Abstract
Fragile X mental retardation syndrome, the most common form of inherited mental retardation, is caused by the absence of the fragile X mental retardation protein (FMRP). FMRP has been shown to use its arginine–glycine–glycine (RGG) box to bind to a subset of RNA targets that form a G quadruplex structure. We performed a detailed analysis of the interactions between the FMRP RGG box and the microtubule associated protein 1B (MAP1B) mRNA, a relevant in vivo FMRP target. We show that MAP1B RNA forms an intramolecular G quadruplex structure, which is bound with high affinity and specificity by the FMRP RGG box. We determined that hydrophobic interactions are important in the FMRP RGG box-MAP1B RNA association, with minor contributions from electrostatic interactions. Our findings that at low protein:RNA ratios the RNA G quadruplex structure is slightly stabilized, whereas at high ratios is unfolded, suggest a mechanism by which the FMRP concentration variation in response to a neurotransmitter stimulation event could act as a regulatory switch for the protein function, from translation repressor to translation activator.
Keywords: FMRP, fragile X syndrome, G quadruplex, MAP1B RNA, RGG box
The lack of fragile X mental retardation protein (FMRP) results in the fragile X syndrome, the most prevalent inherited mental disorder (Crawford et al. 2001). The absence of FMRP is due to the transcriptional inactivation of the fmr1 gene, caused by an unstable expansion of a CGG trinucleotide repeat in its 5′-untranslated region (UTR) (Jin and Warren 2000; O'Donnell and Warren 2002). The function of FMRP has been extensively studied, however its cellular role and how its loss causes mental retardation is still not fully understood. It is believed that FMRP is a translational regulator for specific messenger RNAs (mRNAs), and it has been shown that it is associated with actively translating ribosomes (Antar and Bassell 2003; Jin and Warren 2003; Massimiliano et al. 2004). Nucleic acid chaperone properties have also been attributed to this protein (Gabus et al. 2004). This 632 amino acid protein has two types of RNA binding domains: two K-homology (KH) domains and one arginine–glycine–glycine (RGG) box, suggesting that FMRP exerts its function through RNA binding (Siomi et al. 1993).
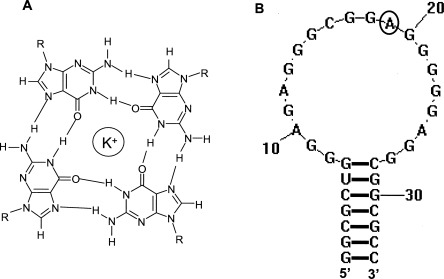
Several studies reported that FMRP uses its RGG box to bind with high affinity to mRNAs rich in G content that could adopt more complex G quadruplex structures (Brown et al. 2001; Darnell et al. 2001; Schaeffer et al. 2001; Ramos et al. 2003). Formed from four guanine residues, a G quartet has a planar conformation stabilized by Hoogsteen base pairs (Fig. 1A). Several such planar structures can stack to form G quadruplexes, which are stabilized by potassium or sodium cations, but are disrupted in the presence of lithium cations (Davis 2004; Hazel et al. 2004; Mergny et al. 2005). It has also been shown that FMRP uses its KH2 domain to bind to a synthetic RNA that harbors a loop–loop kissing complex (Darnell et al. 2005). However, despite these extensive efforts, the in vivo FMRP mRNA targets are still not clearly characterized. This study contributes to the understanding of the principles of recognition between FMRP and its G quadruplex forming mRNA targets, by analyzing at the molecular level its interactions with the microtubule associated protein 1B (MAP1B) mRNA. This RNA encodes for MAP1B, a protein involved in modulating microtubule dynamics, neurite extension, and synapse development (Gonzalez-Billault et al. 2004). A body of evidence supports the proposal that MAP1B mRNA is a relevant in vivo FMRP target: FMRP binds MAP1B mRNA in vitro (Darnell et al. 2001; Ramos et al. 2003), and the Drosophila homolog of FMRP reduces the levels of Futsch, a microtubule-associated protein homolog of the mammalian MAP1B (Zhang et al. 2001). Moreover, two in vivo studies show that FMRP negatively regulates the translation of MAP1B and controls the microtubule stability in brain neuron development (Zalfa et al. 2003; Lu et al. 2004). In addition, FMRP has been shown to co-localize with ribosomes, ribosomal RNA, and MAP1B mRNA (Antar et al. 2005).
FIGURE 1.
(A) Structure of a G quartet. (B) Secondary structure of the MAP1B RNA fragment (34-mer) used in this study, which was generated using Zuker's algorithm (2003). The adenine at position 19 (circled) in the MAP1B RNA structure was replaced with 2-aminopurine in MAP1B-19AP RNA.
We show that MAP1B RNA adopts an intramolecular G quadruplex structure which is bound with high affinity and specificity by the FMRP RGG box. We worked with the FMRP RGG box domain; however, the findings of this study should be relevant to the full-length FMRP, since the full-length FMRP and its isolated RGG box domain bind with the same affinity to G quadruplex forming RNA (Darnell et al. 2001). Our thermodynamic analysis reveals that the association between the FMRP RGG box and MAP1B RNA is dominated by hydrophobic interactions, with minor contributions from electrostatics. Interestingly, we find that superstoichiometric amounts of the FMRP RGG box unfold the MAP1B G quadruplex structure, suggesting a mechanism by which the control of the FMRP concentration might facilitate its translation regulator function.
In addition to its relevance in the context of the fragile X syndrome, this study contributes to our understanding of the specific interactions of the RGG box domain with RNA, since previously it was thought that this RNA binding domain interacts with RNA in a nonspecific manner.
RESULTS AND DISCUSSION
MAP1B RNA forms an intramolecular G quadruplex structure
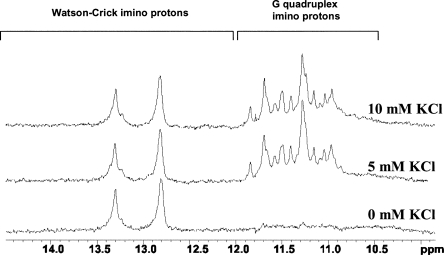
MAP1B mRNA contains in its 5′-UTR a guanine-rich stretch proposed to fold into a G quadruplex stem-capped structure recognized by the FMRP RGG box (Darnell et al. 2001). In this study we have used a 34-mer MAP1B RNA fragment that retains the sequence proposed to form the G quadruplex stem-capped structure, but has a point mutation at position 34 (G → C) to allow for the formation of a perfect seven base pair stem (Fig. 1B). We did not expect this mutation to impact FMRP binding, since it has been reported that although the presence of a stem is necessary for FMRP binding to G quadruplex forming RNA, its specific sequence is not important (Darnell et al. 2001). To investigate whether the 34-mer MAP1B RNA maintains the ability to form a G quadruplex structure, we have employed one-dimensional (1D) 1H NMR spectroscopy. The imino proton resonance region of the 1D 1H NMR spectrum of MAP1B RNA changes upon the titration of increasing K+ ion concentrations (Fig. 2). At 0 mM KCl only resonances corresponding to Watson–Crick imino protons are observed, indicating the presence of a stem structure in MAP1B RNA. At 5 mM KCl a new set of resonances appears in the region 10.8–12 ppm, corresponding to imino protons of guanines involved in G quartet formation, indicating that this MAP1B RNA fragment forms a G quadruplex structure. At KCl concentrations higher than 10 mM, there are additional changes in the 10.5–12 ppm region: the intensity of the existing resonances changes and they become broader, while new resonances appear, indicating that MAP1B RNA is in slow exchange between two different conformations (Supplemental Fig. 1). Thus, to ensure the formation of a single MAP1B RNA conformer, all further experiments were performed in the presence of 10 mM KCl.
FIGURE 2.
Changes of the imino proton resonance region of the 1D 1H NMR spectrum of MAP1B RNA (500 μM in 10 mM Tris, pH 7.5, 29°C) upon titrating increasing concentrations of KCl in the range of 0–10 mM.
To determine if the MAP1B RNA G quadruplex structure is intramolecular or intermolecular, we used thermodynamic methods. For intermolecular species with n number of strands the melting point of the G quadruplex structure, Tm, depends on the total RNA concentration, cT:
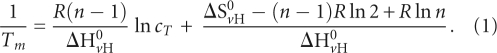 |
For intramolecular species (n = 1), and Tm is independent of CT:
 |
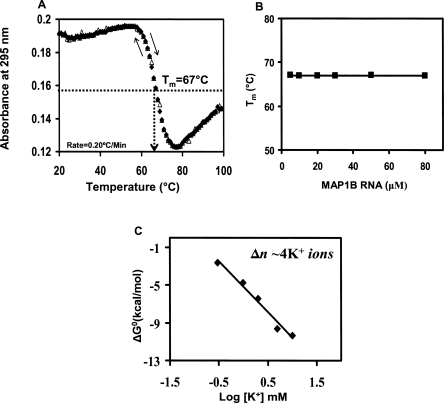
The melting temperature of the MAP1B RNA G quadruplex structure was measured by UV spectroscopy at various RNA concentrations in the range 3–100 μM. First, we have determined that the heating rate of 0.2°C/min used in all thermal denaturation experiments allowed for the measurement of true equilibrium curves since the heating (20°C–99°C) and cooling (99°C–20°C) curves of MAP1B RNA are superimposable (Fig. 3A; Mergny et al. 2005). The UV thermal denaturation profile of MAP1B RNA contains a hypochromic transition with a Tm of ∼67°C, which is attributed to the dissociation of its G quadruplex structure (Mergny et al. 1998). As expected, this transition is absent in the thermal denaturation profile of MAP1B RNA folded in the presence of Li+ ions, that do not support the formation of a G quadruplex structure (Supplemental Fig. 2). The UV melting curve of MAP1B RNA also contains a hyperchromic transition starting around 77°C (Fig. 3A), which we initially attributed to the melting of the MAP1B RNA stem structure. To verify this hypothesis we have monitored the imino proton resonance region of the 1D 1H NMR spectrum of MAP1B RNA (in which the stem and quadruplex imino protons give rise to distinct resonances), as a function of temperature (Supplemental Fig. 3). The MAP1B RNA stem structure was completely melted above 65°C, ruling out the possibility that the UV hyperchromic transition starting at 77°C originates from the melting of the stem structure. The analysis of the G quadruplex forming sequence of MAP1B RNA shows the presence of a stretch of twenty purines (starting at G7) interrupted by a single C; thus, it is possible that upon the melting of the quadruplex structure, the liberated rG residues can stack on their nearest rA neighbors and the melting of these rG-rA stacks could give rise to the observed UV hyperchromic transition (Dolinnaya and Fresco 1992; Shiber et al. 1996; Menon and Mihailescu 2007).
FIGURE 3.
(A) The heating and cooling UV thermal denaturation profiles of MAP1B RNA (10 μM in 10 mM cacodylic acid, pH 6.5 and 10 mM KCl) (black diamonds 20°C–99°C, open triangles 99°C–20°C). (B) Plot of the MAP1B RNA G quadruplex melting temperature as a function of the RNA concentration. (C) Plot of ΔG0 as a function of the logarithm of K+ ion concentration. The number Δn of K+ ion equivalents released upon the unfolding of MAP1B RNA G quadruplex structure was determined from the slope ΔΔG0/Δ log [K+].
As shown in Figure 3B, the Tm of the MAP1B RNA G quadruplex remains constant at all RNA concentrations investigated, indicating the formation of an intramolecular G quadruplex structure (see Equation 2).
After we established that the MAP1B RNA G quadruplex is intramolecular, the UV hypochromic transition was fitted with Equation 7 (Materials and Methods) to obtain the thermodynamic parameters for the G quadruplex structure formation. The values of the enthalpy and free energy of G quadruplex formation ΔH0 = −96.9 ± 0.1 kcal/mol and ΔG0 = −12.1 ± 0.1 kcal/mol are consistent with the presence of four to five G quartet planes in the structure of MAP1B RNA (the enthalpy of formation of a single G quartet plane in an intramolecular G quadruplex, measured in similar experimental conditions, ranges from −18 to −25 kcal/mol, [Hardin et al. 2001]). The analysis of the MAP1B RNA sequence does not immediately reveal the fold of such a structure, since there are not enough uninterrupted stretches of four guanines (Fig. 1B) to complete a structure containing four G quartet planes. Thus, it is possible that this structure might contain mixed purine (adenine and guanine) quartets. This is the second report that FMRP binds to a structure that might contain mixed purine quartets, as these structural elements have also been proposed to exist in the high affinity and specificity FMRP binding site identified in the RGG-coding region of its own mRNA (Schaeffer et al. 2001). Alternately, it has been proposed that repeating GGAGG sequences are involved in the formation of a hexad or pentad in which the amino group of one guanine forms hydrogen bonds within the G quartet, as well as with the N7 of the adenine (Liu et al. 2002). In this arrangement, the adenine interrupting the four G stretch would be pushed away, becoming involved in the hexad or pentad formation and allowing for the formation of four sequential stacking G quartet planes by these sequences. In this case, sharp guanine amino proton resonances originating from guanine amino groups that have both protons involved in hydrogen bonds (one within the G quartet and one with the N7 of the adenine) should be observed in the 1D 1H NMR spectrum of the RNA. However, this is not the case for the GGAGG sequences present in MAP1B RNA, since such sharp guanine amino proton resonances are not observed.
To obtain additional information about the MAP1B RNA G quadruplex structure we determined the number of specifically bound K+ ion equivalents. We assumed a simple model for the folded to unfolded G quadruplex structure, in which Δn K+ ion equivalents are released upon unfolding. Δn can be determined from the slope of the plot of ΔG 0 as a function of logarithm of K+ ion concentration (Cantor and Schimmel 1980):
 |
where ln Keq = −ΔG0/RT and ΔΔG0 /Δlog[K+] is the slope of the plot of ΔG0 as a function of the logarithm of K+ ion concentration (Fig. 3C). We have determined a Δn value of 3.6 ± 0.2, indicating that the MAP1B G quadruplex structure coordinates approximately four K+ ion equivalents.
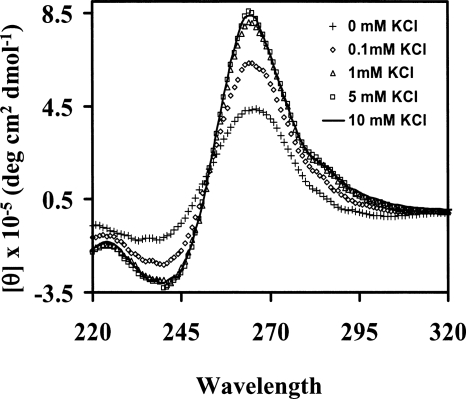
Circular dichroism (CD) spectroscopy was used to obtain information about the G quadruplex fold of MAP1B RNA, by analyzing the CD spectral changes in the presence of various KCl concentrations in the range of 0–10 mM (Fig. 4). In the presence of 10 mM KCl, a positive band is observed at 265 nm and a negative one at 240 nm, corresponding to a type I G quadruplex CD spectrum (Williamson 1994). For intermolecular G quadruplexes there is a strong correlation between the parallel G quadruplex structure and type I CD spectrum (Gaffney et al. 1992; Jin et al. 1992). Although there are examples to support this relationship for intramolecular G quadruplexes as well (Matsugami et al. 2001, 2003; Seenisamy et al. 2004; Ambrus et al. 2005), a generalization cannot be made since there are not enough intramolecular parallel type I high-resolution G quadruplex structures available. In addition to the positive band around 265 nm and the negative one around 240 nm, the CD spectrum of MAP1B RNA shows a positive shoulder around 290 nm, spectral features that have been previously reported for intramolecular G-quadruplexes with anti-glycosidic bonds and external or “propeller” loops (Xu et al. 2006; Pedroso et al. 2007).
FIGURE 4.
CD spectra showing the type I G quadruplex formation by MAP1B RNA (10 μM RNA in 10 mM cacodylic acid, pH 6.5 in the presence of increasing concentrations of KCl ranging from 0 to 10 mM). The KCl concentrations are indicated in the figure inset.
Thus, our thermodynamic and CD spectroscopy results suggest that MAP1B RNA forms an intramolecular G quadruplex structure of parallel nature, which contains four to five G quartet planes, coordinated by ∼4K+ ion equivalents. However, the molecular details of such a structure need to be confirmed by high-resolution structural studies, which we are currently pursuing.
MAP1B RNA interactions with the FMRP RGG box
A detailed thermodynamic study of the interactions between the FMRP RGG box and MAP1B RNA was performed by fluorescence spectroscopy. The MAP1B RNA used in this study has been labeled by the fluorescent analog 2 aminopurine (2AP) at position 19 (circled in Fig. 1B), constructing MAP1B-19AP RNA. The quantum yield of 2AP is very sensitive to its environment, making it a useful spectroscopic probe in DNA and RNA. We hypothesized that the environment of the 2-AP reporter will be different when MAP1B RNA is folded in the presence of Li+ ions that do not promote G quadruplex formation, versus K+ ions (single-stranded loop versus G quadruplex surrounding loop). A twofold change in the 2AP steady-state fluorescence was observed when MAP1B-19AP RNA was folded in the presence of K+ ions, as opposed to Li+ (Supplemental Fig. 4A). To ensure that this change was not due to an effect of the different ions (Li+ versus K+) on the intrinsic fluorescence of 2AP, in a control experiment, we have used a 16 nt RNA named DLS_9AP, which has a 2AP incorporated between two guanines, but which cannot form a G quadruplex structure. As shown in Supplemental Figure 4B, the steady-state fluorescence of the 2AP incorporated in DLS_9AP is the same in the presence of 10 mM LiCl and 10 mM KCl. Thus, our results indicate that the incorporated 2AP in MAP1B-19AP reports on the formation of its G quadruplex structure.
In additional control experiments we showed that the UV thermal profiles and CD spectra of MAP1B and MAP1B-19AP are very similar, indicating that the 2AP insertion does not affect the secondary structure and stability of the RNA (Supplemental Fig. 4C,D). We have also determined by electromobility shift assay that the 2AP insertion in MAP1B-19AP did not affect its ability to bind to the FMRP RGG box (Supplemental Fig. 4E).
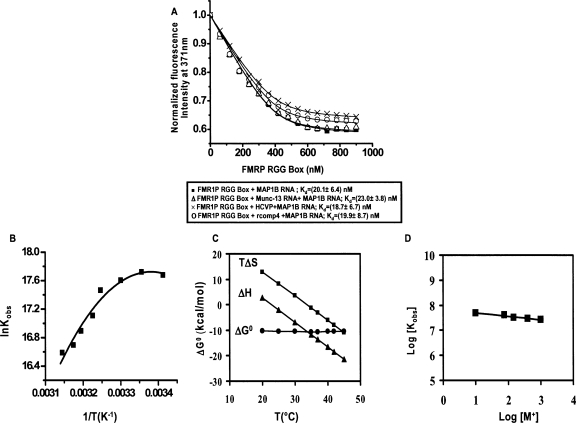
The FMRP RGG box binding to MAP1B-19AP RNA was measured by monitoring the 2AP steady-state fluorescence changes upon the titration of increasing amounts of the FMRP RGG peptide (Fig. 5A, black squares). We first determined that the observed decrease of 2AP fluorescence intensity in MAP1B-19AP is due to the FMRP RGG box binding, and not due to the exposure of 2AP to the intensity of the lamp. Thus, in a control experiment we have tested the fluorescence intensity of the 2AP probe incorporated in DLS_9AP, and determined that there was no signal loss after the same number of measurements that were done in the MAP1B-19AP-FMRP RGG box binding experiment (data not shown). The dissociation constant for the MAP1B RNA:FMRP RGG complex, Kd = 20.1 ± 6.4 nM, which was determined by fitting the binding curve with Equation 8 (Materials and Methods), corresponds to a free energy of binding ΔG0 obs = −10.4 ± 0.2 kcal/mol. This Kd is significantly lower than the value (Kd = 500 nM) reported by Darnell et al. (2001) for the binding of either the full-length FMRP or of the isolated RGG box domain to MAP1B RNA. This discrepancy could be due to the different experimental conditions of the binding essay, as their binding measurements were performed in the presence of 200 mM K+, conditions in which we show that MAP1B RNA exists in more than one conformation. Thus, the 500 nM value is likely an average for the binding of FMRP to these different MAP1B RNA conformers. Our binding experiments were done in the presence of 10 mM K+, conditions in which MAP1B exists as a single conformer.
FIGURE 5.
(A) Binding curves of the FMRP RGG box to MAP1B-19AP RNA in the absence (black squares) and presence of a 10-fold excess of a nonspecific Munc-13 site 1 RNA (black triangles) or of a nonspecific HCV peptide (black crosses) or of rComp4 RNA sequence (black circles). These experiments were carried out at 25°C. (B) Nonlinear van't Hoff plot, showing the temperature dependence of the association constant, Kobs = 1/Kd, for the MAP1B-19AP RNA:FMRP RGG complex, in the range of 20°C–45°C. The ΔC°pobs = −(1.0 ± 0.2) kcal/ mol K, T H (K) = (296 ± 2); T S (K) = (307 ± 1) values were determined by fitting this plot with Equation 5. (C) Dependence of the entropy, enthalpy, and free energy of the MAP1B-19AP RNA:FMRP RGG complex formation on temperature. (D) The binding of FMRP RGG box to MAP1B-19AP was measured at 25°C in the presence of different salt concentrations [M+] (Salt composition: 10 mM KCl + variable mM LiCl mM) in the range 10 mM to 1000 mM [M+]. A value of (−∂logKobs/∂log[M+]) of ∼0.2 was determined from the linear fit of the plot.
To determine if the FMRP RGG box binds specifically to MAP1B RNA, we have measured the Kd values of the MAP1B RNA:FMRP RGG complex in the presence of a 10-fold excess of nonspecific RNAs, as well as in the presence of a 10-fold excess of a nonspecific peptide. The 58-mer nonspecific peptide used in the binding assay is derived from the hepatitis C virus (HCV) core protein. As illustrated in Figure 5A (black crosses), the presence of a 10-fold excess of the HCV core peptide does not affect the binding of the FMRP RGG box to MAP1B RNA (Kd = 18.7 ± 6.7 nM). The two nonspecific RNAs used in the binding assay were the G rich Munc 13 site 1 RNA (Darnell et al. 2001), and the single-stranded rComp4 RNA (Materials and Methods). The dissociation constant values of the MAP1B RNA-FMRP RGG complex measured in the presence of a 10-fold excess of Munc 13 site 1 RNA (Kd = 23.0 ± 3.8 nM) or rComp4 RNA (Kd = 19.0 ± 8.6 nM) are similar to those measured in the absence of the nonspecific RNAs. Taken together, these results indicate that the FMRP RGG box binds specifically and with high affinity to the MAP1B RNA G quadruplex structure. Interestingly however, were the findings that Munc 13 site 1 RNA adopts a G quadruplex structure in the presence of 10 mM K+, as evidenced by the presence of guanine imino proton resonances in the 10–11.5 ppm region of its 1D 1H NMR spectrum (Supplemental Fig. 5A). Moreover, based on its CD signature, it is likely that the Munc 13 site 1 G quadruplex structure has a type I parallel fold (Supplemental Fig. 5B). Thus, the finding that the FMRP RGG box binds specifically to the MAP1B RNA G quadruplex, in the presence of a 10-fold excess of the Munc 13 site 1 RNA G quadruplex structure, indicates that the RGG box is able to discriminate among different type I parallel fold G quadruplex structures. It is not known whether the FMRP recognition of its G quadruplex forming mRNA targets involves a structure specific mechanism (FMRP would recognize the unusual G quadruplex “shape” of the RNA) or a sequence specific mechanism (the G quadruplex would function to properly expose the nucleotides located in its surrounding loops to FMRP) or a combination of both mechanisms. Our findings that the FMRP RGG box binds with high affinity to the MAP1B RNA G quadruplex but not to the Munc 13 site 1 RNA G quadruplex, although both have a type I parallel fold, indirectly suggests a sequence specific mechanism of recognition. However, only the high resolution structure of the FMRP RGG box-MAP1B RNA complex will reveal the molecular details of their association.
To obtain a more complete description of the forces driving the association between MAP1B-19AP RNA and the FMRP RGG box, we have determined the enthalpic and entropic contributions to the free energy of binding by measuring the binding association constant, Kobs = 1/Kd, as a function of temperature, in the range of 20°C–45°C. If the standard enthalpy, ΔH0 obs, and entropy, ΔS0 obs, do not depend on temperature, the van't Hoff plot, which shows the dependence of ln(Kobs) upon 1/T, is linear. However, as shown in Figure 5B, the van't Hoff plot for the FMRP RGG box:MAP1B RNA complex is nonlinear, indicating that ΔH0 obs and ΔS0 obs are not constant. In such a case, the entropy and enthalpy changes at different temperatures, can be obtained if the heat capacity change of the system, ΔC0 p obs, and the temperatures, T H and T S, at which the enthalpy and entropy do not make any contributions to the energy of the system are known:
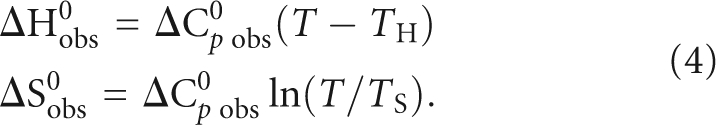 |
The heat capacity change, ΔC0 p obs, and the T H and T S temperatures at which ΔH0 obs = 0 and ΔS0 obs = 0, were obtained by fitting the nonlinear van't Hoff plot with the following equation (Becktel and Schellman 1987; Ha et al. 1989):
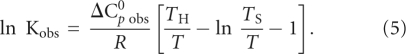 |
The values, ΔC0 p obs = −1.0 ± 0.2 kcal/mol K, T H = 296 ± 2 K (23°C) and T S = 307 ± 1 K (34°C) were then used in Equation 4 to obtain ΔH0 obs and ΔS0 obs at various temperatures. Both the standard entropy and enthalpy vary drastically with temperature, while the binding free energy remains constant (Fig. 5C). This entropy–enthalpy compensation indicates a change in the nature of the forces driving the complex formation: below 23°C entropy-driven (ΔS0 obs > 0) and enthalpy-opposed (ΔH0 obs > 0), between 23°C and 34°C entropy- and enthalpy-driven (ΔH0 obs < 0 and ΔS0 obs > 0), and above 34°C enthalpy-driven (ΔH0 obs < 0) and entropy-opposed (ΔS0 obs < 0). Thus, at the physiological temperature, the FMRP RGG box-MAP1B RNA association is enthalpically driven with an unfavorable entropic contribution, similarly to what has been reported for two other G quadruplex forming RNAs bound by the FMRP RGG box, semaphorin 3F RNA and Sc1 RNA (Zanotti et al. 2006; Menon and Mihailescu 2007).
The large negative value of ΔC0 p obs = −1.0 ± 0.2 kcal/mol K suggests that hydrophobic interactions are significant in the FMRP RGG box-MAP1B RNA association. ΔC0 p obs has been related to the change in polar (ΔAp) and nonpolar (ΔAnp) surface area (Å2) occurring during a complex formation (Spolar and Record 1994):
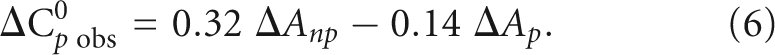 |
A large negative value for ΔC0 p obs can be obtained from a large (−ΔAnp) value, which results from hydrophobic interactions that bury the nonpolar surface of the forming complex and reduce exposure to water (Ha et al. 1989; Record et al. 1991; Spolar and Record 1994). However, in this process the expulsion of ordered water molecules from the binding surfaces typically contribute favorable changes in entropy (Spolar and Record 1994). The finding that unfavorable changes of ΔS0 obs are observed for the FMRP RGG box:MAP1B RNA complex formation at the physiological temperature indicates that structural changes in the peptide and/or RNA are likely associated with the binding event. Such structural changes could result in the restriction of molecular motion, and/or could affect the ionic and/or hydration shells around the complex by enhancing the association of ions and additional water molecules with the complex. The sequence analysis of the FMRP RGG box (Materials and Methods) does not reveal obvious amino acid residues that could become involved in stacking interactions with the MAP1B RNA bases. One possible candidate is Phe549, or alternately, the guanidino group of some of the RGG box Arg residues might stack with bases located in the G quadruplex surrounding loops.
Considering that the FMRP RGG box is rich in basic residues and that the RNA is an anionic polymer, we evaluated the role played by electrostatic interactions in the FMRP RGG box-MAP1B RNA complex formation. Quantitative information about the electrostatic interactions is obtained by measuring the association constant of the complex, Kobs, in the presence of increasing cation [M+] concentrations. The slope (−∂log Kobs / ∂log[M+]), known as the salt dependence, is related to the number of electrostatic interactions that stabilize the peptide-RNA complex (Draper 1995). Since we determined that increasing the K+ concentration above 10 mM promotes the formation of alternate species in MAP1B RNA, the cation concentration, [M+], was varied in the range 10 to 1000 mM by the addition of increasing concentrations of Li+ ions. The dissociation constant of the FMRP RGG box:MAP1B-19AP RNA complex changes from 20 nM in the presence of 10 mM [M+] to 38 nM in the presence of 1000 mM [M+], corresponding to a value of (−∂log Kobs / ∂log[M+]) of ∼0.2. (−∂log Kobs / ∂log[M+]) values between 0 and 2 have been measured for other protein–RNA complexes in which electrostatic interactions are not significant (Draper 1995; Baumann et al. 1996; Fisher et al. 1998; Wilson et al. 2001). We calculated a difference in the free energy ΔΔG0 obs in the presence of 10 mM and 1000 mM [M+] of ∼0.4 kcal/mol, which represents a mere 3% fraction of the total binding free energy. Thus, we conclude that electrostatic interactions play a minor contribution in the association between the FMRP RGG box and MAP1B RNA. Similar findings have also been reported for the binding of the FMRP RGG box to two other G quadruplex forming RNAs, Sc1 RNA and Semaphorin 3F RNA (Zanotti et al. 2006; Menon and Mihailescu 2007). However, it is not clear if these findings are a general property of the RGG box RNA binding domain, or if they are unique to the FMRP RGG box binding to G quadruplex RNA, since there are no other studies available about the thermodynamics of the RGG box domain specific binding to RNA.
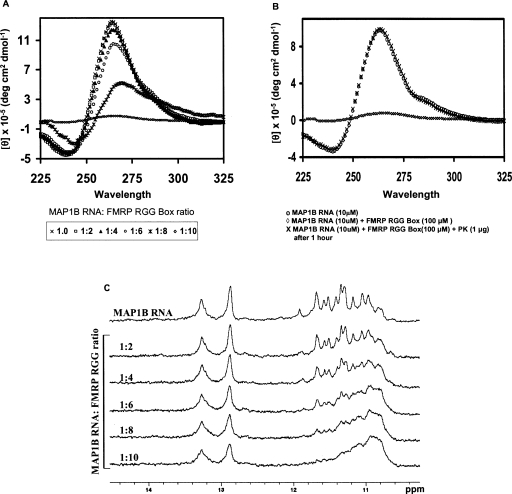
Next we investigated if the FMRP RGG box binding has any effect upon the stability and structure of MAP1B RNA. We have determined by UV spectroscopy that the melting temperature of the G quadruplex structure in the MAP1B-FMRP RGG box complex is ∼71°C, which corresponds to a free energy of G quartet formation of ΔG0 = −13.3 ± 0.1 kcal/mol (data not shown). Thus, the binding of the FMRP RGG box in a 1:1 ratio to MAP1B RNA induces a slight stabilization of the RNA G quadruplex structure. Interestingly however, our CD spectroscopy results showed that an excess of the FMRP RGG box (RNA:peptide ratios higher than 1:8) induces a reverse effect, in which the G quadruplex structure is unfolded (Fig. 6A). To ensure that the reduction of the 265 nm band intensity observed in the presence of an excess of the RGG box peptide was not due to the RNA degradation, we have treated the sample containing a ratio of 1:10 MAP1B RNA-FMRP RGG peptide with proteinase K, which degrades the RGG box peptide. As seen in Figure 6B, upon the degradation of the FMRP RGG peptide, the MAP1B G quadruplex structure refolds, giving rise to the same CD spectrum as that of the free MAP1B RNA. The same result was confirmed by NMR spectroscopy (Fig. 6C): Upon increasing the FMRP RGG box concentration, the sharp resonances centered ∼11.5 ppm (corresponding to guanine imino protons involved in Hoogsteen hydrogen bonds in the G quadruplex structure) disappear, indicative of the unfolding of the G quadruplex structure. Interestingly, the MAP1B RNA stem structure remains unaffected, suggesting that the interactions of the FMRP RGG box are limited to the G quadruplex structure. A similar effect of G quadruplex unfolding at high RNA:FMRP RGG peptide ratio was observed for the binding of the FMRP RGG box to the G quadruplex forming Semaphorin 3F RNA and Sc1 RNA (Menon and Mihailescu 2007; data not shown). It is tempting to speculate on a possible biological function for the G quadruplex RNA structure stabilization at low FMRP RGG box concentrations and its unwinding at high FMRP RGG box concentrations. Several studies show that FMRP represses mRNA translation (Li et al. 2001; Zhang et al. 2001; Lu et al. 2004), supporting its proposed function of translational repressor. Moreover, consistent with this proposal are the findings that the basal levels of two proteins whose mRNAs are proposed FMRP targets (MAP1B, α-CaMKII), are increased in hippocampal area CA1 of Fmr1 knock-out mice as compared to the wild-type mice (Hou et al. 2006). One possible mechanism by which FMRP might achieve the translational repressor function is to bind to and stabilize a G quadruplex structure present in the 5′-UTR of a subclass of its specific mRNA targets (Fig. 7A). In agreement with this model is the data showing that when a G quadruplex structure is placed in the 5′-UTR of a reporter gene, the FMRP binding to this structure is strong enough to repress significantly the translation initiation of the reporter gene (Schaeffer et al. 2001).
FIGURE 6.
(A) CD spectra of 10 μM MAP1B RNA in 10 mM cacodylic acid, pH 6.5, 25°C in the presence of increasing concentrations of the FMRP RGG box. (B) 10 μM MAP1B RNA+100 μM FMRP RGG box was incubated with proteinase K (1 μg) for 1 h at 25°C to check whether the addition of an excess of the FMRP RGG box causes the RNA degradation. (C) Imino proton resonance region of the 1D 1H NMR spectra of MAP1B RNA in the presence of increasing concentrations of the FMRP RGG box.
FIGURE 7.
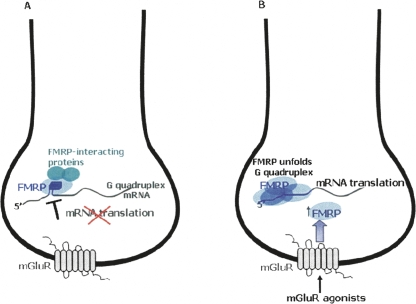
Proposed model for a regulatory switch of FMRP function from translation repressor to activator, in response to a neurotransmitter stimulation event. (A) In resting state when the FMRP:RNA ratio is low, FMRP represses the translation of specific mRNA targets by stabilizing a G quadruplex structure present in their 5′-UTR. (B) A stimulation of the mGluR by agonists leads to an increase in the FMRP concentration, unfolding the RNA G quadruplex structure, and allowing translation to occur.
We demonstrated that at high protein:RNA ratios, the FMRP RGG box unwinds the G quadruplex structure of several of its proposed mRNA targets, namely semaphorin 3F RNA (Menon and Mihailescu 2007), Sc-1 RNA (data not shown), and MAP1B RNA (this paper). If the stabilization of the mRNA G quadruplex structure is associated with translation inhibition, then its unwinding could allow mRNA translation. Thus, a variation in the cellular concentration of FMRP, for example, in response to a neurotransmitter stimulation event, could act as a regulatory switch for FMRP function, from translation repressor at low protein:RNA ratios, to translation promoter at high protein:RNA ratios (Fig. 7B). FMRP is proposed to be involved in the transport of specific mRNA targets to distant sites (as part of a large mRNP complex) where these mRNAs have to be maintained in a repressed state until synaptic input triggers their translation. Our model is consistent with this proposal: FMRP could bind to and stabilize the G quadruplex structure present in a subset of its specific mRNA targets, maintaining them in a translationally repressed state (Fig. 7A). An increase in the FMRP concentration triggered by synaptic input, would lead to the unwinding of the G quadruplex structure of these mRNA targets, promoting their translation (Fig. 7B). This model is supported by several studies that showed a rapid increase in FMRP synthesis locally in synaptoneurosomes in response to application of glutamate or metabotropic glutamate receptor (mGluR) agonists (Weiler et al. 1997; Greenough et al. 2001; Hou et al. 2006), indicating a regulation of FMRP concentration in response to synaptic stimulation. Moreover, this local protein synthesis was followed by a rapid decrease in FMRP levels back to baseline, suggesting a dynamic regulation of the FMRP concentration (Hou et al. 2006).
According to our model, the synaptically triggered local protein synthesis from a G quadruplex containing class of FMRP mRNA targets would depend on the presence of a high FMRP concentration. Thus, the model predicts that in the absence of FMRP, the levels of these proteins will be unaffected by a neurotransmitter stimulation event. Consistent with this prediction, several studies have showed that the Fmr1 knockout mice were deficient in their ability to induce local-protein synthesis of FMRP mRNA targets after stimulation of glutamate receptors (Todd et al. 2003; Hou et al. 2006; Muddashetty et al. 2007) or of the NMDA-receptor (Muddashetty et al. 2007).
Very recently another group has put forward a model which proposes for the first time the existence of a regulatory switch for the FMRP function from translation repressor to activator in response to the activation of mGluR (Ronesi and Huber 2008). This model differs from ours in that it proposes that the rapid degradation of FMRP observed after the stimulation of the mGluR, leads to the translation of the FMRP mRNA targets. However, this model (Ronesi and Huber 2008) does not propose a role for the observed initial surge in the FMRP concentration (prior to its degradation and return to basal levels) in response to the stimulation of the mGluR. The authors acknowledge that “the purpose of the rapid and bidirectional regulation of FMRP by mGluRs is unclear.” Our experimental results suggest a functional role for the observed increase, followed by a decrease in the FMRP concentration in response to neurotransmitter receptor stimulation, and the model we propose explains how such a switch of FMRP function could operate, at least for a subset of FMRP RNA targets that contain a G quadruplex structure.
SUMMARY
In summary we have determined that MAP1B RNA adopts in vitro an intramolecular G quadruplex structure bound with high affinity and specificity by the FMRP RGG box. At the physiological temperature the complex formation is enthalpically driven with an unfavorable entropic contribution. The thermodynamic analysis of the FMRP RGG box binding to MAP1B RNA reveals that hydrophobic effects are significant, whereas electrostatic interactions have only a minor contribution to the complex formation. We found that at low FMRP RGG box:MAP1B RNA ratios the peptide binding stabilizes slightly the RNA G quadruplex structure, whereas at high ratios, the peptide completely unfolds it. Based on these results we propose a model according to which the variation of the FMRP concentration in response to a neurotransmitter stimulation event acts as a switch for the protein function from repressor to promoter of translation.
MATERIALS AND METHODS
RNA synthesis
The unlabeled MAP1B and Munc 13 site 1 RNA oligonucleotides were synthesized by in vitro transcription reactions off synthetic DNA templates (Trilink Biotechnologies, Inc.) using T7 RNA polymerase (produced in-house following the procedure by Milligan and Uhlenbeck [1989]). The Munc 13 site 1 RNA has the following sequence (Darnell et al. 2001): 5′-GGGUUUGAGGCCAUGGGAGGCAAGGAGCUGGAC-3′. The rComp4 single stranded RNA, which has the sequence 5′-AUUAUUAUUAUUA-3′, was a generous gift from Dr. Bruce Armitage (Carnegie Mellon University). The RNA oligonucleotides were purified by 15% denaturing polyacrylamide/urea gels and electrophoretic elution and were extensively dialyzed against 10 mM cacodylic acid (pH 6.5) or 10 mM Tris (pH 7.5). For the fluorescence spectroscopy studies, we constructed MAP1B-19AP RNA which had the fluorescent analog 2-aminopurine (2AP) (Dharmacon, Inc.) incorporated at the 19th position (Fig. 1B, circled nucleotide). All experiments were performed with RNA samples that were annealed by heating at 95°C followed by slow cooling at room temperature for 20 min. The control 16 nt RNA DLS_9AP used in the fluorescence spectroscopy experiments has the following sequence: UCACGGCG(2AP)GCUGUGA.
Peptide synthesis
The FMRP RGG box, which has the following sequence: 528- RRGDGRRRGGGGRGQGGRGRGGGFKGNDDHSR-560 was chemically synthesized and purified by the Peptide Synthesis Unit at the University of Pittsburgh, Center for Biotechnology and Bioengineering.
UV spectroscopy
The melting curves of the MAP1B and the MAP1B-19AP RNA were collected on a Varian Cary 3E spectrophotometer equipped with a Peltier cell. The experiments were carried out in 10 mm path-length quartz cuvettes, using annealed RNA samples in 10 mM cacodylic acid, pH 6.5 or 10 mM Tris, pH 7.5, containing 10 mM KCl, in a final volume of 200 μl. The RNA samples were heated from 20°C to 99°C at a rate of 0.2°C/min, recording points every 1°C, and to prevent evaporation, a layer of mineral oil (Sigma-Aldrich) was added to the cuvettes. Blank samples were treated in the same manner. Depending upon the RNA concentration, the spectral absorbance was measured at either 295 or 305 nm, wavelengths which have been previously identified to be sensitive to G quadruplex dissociation (Mergny et al. 1998). For all UV spectroscopy experiments, the measured OD range at either 295 nm or 305 nm was between 0.1–0.8. All UV spectroscopy thermal denaturation curves were measured in duplicate. To determine if MAP1B RNA folds into an intermolecular or intramolecular conformation, the melting temperature of the G quadruplex structure was determined at different RNA concentrations in the range 3–100 μM.
The thermodynamic parameters of G quadruplex formation in MAP1B or MAP1B-19AP RNA were determined by fitting the G quadruplex dissociation hypochromic transition assuming an independent two state model:
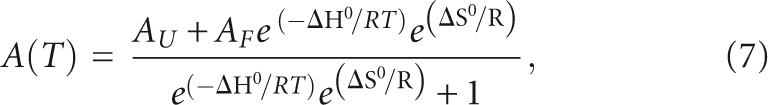 |
where AU and AF represent the absorbance of the unfolded and native G quadruplex RNA, respectively, and R is the universal gas constant.
The melting temperature of the MAP1B RNA complex with the FMRP RGG box was determined using the same parameters described above for the free RNA. The FMRP RGG box was added to MAP1B RNA (in 10 mM cacodylic acid, at pH 6.5, and 10 mM KCl) in a 1:1 ratio and allowed to equilibrate for 30 min for the RNA:peptide complex formation. The UV melting curve of the MAP1B RNA:FMRP RGG box complex was corrected for the peptide contribution, by subtracting the melting curve of the free peptide.
To determine the potassium ion equivalents released upon the melting of the MAP1B G quadruplex structure, increasing concentrations of KCl, in the range 0.1 mM–10 mM were added to the MAP1B RNA in 10 mM Tris (pH 7.5) and the samples were equilibrated for 20 min at room temperature before the recording of the melting curves.
Circular dichroism spectroscopy
The CD spectroscopy experiments were performed on a J-810 spectropolarimeter at 25°C, using a 1 mm path-length quartz cuvette (Starna Cells). The G quadruplex formation in MAP1B RNA (10 μM in 10 mM cacodylic acid, pH 6.5, 200 μL final volume) was monitored by titrating increasing amounts of KCl from a 4 M stock solution to a final concentration of 10 mM KCl. Each spectrum was scanned three times between 200 and 350 nm, with a 1 s response time and a 2 nm bandwidth. For the binding studies, increasing amounts of the FMRP RGG box (0–100 μM) were titrated into a fixed concentration of MAP1B RNA (10 μM) in 10 mM cacodylic acid, pH 6.5, and 10 mM KCl, monitoring the change in the molar ellipticity of the RNA at 264 nm. The CD spectra were corrected for the peptide contribution and dilution after each addition of the peptide.
To determine if MAP1B RNA degrades upon the excess addition of the FMRP RGG peptide, the sample containing a 1:10 ratio of RNA:peptide was treated with proteinase K, which degrades the RGG peptide (1 μg) for 1 h at 25°C. Subsequently, the CD spectrum was re-recorded and corrected for the presence of proteinase K.
Fluorescence spectroscopy
Steady-state fluorescence spectroscopy measurements of MAP1B-19AP RNA and the binding studies with the FMRP RGG peptide were performed on a J.Y. Horiba Fluoromax-3 with a 150 W ozone-free Xenon arc-lamp, equipped with variable temperature control in the sample chamber. The fluorescence spectroscopy experiments were carried out in 3 mm path-length quartz cuvettes (Starna Cells), on samples having typically a final volume of 150 μL. The excitation wavelength was set at 310 nm, the emission spectrum was recorded in the range of 330–450 nm, and the bandpasses for the excitation and emission monochromators were set at 5 nm. Increasing concentrations of the FMRP RGG box (60 nM increments from a 10 μM stock solution) were titrated to a fixed concentration of MAP1B-19AP RNA (400 nM), the sample being equilibrated for 15 min after each peptide addition. The volume changes in the sample due to each peptide addition varied from 0.6% for the first point to 6% for the last point. The binding curves were obtained by plotting the normalized 2AP steady-state fluorescence intensity measured at 371 nm as a function of the FMRP RGG box concentration. The binding dissociation constant, Kd, was determined by fitting the binding curves to the equation:
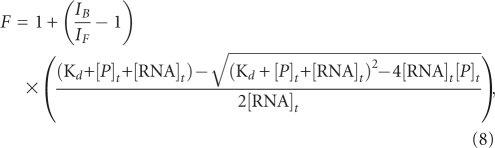 |
where IF and IB represent the steady-state fluorescence intensities of the free and bound MAP1B-19AP, [RNA]t is the total concentration of MAP1B-19AP, and [P]t is the total FMRP RGG box concentration. The experiments were performed in duplicate or triplicate, the reported errors are the standard uncertainties of the data from the best-fit theoretical curves, since they are larger than the standard uncertainty of the measurements.
Competition experiments were performed in the presence of a 10-fold excess of unlabeled nonspecific G quadruplex forming Munc 13 site 1 RNA, or in the presence of nonspecific single-stranded rComp4 RNA. Competition experiments were also performed in the presence of a 10-fold excess of the nonspecific Hepatitis C virus peptide (HCV).
The thermodynamic parameters for the binding of the FMRP RGG box to MAP1B-19AP were determined by measuring the binding association constant, Kobs = 1/Kd at different temperatures in the range of 20°C–45°C.
NMR spectroscopy
The 1D 1H NMR spectra of MAP1B RNA or Munc 13 site 1 RNA were acquired at 29°C on a 500 MHz Varian Unity Plus spectrometer or on a 500 MHz Bruker AVANCE spectrometer. The water suppression was accomplished using the jump-and-return pulse sequence (Plateau and Gueron 1982) with the maximum of excitation set at 11 ppm or by using the Watergate pulse sequence (Piotto et al. 1992). MAP1B RNA (500 μM) or Munc 13 site 1 RNA (263 μM) were prepared in 10 mM Tris (pH 7.5) in a 90% H2O/10%D2O ratio. To observe the G quadruplex formation, each sample was titrated, with increasing concentrations of KCl up to 10 mM from a 4 M stock solution. For the binding studies, the FMRP RGG box was added to MAP1B RNA in increasing ratios up to 1:10, allowing the sample to equilibrate for 20 min after each peptide addition.
Electrophoretic mobility gel shift assay (EMSA)
The RNA:peptide complexes were prepared by preincubating MAP1B or MAP1B-19AP RNA (10 μM) for 20 min at room temperature with the FMRP RGG box in a 1:2 RNA:peptide ratio. The complexes were then resolved on 15% nondenaturing acrylamide gels in the presence of 10 mM KCl at 35 V and 4°C. The gels were visualized by UV-shadowing at 254 nm using an AlphaImager HP (AlphaInnotech, Inc.).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Bruce Armitage (Carnegie Mellon University) for his generous gift of rComp4 RNA and Dr. John Marino (CARB/NIST) for providing us with the plasmid for T7 RNA polymerase expression. This work was supported by a FRAXA Research Foundation grant to M.-R.M. An equipment grant from the National Science Foundation (NMR: CHE 0614785) is gratefully acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1100708.
REFERENCES
- Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- Antar L.N., Bassell G.J. Sunrise at the synapse: The FMRP mRNP shaping the synaptic interface. Neuron. 2003;37:555–558. doi: 10.1016/s0896-6273(03)00090-4. [DOI] [PubMed] [Google Scholar]
- Antar L.N., Dictenberg J.B., Plociniak M., Afroz R., Bassell G.J. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Baumann C., Otridge J., Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 1996;271:12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- Becktel W.J., Schellman J.A. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Cantor C.R., Schimmel P.R. W.H. Freeman; New York: 1980. Biophysical Chemistry, Parts I-III. [Google Scholar]
- Crawford D.C., Acuna J.M., Sherman S.L. FMR1 and the fragile X syndrome: Human genome epidemiology review. Genet. Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes & Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.T. G-quartets 40 years later: From 5′GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- Dolinnaya N.G., Fresco J.R. Single-stranded nucleic acid helical secondary structure stabilized by ionic bonds: d(A+-G)10. . Proc. Natl. Acad. Sci. 1992;89:9242–9246. doi: 10.1073/pnas.89.19.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. Protein–RNA recognition. Annu. Rev. Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- Fisher B.M., Ha J.-H., Raines R.T. Coulombic forces in protein–RNA interactions: Binding and cleavage by ribonuclease A and variants at Lys7, Arg10, and Lys66. Biochemistry. 1998;37:12121–12132. doi: 10.1021/bi980743l. [DOI] [PubMed] [Google Scholar]
- Gabus C., Mazroui R., Tremblay S., Khandjian E.W., Darlix J.L. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;7:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney B.L., Wang C., Jones R.A. Tetraplex formation of d[G(15N7)GTTTTTGG] and d[T(15N7)GGGT] monitored by proton detected 15N NMR. J. Am. Chem. Soc. 1992;114:4047–4050. [Google Scholar]
- Gonzalez-Billault C., Jimenez-Mateos E.M., Caceres A., Diaz-Nido J., Wandosell F., Avila J. Microtubule-associated protein 1B function during normal development, regeneration, and pathological conditions in the nervous system. Neurobiol. 2004;58:48–59. doi: 10.1002/neu.10283. [DOI] [PubMed] [Google Scholar]
- Greenough W.T., Klintsova A.Y., Irwin S.A., Galvez R., Bates K.E., Weiler I.J. Synaptic regulation of protein synthesis and the fragile X protein. Proc. Natl. Acad. Sci. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J.-H., Spolar R.S., Record M.T. Role of the hydrophobic effect in stability of site-specific protein–DNA complexes. J. Mol. Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Hardin C.G., Perry A.G., White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2001;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Hazel P., Huppert J., Balasubramaniam S., Neidle S. Loop-length-dependent folding of G quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- Hou L., Antion M.D., Hu D., Spencer C.M., Paylor R., Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Jin P., Warren S.T. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet. 2000;6:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- Jin P., Warren S.T. New insights into fragile X syndrome: From molecules to neurobehaviors. Trends Biochem. Sci. 2003;28:152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- Jin R., Gaffney B.L., Wang C., Jones R.A., Breslauer K.J., Kenneth J. Thermodynamics and structure of a DNA tetraplex: A spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl. Acad. Sci. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Ku L., Wilkinson K.D., Warren S.T., Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Matsugami A., Katahira M., Uesugi S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J. Mol. Biol. 2002;322:955–970. doi: 10.1016/s0022-2836(02)00876-8. [DOI] [PubMed] [Google Scholar]
- Lu R., Wang H., Liang Z., Ku L., O'Donnell W.T., Li W., Warren S.T., Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl. Acad. Sci. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimiliano V., Zalfa F., Bagni C. FMRP and its target RNAs: Fishing for the specificity. Neuroreport. 2004;16:2447–2450. doi: 10.1097/00001756-200411150-00002. [DOI] [PubMed] [Google Scholar]
- Matsugami A., Ouhashi K., Kanagawa M., Liu H., Kanagawa S., Uesugi S., Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- Matsugami A., Okuizumi T., Uesugi S., Katahira M. Intramolecular higher-order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- Menon L., Mihailescu M.R. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny J.-L., Phan A.-T., Lacroix L. Following G-quartet formation by UV- spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- Mergny J.L., Cian A.N., Ghelab A., Sacca B., Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Muddashetty R.S., Kelic S., Gross C., Xu M., Bassell G.J. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell W.T., Warren S.T. A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Pedroso I.M., Duarte L.F., Yanez G., Burkewitz K., Fletcher T.M. Sequence specificity of inter- and intramolecular G-quadruplex formation by human telomeric DNA. Biopolymers. Biopolymers. 2007;87:74–84. doi: 10.1002/bip.20790. [DOI] [PubMed] [Google Scholar]
- Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Plateau P., Gueron M. Exchangeable protons without base line distortion using a new strong pulse sequence. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- Ramos A., Hollingworth D., Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M.T., Ha J.H., Fisher M. Use of equilibrium and kinetic measurements to determine the thermodynamic origins of stability and specificity and mechanism of formation of site specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- Ronesi J.A., Huber K.M. Metabotropic glutamate receptors and fragile X mental retardation protein: Partners in translational regulation at the synapse. Sci. Signal. 2008;1:pe6. doi: 10.1126/stke.15pe6. [DOI] [PubMed] [Google Scholar]
- Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenisamy J., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- Shiber M.C., Braswell E.H., Klump H., Fresco J.R. Duplex–tetraplex equilibrium between a hairpin and two interacting hairpins of d(A-G)10 at neutral pH. Nucleic Acids Res. 1996;24:5004–5012. doi: 10.1093/nar/24.24.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Siomi M.C., Nussbaum R.L., Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;2:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Spolar R.S., Record M.T. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Todd P.K., Mack K.J., Malter J.S. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl. Acad. Sci. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler I.J., Irwin S.A., Klintsova A.Y., Spencer C.M., Brazelton A.D., Miyashiro K., Comery T.A., Patel B., Eberwine J., Greenough W.T. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- Wilson G.M., Sutphen K., Bolikal S., Chuang K.Y., Brewer G. Thermodynamics and kinetics of Hsp70 association with A+U-rich mRNA-destabilizing sequences. J. Biol. Chem. 2001;276:44450–44456. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- Xu Y., Noguchi Y., Sugiyama H. The new models of the human telomere d [AGGG (TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Surya R., Oostra B., Bagni C. The fragile X syndrome protein FMRP regulates the translation of specific mRNAs at the synspes via BC1 RNA. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zanotti K.J., Lackey P.E., Evans G.L., Mihailescu M.R. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- Zhang Y.Q., Bailey A.M., Matthies H.J., Renden R.B., Smith M.A., Speese S.D., Rubin G.M., Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]