important for catalysis, including the involvement of divalent metal ions, or functional groups on the RNA backbone or nucleobases, that might provide electrostatic stabilization or act as general acid and base catalysts (Lönnberg and Lönnberg 2005; Sigel and Pyle 2007). Nonetheless, there currently exists no general consensus as to the origin of the catalytic proficiency exhibited by ribozymes nor any detailed mechanism that has been unambiguously determined. The hairpin ribozyme (HPR) (Walter and Burke 1998; Lilley 1999), which catalyzes the reversible, site-specific phosphodiester bond cleavage of an RNA substrate, is unique in that the chemical steps of the reaction do not require a specific metal ion in the active site (Walter and Burke 1998; Lilley 1999; Doherty and Doudna 2001; Rupert et al. 2002; Bevilacqua 2003) but is able to achieve rate acceleration similar to other metal-dependent ribozymes (Fedor 2000). There is an active debate regarding the origins of the catalytic proficiency of HPR, focusing mostly on the role of active site nucleobases in general acid and base catalysis (Hampel and Cowan 1997; Nesbitt et al. 1999; Fedor 2000; Pinard et al. 2001; Ryder and Strobel 2002; Kuzmin et al. 2005; Cottrell et al. 2007; Tang et al. 2007). X-ray crystallographic structures of HPR provide valuable insight into the active site conformation in the reactant state (Rupert and Ferré-D'Amaré 2001; Salter et al. 2006), transition state, and product state mimic structures (Rupert et al. 2002; Torelli et al. 2007). As yet, there are no studies that focus on the effects of the solvated electrostatic environment of the ribozyme itself on catalysis. The lack of an explicit metal ion requirement (Fedor and Williamson 2006) makes the HPR an ideal target for theoretical studies aimed to characterize the contribution of “generalized solvation” on catalysis.
In this report, we show that the electrostatic environment provided by solvated HPR active site lowers the cleavage activation barrier up to 9 kcal/mol relative to the uncatalyzed transphosphorylation barrier in water. We also explore the mechanistic scenarios whereby G8 acts as a general base to activate the 2′-hydroxyl nucleophile, as implicated by experiment (Pinard et al. 2001; Rupert et al. 2002; Wilson et al. 2006; Cottrell et al. 2007). The results suggest that the electrostatic environment of the solvated ribozyme active site contributes significantly in achieving 106–107-fold rate enhancement of the phosphodiester cleavage (Nesbitt et al. 1999; Fedor 2000; Kuzmin et al. 2005) relative to the uncatalyzed, but spontaneous, cleavage of RNA molecule in aqueous solution (Hertel et al. 1997; Li and Breaker 1999). Without the aid of a divalent metal ion or direct participation of nucleobase functional groups as a general acid or a general base, the majority of the observed rate enhancement can be realized through specific hydrogen-bonding interactions (provided from G8 and other nucleobases) and nonspecific electrostatic interactions of the solvated ribozyme active site. In the discussion that follows, the term “electrostatic solvation” is used to discuss the electrostatic component of the “generalized solvation” provided by the ribozyme environment.
RESULTS AND DISCUSSION
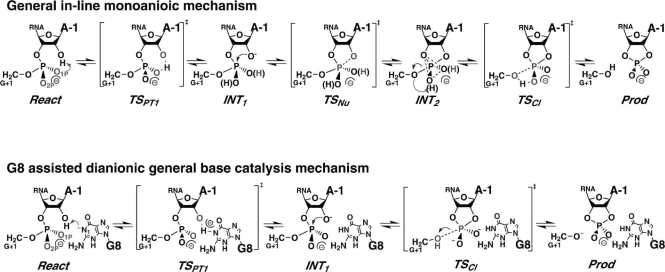
The HPR mechanisms considered in the present work are depicted in Scheme 1 for the in-line monoanionic catalysis mechanism and the dianionic general base catalysis mechanism. The in-line monoanionic reaction mechanism involves three reaction steps: (1) an initial intramolecular proton transfer from the 2′-hydroxyl group to either the pro-R (O1P) or the pro-S (O2P) nonbridging oxygen atoms of the scissile phosphate group, (2) a nucleophilic attack from the 2′-hydroxyl oxygen at the phosphate center, and (3) an exocyclic bond-cleavage of the leaving group from the phosphate center along with a second intramolecular proton transfer from the phosphate nonbridging oxygen to the leaving group. The in-line monoanionic mechanism only invokes generalized solvation and specific hydrogen-bond interactions for both catalyzed and uncatalyzed reactions. This mechanism is ideally suited to address the role of the ribozyme environment as a general solvent in the catalysis, which is the main focus of this report. The dianionic general base mechanism tests the effect of G8 acting as a general base by abstracting the proton from the 2′-hydroxyl group to activate the nucleophile and extends the scope of previous work (Nam et al. 2008). In these two mechanisms, the proton transfer reactions and the nucleophilic substitution step (nucleophilic attack and cleavage) can be either concerted or stepwise, and it is a goal in mechanistic enzymology to characterize the nature of the reaction sequence. This can be accomplished computationally by the determination of two-dimensional reaction free energy profiles for the proton transfer and nucleophilic substitution reaction coordinates. In this report, we perform molecular dynamics (MD) free energy simulations with a combined quantum mechanics/molecular mechanics (QM/MM) potential along with density functional theory (DFT) calculations to determine two-dimensional free energy surfaces from which the stepwise/concerted nature of each step can be ascertained. In addition, the electrostatic solvation free energies are determined for the reactant state, intermediate states, transition states, and product state for both catalyzed and uncatalyzed reactions to address the effects of electrostatic environment provided by the ribozyme on the reaction. Details are provided in Materials and Methods.
SCHEME 1.
General in-line monoanionic mechanism (top) and G8-assisted dianionic general base catalysis mechanism (bottom) of phosphodiester cleavage transesterification catalyzed by the hairpin ribozyme. In the in-line monoanionic mechanism, the first proton transfer (PT1), the nucleophilic attack (Nu), and the exocyclic cleavage (Cl) steps are shown. For the uncatalyzed model reaction in solution, the O1P and O2P pathways are energetically equivalent. In the G8-assisted dianionic general base catalysis mechanism, the deprotonated G8 acts as a general base to activate 2′-hydroxyl group, and the reaction proceeds to the product without forming INT2.
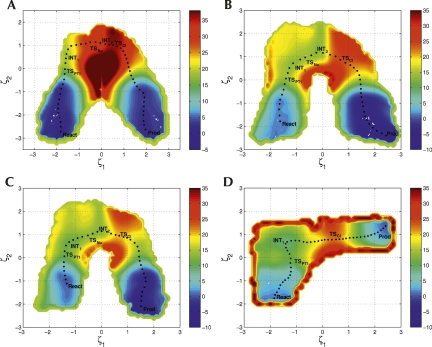
Figure 1, A–C, shows the two-dimensional free energy profiles for the uncatalyzed model transphosphorylation reaction in aqueous solution, and catalyzed by the HPR along O1P and O2P pathways (Scheme 1), and Figure 1D shows the two-dimensional free energy profile for the dianionic general base catalytic reaction. Table 1 lists free energy values corresponding to stationary points along the minimum free energy path on the surface, determined from the QM/MM free energy simulations, along with corrections at the DFT level to the activation and reaction free energy values.
FIGURE 1.
Two-dimensional free energy surfaces for in-line monoanionic mechanisms for the (A) uncatalytic model reaction in solution, the catalytic (B) O1P and (C) O2P pathways in the hairpin ribozyme (HPR), and the (D) G8-assisted dianionic general base catalytic mechanism of the HPR, in which the 2′-proton is transferred to the N1 of a deprotonated G8. For both mechanisms, ζ1 is defined as R P − O5′ − R P − O2′. In the in-line monoanionic mechanism, ζ2 is R O2′ − H2′ − R ONB − H2′ for ζ1 < 0.0 Å and R O5′ − H2′ − R ONB − H2′ for ζ1 > 0.0 Å, where ONB is for the O1P proton transfer in (B), and for the O2P proton transfer in (A) and (C), respectively; in the G8-assisted general base catalysis mechanism, ζ2 is R O2′ − H2′ − R N 1 − H2′. The units for free energies and distances are kcal/mol and Å, respectively.
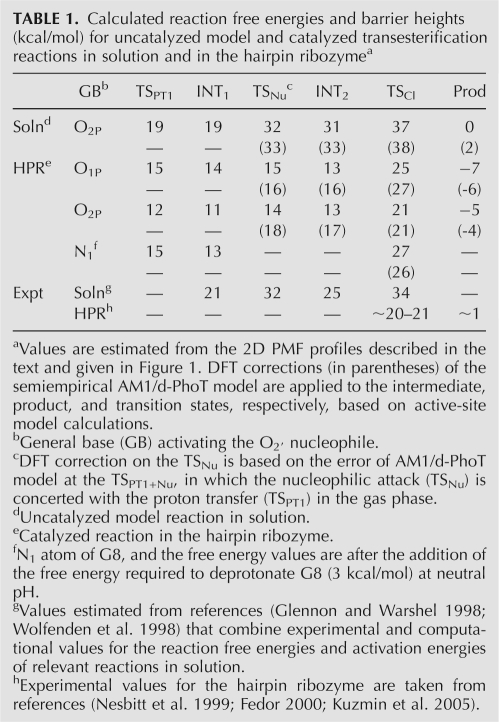
TABLE 1.
Calculated reaction free energies and barrier heights (kcal/mol) for uncatalyzed model and catalyzed transesterification reactions in solution and in the hairpin ribozymea
In the in-line monoanionic reaction mechanism, the overall reaction may be characterized by a sequence of proton transfer, nucleophilic attack, exocyclic cleavage, and proton transfer reactions (see Fig. 1A–C); a shallow minimum exists prior to both the second and third steps. Nevertheless, it is difficult to characterize the first two steps (the proton transfer to the either nonbridging oxygens and the nucleophilic attack) as purely stepwise because the barriers for the reverse process are expected to lie below the zero-point vibrational energies if nuclear quantum effects are considered. The rate-limiting step is the exocyclic bond cleavage of the leaving group from the phosphorus atom, followed by a barrierless proton transfer to the departing O5′ alkoxide anion. The same trend is found both for the catalyzed reaction in the ribozyme and the uncatalyzed model reaction in water, but the HPR markedly lowers the reaction barriers for each of the three reaction steps and the free energies of the resulting intermediates. The greatest barrier reduction occurs in the nucleophilic attacking step (net decreases of 17 kcal/mol for the O1P pathway and 18 kcal/mol for the O2P pathway relative to the reaction in water). For the rate-limiting step, the free energy barriers are lowered by 12 and 16 kcal/mol along the O1P and O2P pathways, respectively, while experimental estimation of barrier reduction is 13–14 kcal/mol (Table 1). After the density functional correction at the B3LYP/6-311++G(3df,2p) (Lee et al. 1988; Becke 1993) level, the overall free energy barrier becomes 27 kcal/mol for the O1P pathway and 21 kcal/mol for the O2P pathway. These results are in accord with the experimental estimate of 20–21 kcal/mol (Nesbitt et al. 1999; Fedor 2000; Kuzmin et al. 2005). Nonetheless, this does not preclude alternate mechanisms with explicit nucleobase involvement, such as A38 and G8 acting as general acid and general base catalysts (Rupert et al. 2002; Ferré-D'Amaré 2004; Cottrell et al. 2007), respectively, which could further lower the barrier.
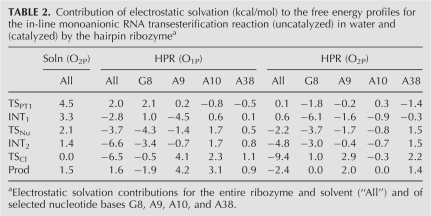
In order to address the question as to what extent the electrostatic environment and nonspecific interactions alone can account for the observed rate enhancement, electrostatic solvation free energies are computed at various stationary points on the free energy surface and are compared to the values at the reactant state. Table 2 lists the free energy contribution of the electrostatic solvation provided by the ribozyme and aqueous solution environment in stabilizing the intermediates and transition states. We also compare the free energy contribution to solvation stabilization of selected nucleotide bases at each stationary point. For the uncatalyzed reaction, water solvation does not significantly affect the exocyclic cleavage barrier (TSCl), while the same barriers are lowered significantly by the electrostatic solvation at the active site of the HPR (−6.5 kcal/mol for the O1P pathway and −9.4 kcal/mol for the O2P pathway). The results suggest that the electrostatic environment of the ribozyme active site can account for the majority of the experimentally observed rate enhancement without chemical participation by active site nucleobases. Each of the four nucleobases (G8, A9, A10, and A38) at the HPR active site individually contributes a relatively small amount in the direct lowering of the exocyclic cleavage barrier via electrostatic solvation. However, this does not imply a minor role for these nucleobases in the reaction. As previous studies have indicated they interact strongly with the scissile phosphate group to provide a proper solvation (Nam et al. 2008), which is critical to achieve stabilization of the exocyclic cleavage step. It is likely that these interactions are not fully compensated by the solvent in the abasic substitution experiments at these positions.
TABLE 2.
Contribution of electrostatic solvation (kcal/mol) to the free energy profiles for the in-line monoanionic RNA transesterification reaction (uncatalyzed) in water and (catalyzed) by the hairpin ribozymea
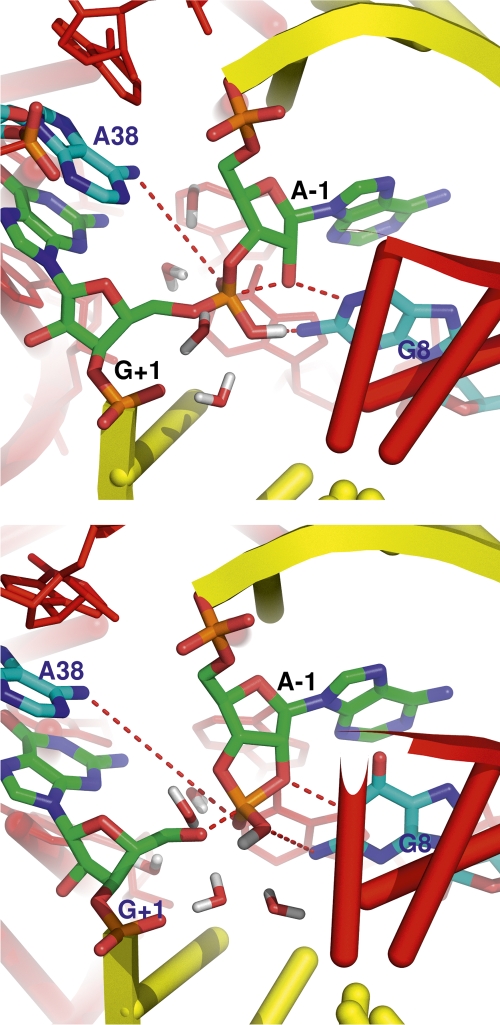
In the HPR-catalyzed reaction, the overall barrier for the in-line monoanionic O2P pathway is lower than that for the O1P pathway by 4 and 6 kcal/mol at the AM1/d-PhoT and density functional B3LYP/6-311++G(3df,2p)-corrected levels of theory, respectively. This difference is statistically significant, and two factors appear to contribute to this difference: (1) the asymmetric interactions provided by the ribozyme and surrounding waters to the two nonbridging oxygens and the leaving O5′ oxygen, and (2) an intramolecular dipole–dipole interactions at the exocyclic cleavage step. The asymmetric interactions are expected from inspection of X-ray crystallographic data (Rupert et al. 2002; Torelli et al. 2007) that exhibit an asymmetry of binding interactions of the scissile phosphate with the surrounding nucleobases, which, in turn, affect the water solvation at the O5′ leaving group. In the O2P pathway, the O5′ and O1P atoms, which share a −1 formal charge at the exocyclic cleavage transition state, point away from each other; thus, surrounding water molecules can solvate them effectively. In the O1P pathway, the O5′ and O2P atoms are oriented in the same direction and compete for interactions with surrounding water molecules. Figure 2 shows a network of water molecules in the active site that forms hydrogen bonds with the nonbridging phosphoryl oxygens and leaving group in O2P pathway, that, together with nucleobases in the active site that form hydrogen bonds with the scissile phosphate, stabilize the transition state. Similar roles for water-mediated hydrogen-bond networks in ribozyme active site have been previously suggested (Rhodes et al. 2006; Torelli et al. 2007). For the intramolecular contribution, the proton that has been transferred to the O2P at earlier step is pointing toward the O5′ leaving group at the exocyclic cleavage transition state in the O2P pathway so as to form an antiparallel dipole pair (see Fig. 2), while such dipole alignment is prevented in the O1P pathway due to steric hindrance with C5′.
FIGURE 2.
Snapshots of the active-site structures near the transition state of (top) the nucleophilic attack and (bottom) the exocyclic cleavage for the in-line monoanionic O2P mechanism of cleavage transesterification in the hairpin ribozyme. The yellow- and red-colored cartoons represent the substrate and ribozyme strands, respectively, and water molecules interacting with nonbridging oxygens and O5′ are also shown.
For the dianionic general base catalysis mechanism, we assume here that a deprotonated G8 abstracts a proton from the 2′-hydroxyl nucleophile (2′-OH) as a general base (GB), and the leaving 5′-alkoxide group is not yet protonated after the exocyclic cleavage step (see Fig. 1D). The reaction steps are similar to the in-line monoanionic reaction, except no stable intermediate (INT2) is formed and the reaction proceeds directly to the exocyclic cleavage in this mechanism. This is due to a nature of the dianionic phosphorane that is less stable than the monoprotic phosphorane (Perreault and Anslyn 1997; Zhou and Taira 1998). Although there exists a possibility that G8 is tautomerized to act as a general base, the results from our previous study (Nam et al. 2008) do not support this scenario. Since the starting point in the general base catalysis mechanism is a deprotonated G8, the free energy required for the deprotonation of G8 must be taken into account. By assuming a pK a of 9.2 of the free guanine base (Bloomfield et al. 2000), a rough estimate of 3 kcal/mol is obtained for the free energy required to deprotonate G8 at neutral pH. The free energy values for the general base (N1) mechanism shown in Table 1 include this estimated free energy correction. Overall, the in-line monoanionic O2P pathway has lower free energy barrier compared to the dianionic general base catalysis mechanism (5 kcal/mol for the TSCl step), while the step to activate the nucleophile shows similar free energies and barrier heights for both catalytic mechanisms. The preference for the in-line monoanionic O2P pathway relative to the general base pathway cannot be considered quantitative due to the need for a more accurate determination of the pK a of G8 in the ribozyme environment.
The present study explores the effects by which the change of the electrostatic environment provided by the HPR relative to that of aqueous solution affects the rate of the transphosphorylation reaction. Since the specific mechanisms explored here do not involve direct intervention of any nucleobases as a general base or general acid in the catalysis, the computed change in the free energy barriers is mostly due to the change of the heterogeneous electrostatic environment in the HPR active site relative to that of bulk solvation by water. The direct electrostatic solvation by the ribozyme and water lowers the overall free energy barrier by 7 and 9 kcal/mol, respectively, for the two reaction paths, corresponding to an initial proton transfer to either of the two nonbridging phosphate oxygen atoms. The results suggest that the nonspecific interactions in HPR are sufficient to account for the majority of the observed change of barrier heights without the involvement of a metal ion and general acid–base catalysis by active site nucleobases. Of course, the specific arrangements of the nucleobases as well as the phosphoribose backbones and active site water molecules are essential to provide the electrostatic effects that ultimately lower the free energy barrier in the ribozyme. Specific deletions of key nucleobases would disrupt this environment and, consequently, affect the reaction rate as observed experimentally (Kuzmin et al. 2004, 2005; Cottrell et al. 2007). For example, deletion of the nucleobase A38 reduces the self-cleavage reaction rate of HPR more than 104-fold (Kuzmin et al. 2005), which has been tested from our previous computational study (Nam et al. 2008). It has been proposed that a protonated A38 may act as a general acid in the rate-limiting step (Rupert et al. 2002; Bevilacqua 2003; Ferré-D'Amaré 2004; Kuzmin et al. 2005), in which case in the absence of the availability of an alternate reaction path excluding the direct involvement of protonated A38 this might imply an even greater rate reduction. The present results indicate that in the absence of a protonated A38, the HPR can exploit an alternative reaction path that involves specific hydrogen bonding interactions with active site nucleobases to achieve catalysis.
The in-line monoanionic mechanism studied in the present work establishes a baseline mechanism that invokes only the generalized solvation and specific hydrogen bonding interactions provided by the ribozyme environment and provides a departure point for the exploration of alternate mechanisms where participation of nucleobases in the active site play an active chemical role. The results underscore the importance of hydrogen bonding interactions provided by the nucleobases and network of solvent molecules in the active site. The elucidation of the mechanism of HPR represents an important step toward the understanding of the general principles that govern catalysis of more complex RNA systems.
MATERIALS AND METHODS
Simulations are based on the second transition state analog crystal structure (PDB code 1M5O) (Rupert et al. 2002) and performed using CHARMM (version c32a2) (Brooks et al. 1983). Nucleotides, crystal waters, and Ca2+ ions 25 Å away from the scissile phosphate are removed, leaving 77 nucleotides, three Ca2+, and 18 waters. A 25-Å sphere of 1607 water molecules solvates the ribozyme centered at the scissile phosphate, and 41 Na+ ions are included to partially neutralize the system. In the QM/MM calculations, we used the all-atom CHARMM27 nucleic acid force field (Foloppe and MacKerell 2000) and TIP3P water model (Jorgensen et al. 1983), with the AM1/d-PhoT quantum model (Nam et al. 2007) and generalized hybrid orbital method (Gao et al. 1998) for treatment of the QM/MM boundary. After initial energy minimizations, stochastic boundary molecular dynamic simulations (Brooks et al. 1985) are performed (Langevin dynamics is applied to atoms in the radial range between 21 and 25 Å) at 300 K and a 1-fs integration time step, and the SHAKE algorithm (Ryckaert et al. 1977) is used to constrain all bond lengths involving hydrogen atoms in the molecular mechanical region. Free energy profiles are constructed from the sampled configurations from two-dimensional umbrella sampling (Torrie and Valleau 1977) MD simulations by using the weighted histogram analysis method (Kumar et al. 1992). In the umbrella sampling, we define the reaction coordinate for the nucleophilic process as the difference between the breaking and forming bonds with the phosphorus atom, i.e., ζ1 = R P − O5′ − R P − O2′, for both catalytic mechanisms tested. In the in-line monoanionic reactions, the proton transfer reaction coordinates are defined as ζ2 = R OX′ − H2′ − R ONB − H2′, where ONB is either O1P or O2P, and OX ′ is O2′ in the first proton transfer step and O5′ in the second proton transfer step, respectively. The total number of umbrella sampling windows is 395 for the O1P pathway and 344 for the O2P pathway, each simulation of which is run for 17 ps for equilibration and 50 ps for configuration sampling. The total simulation times are 26.5 ns and 23.0 ns for the O1P and O2P pathways, respectively. In the dianionic general base catalytic reaction, the proton transfer reaction coordinate is defined as ζ2 = R O2′ − H2′ − R N1 − H2′, where N1 is the N1 atom of G8. The total number of umbrella sampling windows is 269, and each window is sampled for 50 ps after the 17 ps of equilibration. The total simulation time is 18.0 ns. For the uncatalyzed model reaction, the reactant molecule, 2′-hydroxyethyl methyl phosphate, is solvated with a 40-Å3 cubic box of 2038 water molecules and one Na+ ion. Simulations are carried out using QM/MM-Ewald scheme (Nam et al. 2005) at 1 atm and 300 K. To simulate the chemical reaction, a total of 338 umbrella sampling MD simulations are run each for 5 ps of equilibration and 30 ps of configuration sampling, resulting a total of 11.8 ns of MD simulations.
The computed free energy values are further refined by density functional quantum chemical calculations on the active site reaction models. The density functional adiabatic energies are computed at the B3LYP/6-311++G(3df,2p)//B3LYP/6-31++G(d,p) level, in which “//” separates the level for the refined single point energy from the level for geometry optimization. The same geometry optimizations are carried out also at the AM1/d-PhoT level in order to derive the correction factors. The correction factors for the rate-controlling transition states are in general small, ranging in magnitude between 0.2 and 2.2 kcal/mol (a full comparison of the computed reaction energies and geometrical quantities calculated at the DFT and AM1/d-PhoT levels is provided as supplemental research data). The active site model reactants for the uncatalyzed and catalyzed reactions include the nucleophilic ribose, the scissile phosphate, and the leaving ethyl group.
The free energy of electrostatic solvation is computed by using free energy perturbation (FEP) approach (Gao 1992), in which the electrostatic interactions between quantum mechanical and molecular mechanical regions are scaled from 1 to 0. The scaling value of 1 represents the state of full solute–solvent interactions, and the value of 0 represents the state of no solute–solvent interactions. In order to mimic the electrostatic solvation outside the 25-Å water sphere, we have applied a variational electrostatic projection method (Gregersen and York 2005) with a conductor-like screening solvation model reaction field (Gregersen et al. 2005; Khandogin et al. 2005), in which the component for the QM-reaction field interaction is scaled in accord with other QM/MM electrostatic interactions during FEP MD simulations. A series of FEP MD simulations are carried out at each stationary point along the minimum free energy path for the uncatalyzed and catalyzed in-line monoanionic reactions, respectively; at a given scaling value, 100-ps FEP MD simulations are carried out for statistical averaging.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was partially supported by grants from the National Institutes of Health (GM62248 to D.M.Y. and GM46736 to J.G.), the IBM–Rochester Life Sciences Group (to D.M.Y), the University of Minnesota Biomedical Informatics and Computational Biology program (to D.M.Y.), and the Office of Naval Research (ONR, grant N00012-05-01-0538). Computational resources were provided by the Minnesota Supercomputing Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.863108.
REFERENCES
- Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- Bevilacqua P.C. Mechanistic considerations for general acid–base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bloomfield V.A., Crothers D.M., Tinoco I., Jr . Nucleic acids: Structures, properties, and functions. University Science Books; Sausalito, CA: 2000. [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- Brooks C.L., III, Brunger A., Karplus M. Active site dynamics in protein molecules: A stochastic boundary molecular dynamics approach. Biopolymers. 1985;24:843–865. doi: 10.1002/bip.360240509. [DOI] [PubMed] [Google Scholar]
- Cottrell J.W., Kuzmin Y.I., Fedor M.J. Functional analysis of hairpin ribozyme active site architecture. J. Biol. Chem. 2007;282:13498–13507. doi: 10.1074/jbc.M700451200. [DOI] [PubMed] [Google Scholar]
- Doherty E.A., Doudna J.A. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- Fedor M.J. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- Fedor M.J., Williamson J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2006;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- Foloppe N., MacKerell A.D., Jr All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21:86–104. [Google Scholar]
- Gao J. Absolute free energy of solvation from Monte Carlo simulations using combined quantum and molecular mechanical potentials. J. Phys. Chem. A. 1992;96:537–540. [Google Scholar]
- Gao J., Amara P., Alhambra C., Field M.J. A generalized hybrid orbital (GHO) method for the treatment of boundary atoms in combined QM/MM calculations. J. Phys. Chem. A. 1998;102:4714–4721. [Google Scholar]
- Glennon T.M., Warshel A. Energetics of the catalytic reaction of ribonuclease A: A computational study of alternative mechanisms. J. Am. Chem. Soc. 1998;120:10234–10247. [Google Scholar]
- Gregersen B.A., Khandogin J., Thiel W., York D.M. Smooth solvation method for d-orbital semiempirical calculations of biological reactions. 2. Application to transphosphorylation thio effects in solution. J. Phys. Chem. B. 2005;109:9810–9817. doi: 10.1021/jp044061l. [DOI] [PubMed] [Google Scholar]
- Gregersen B.A., York D.M. Variational electrostatic projection (VEP) methods for efficient modeling of the macromolecular electrostatic and solvation environment in activated dynamics simulations. J. Phys. Chem. B. 2005;109:536–556. doi: 10.1021/jp0469968. [DOI] [PubMed] [Google Scholar]
- Hampel A., Cowan J.A. A unique mechanism for RNA catalysis: The role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- Hertel K.J., Peracchi A., Uhlenbeck O.C., Herschlag D. Use of intrinsic binding energy for catalysis by an RNA enzyme. Proc. Natl. Acad. Sci. 1997;94:8497–8502. doi: 10.1073/pnas.94.16.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Khandogin J., Gregersen B.A., Thiel W., York D.M. Smooth solvation method for d-orbital semiempirical calculations of biological reactions. 1. Implementation. J. Phys. Chem. B. 2005;109:9799–9809. doi: 10.1021/jp044062d. [DOI] [PubMed] [Google Scholar]
- Kumar S., Bouzida D., Swendsen R.H., Kollman P.A., Rosenberg J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Cottrell J.M., Fedor M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Fedor M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Lee C., Yang W., Parr R.G. Development of the Colle–Savetti correlation energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Li Y., Breaker R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- Lilley D.M.J. Structure, folding, and catalysis of the small nucleolytic ribozymes. Curr. Opin. Struct. Biol. 1999;9:330–338. doi: 10.1016/S0959-440X(99)80044-X. [DOI] [PubMed] [Google Scholar]
- Lönnberg T., Lönnberg H. Chemical models for ribozyme action. Curr. Opin. Chem. Biol. 2005;9:665–673. doi: 10.1016/j.cbpa.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Nam K., Cui Q., Gao J., York D.M. Specific reaction parametrization of the AM1/d Hamiltonian for phosphoryl transfer reactions: H, O, and P atoms. J. Chem. Theory Comput. 2007;3:486–504. doi: 10.1021/ct6002466. [DOI] [PubMed] [Google Scholar]
- Nam K., Gao J., York D.M. An efficient linear-scaling Ewald method for long-range electrostatic interactions in combined QM/MM calculations. J. Chem. Theory Comput. 2005;1:2–13. doi: 10.1021/ct049941i. [DOI] [PubMed] [Google Scholar]
- Nam K., Gao J., York D.M. Molecular simulation study of the mechanism of hairpin ribozyme catalysis. J. Am. Chem. Soc. 2008;130:4680–4691. doi: 10.1021/ja0759141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt S.M., Erlacher H.A., Fedor M.J. The internal equilibrium of the hairpin ribozyme: Temperature, ion and pH effects. J. Mol. Biol. 1999;289:1009–1024. doi: 10.1006/jmbi.1999.2543. [DOI] [PubMed] [Google Scholar]
- Perreault D.M., Anslyn E.V. Unifying the current data on the mechanism of cleavage–transesterification of RNA. Angew. Chem. Int. Ed. Engl. 1997;36:432–450. [Google Scholar]
- Pinard R., Hampel K.J., Heckman J.E., Lambert D., Chan P.A., Major F., Burke J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes M.M., Réblová K., Sponer J., Walter N.G. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc. Natl. Acad. Sci. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert P.B., Ferré-D'Amaré A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert P.B., Massey A.P., Sigurdsson S.T., Ferré-D'Amaré A.R. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- Ryder S.P., Strobel S.A. Comparative analysis of hairpin ribozyme structures and interference data. Nucleic Acids Res. 2002;30:1287–1291. doi: 10.1093/nar/30.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter J., Krucinska J., Alam S., Grum-Tokars V., Wedekind J.E. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel R.K.O., Pyle A.M. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- Takagi Y., Ikeda Y., Taira K. Ribozyme mechanisms. Top. Curr. Chem. 2004;232:213–251. [Google Scholar]
- Tang C.L., Alexov E., Pyle A.M., Honig B. Calculation of pK as in RNA: On the structural origins and functional roles of protonated nucleotides. J. Mol. Biol. 2007;366:1475–1496. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Torelli A.T., Krucinska J., Wedekind J.E. A comparison of vanadate to a 2′–5′ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- Walter N.G., Burke J.M. The hairpin ribozyme: Structure, assembly, and catalysis. Curr. Opin. Chem. Biol. 1998;2:24–30. doi: 10.1016/s1367-5931(98)80032-x. [DOI] [PubMed] [Google Scholar]
- Wilson T.J., Ouellet J., Zhao Z., Harusawa S., Araki L., Kurihara T., Lilley D.M.J. Nucleobase catalysis in the hairpin ribozyme. RNA. 2006;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R., Ridgway C., Young G. Spontaneous hydrolysis of ionized phosphate monoesters and diesters and the proficiencies of phosphatases and phosphodiesterases as catalysts. J. Am. Chem. Soc. 1998;120:833–834. [Google Scholar]
- Zhou D.-M., Taira K. The hydrolysis of RNA: From theoretical calculations to the hammerhead ribozyme-mediated cleavage of RNA. Chem. Rev. 1998;98:991–1026. doi: 10.1021/cr9604292. [DOI] [PubMed] [Google Scholar]