Abstract
The mechanism by which miRNAs inhibit translation has been under scrutiny both in vivo and in vitro. Divergent results have led to the suggestion that miRNAs repress translation by a variety of mechanisms including blocking the function of the cap in stimulating translation. However, these analyses largely only examine the final output of the multistep process of translation. This raises the possibility that when different steps in translation are rate limiting, miRNAs might show different effects on protein production. To examine this possibility, we modeled the process of translation initiation and examined how the effects of miRNAs under different conditions might be explained. Our results suggest that different effects of miRNAs on protein production in separate experiments could be due to differences in rate-limiting steps. This analysis does not rule out that miRNAs directly repress the function of the cap structure, but it demonstrates that the observations used to argue for this effect are open to alternative interpretations. Taking all the data together, our analysis is consistent with the model that miRNAs may primarily repress translation initiation at a late step.
Keywords: miRNA, modeling, repression, translation
INTRODUCTION
MicroRNAs (miRNAs) are 20–22 nucleotide RNAs that regulate the function of eukaryotic mRNAs and play important roles in development, cancer, stress responses, and viral infections. miRNAs function, at least in part, to inhibit translation of mRNAs and promote mRNA deadenylation, decapping, and 5′ to 3′ degradation of the mRNA body (Valencia-Sanchez et al. 2006; Jackson and Standart 2007; Eulalio et al. 2008a; Filipowicz et al. 2008).
An unresolved issue is the mechanism(s) by which miRNAs repress protein synthesis. miRNAs have been proposed to repress protein production by multiple mechanisms including promoting ribosome drop-off (Petersen et al. 2006), degradation of the nascent polypeptide (Nottrott et al. 2006), sequestration of mRNAs in P-bodies or stress granules (Liu et al. 2005; Pillai et al. 2005; Leung et al. 2006), inhibition of translation initiation (Pillai et al. 2005; Wang et al. 2006; Mathonnet et al. 2007; Thermann and Hentze 2007), and inhibition of translation at a step after translation initiation (Olsen and Ambros 1999; Lytle et al. 2007). Despite this confusion, recent experiments using in vitro translation systems have indicated that miRNAs can repress translation, at least in part, by inhibiting translation initiation (Wang et al. 2006; Mathonnet et al. 2007; Thermann and Hentze 2007), although in some cases deadenylation of the mRNA might contribute to the observed repression (Wakiyama et al. 2007).
In vitro experiments have led to the suggestion that miRNAs inhibit translation initiation by interfering with the ability of the m7G-cap structures at the 5′ of the mRNA to recruit the translation initiation complex (Wang et al. 2006; Mathonnet et al. 2007; Thermann and Hentze 2007). This conclusion is based on the following observations. First, mRNAs that are lacking the normal 5′ m7GpppG cap (m7G-cap) structure, and instead have a 5′ ApppG cap (A-cap), which is unable to bind the translation initiation factor eIF4E, are not repressed by miRNAs in vitro (Mathonnet et al. 2007; Thermann; and Hentze 2007). Second, addition of increasing concentrations of exogenous eIF4F, which binds the m7G-cap structure and promotes initiation, prevents miRNA-mediated repression (Mathonnet et al. 2007). Third, initiation of translation from the EMCV internal ribosome entry site (IRES), which promotes translation initiation independent of the 5′ cap structure, can be resistant to miRNA-mediated repression of translation in vitro (Mathonnet et al. 2007).
The conclusion that miRNAs inhibit cap-dependent translation has also been supported by in vivo experiments showing that (1) in some cases, mRNAs lacking the m7G-cap structure are not able to be repressed by miRNAs (Humphreys et al. 2005; Pillai et al. 2005); (2) artificial tethering of the cap binding complex to a mRNA can make it resistant to translation repression by miRNAs (Pillai et al. 2005); (3) in some experiments, messages containing IRES elements are unable to be repressed by miRNAs in vivo (Pillai et al. 2005); and (4) the Argonaute protein has been suggested to have a conserved “cap binding protein” motif required for miRNA-mediated repression (Kiriakidou et al. 2007), although recent results suggest this motif is involved in protein–protein interactions required for repression (Eulalio et al. 2008b). Taken together, the in vitro and in vivo work has led to an emerging view that miRNAs repress translation, at least in part, by affecting an early cap-dependent stage in translation initiation (Standart and Jackson 2007; Eulalio et al. 2008a; Filipowicz et al. 2008).
Despite this apparent consensus, other evidence argues that miRNAs can work independently of the cap structure. For example, transfected ApppG-capped and uncapped mRNAs whose translation is driven by an IRES can be subjected to translation repression by miRNAs (Humphreys et al. 2005; Lytle et al. 2007). In addition, when tethered to the mRNA, the GW182 protein can promote repression even in the absence of Drosophila Ago1, which is the Argonaute protein responsible for miRNA-mediated translational repression in flies (Behm-Ansmant et al. 2006). This latter observation provides evidence that Argonaute proteins, and any proposed direct interaction with the cap, are not required for miRNA-mediated repression. Similarly, mRNAs whose translation is driven by IRES elements expressed in cells or transfected can undergo miRNA-mediated repression (Petersen et al. 2006; Lytle et al. 2007).
These apparently divergent results on how miRNAs repress translation can be interpreted in two possibly overlapping manners. First, it could be that miRNAs act by different mechanisms and the various experimental protocols reveal those multiple mechanisms. Alternatively, the apparently conflicting results might be due to creation of different rate-limiting steps in translation caused by variations in the protocols or substrates, which might mask any effect of miRNAs on protein production. To examine the possibility of altered translational kinetics masking the actual step in which miRNA act, we developed a simple computational model of translational initiation. We then used this model to predict how protein production from m7G-capped mRNAs, A-capped mRNAs, IRESs, and in the presence of high levels of eIF4F, would be affected by possible miRNA-mediated inhibition of different steps in translation initiation. These perturbations were then compared to in vivo and in vitro repression data to assess probable sites of repression. Our results suggest that some apparently different results can be explained by differences in rate-limiting events under different translation protocols. This analysis does not rule out that miRNAs directly repress the function of the cap structure, but it demonstrates that the observations used to argue for this effect are open to alternative interpretation. Moreover, taking all the observations together our analysis is most consistent with miRNAs affecting a late stage in translation initiation such as AUG recognition or 60S joining.
RESULTS
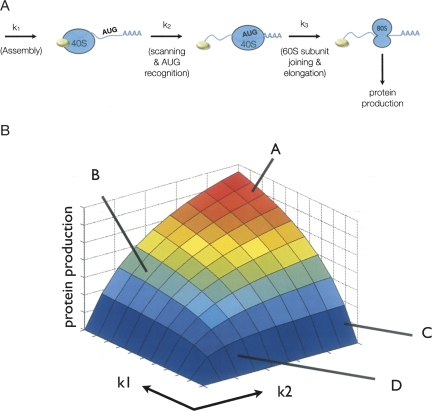
In order to consider mechanistic explanations for observations on how miRNAs repress translation, we created a computational model for the initiation of translation. The model we developed is an extreme simplification of the process of translation initiation, and more complex models could be developed. However, we utilized a simple model in our analyses since it is sufficient to illustrate the relevant kinetic issues and is more easily interpretable. In this model, we consider the process of translation initiation to consist of three steps, each of which affects the overall rate of protein production (Fig. 1A). The first is an assembly process of factors promoting translation initiation resulting in the binding of the 40S subunit to the mRNA (k1). The second is scanning by the small ribosome subunit and recognition of the AUG (k2). The third step is recruitment of the 60S subunit, entry into translation elongation, and recycling of initiation factors (k3).
FIGURE 1.
Details of the computational model. (A) Depiction of the relevant states in the translational model. Each rate and species that is included in the model is indicated as well as their primary function. (B) Protein production predicted from the model plotted as a function of k1 and k2, lines A–D correspond to specific rates described in the text.
The relative value of the rate constants was assigned using the following logic. First, because different mRNAs, particularly those involved in the analysis of miRNA repression, can vary significantly in the recruitment of the translation initiation complex, we allow k1 to vary over a large range. Second, because the efficiency of scanning and AUG recognition can vary between mRNAs, we also consider a range of k2 values. The subunit joining and protein production rate (k3) is faster than k1 and k2 since mRNA–40S complexes bound to the AUG without the 60S subunit are generally not observed in translation initiation unless this step is stalled by experimental methods, and elongation is generally thought to not be rate limiting in protein synthesis.
To compare how changes in the steps affect protein production, we examined the predicted protein output for various values of k1 and k2. These comparisons present several key points. Specifically, when k1 is substantially faster than k2, changes in the rate of k2 have clear impact on the overall amount of protein produced (Fig. 1B). For example, at a fast k1 rate, a two-thirds reduction in k2 leads to a 2.5-fold reduction in protein production (cf. lines A and B in Fig. 1B). However, if the k1 rate is relatively slow, a similar two-thirds reduction in k2 rate only reduces protein production by 3% (cf. lines C and D in Fig. 1B). Thus, the effect on protein production of a given change in a particular rate is dependent on the relative rates of other steps in protein production.
Effects of miRNAs on A-capped mRNAs
One set of observations that have been used to argue for miRNAs targeting a cap-dependent step in translation initiation is that there are differences in how miRNAs repress translation from mRNAs with different cap structures. Specifically, miRNAs in many systems have been documented to repress translation of mRNAs with 5′ m7G caps, which promote the assembly of the translation initiation complex (k1), but not to repress translation from mRNAs with the ApppG cap analog (A-cap), which are limited in their rates of translation complex assembly (represented by reduced k1 in this model). This observation has been interpreted to indicate that miRNAs may interfere with the function of the cap-binding complex and thereby reduce translation initiation (Mathonnet et al. 2007; Thermann and Hentze 2007).
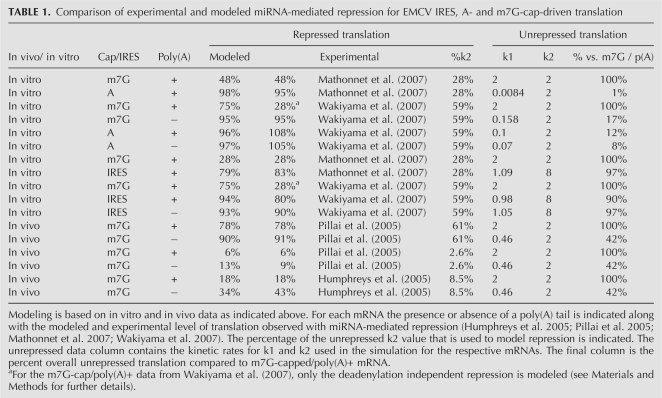
We used our translational model system to examine translational repression of A-capped mRNA. A-capped mRNAs have a reduced recruitment of the initiation complex, which appears as a lower k1 rate in this model. The reduction in k1 can be estimated from published translational data. For example, in vitro translation results from Krebs-2 cell-free extracts (Mathonnet et al. 2007) show a 100-fold decrease in total Renilla luciferase expression from an A-capped mRNA versus an m7G-capped mRNA. This cap-dependent rate effect is also supported by in vivo data from HeLa cells showing approximately the same amount of enhancement of translation by the m7G-cap (Humphreys et al. 2005). Such a 100-fold reduction in protein production corresponds to a 240-fold reduction of k1 in our model, rendering this step much slower than k2 (all modeling results and altered parameters are presented in Table 1.)
TABLE 1.
Comparison of experimental and modeled miRNA-mediated repression for EMCV IRES, A- and m7G-cap-driven translation
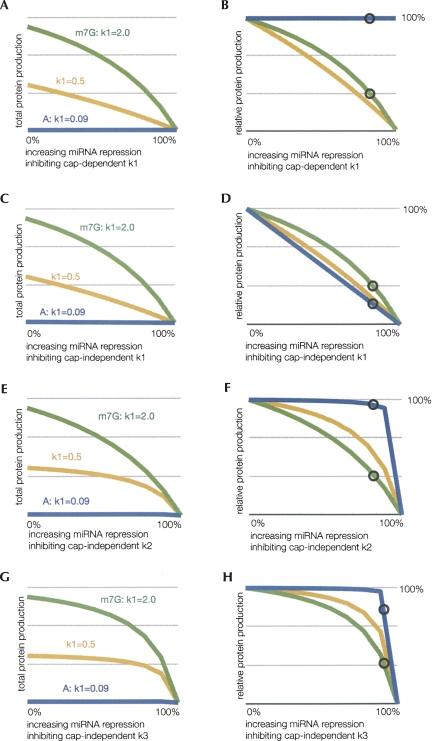
We then modeled how such a decrease in k1 would affect protein production by considering miRNA acting in four potential manners. The four possible scenarios considered were: (1) if miRNAs prevent recognition of the cap structure and therefore would have no effect on changing k1 on an A-capped mRNA, (2) if miRNAs inhibit a cap-independent step in the assembly of the initiation complex (e.g., inhibiting eIF4G–eIF3 interaction, which ultimately is an effect of reducing k1), (3) if miRNAs repress some aspect of scanning and/or AUG recognition (reduced k2), and (4) if miRNAs affect 60S joining or elongation (reduced k3). The experimental and modeled results are displayed in Table 1 along with the kinetic parameters used in the simulations. The results of this analysis are shown in Figure 2. In each case, we plot the absolute amount of protein produced as a function of the extent of hypothetical inhibition of a specific step in the model by miRNAs. Moreover, to illustrate the relative levels of protein production from each mRNA under different hypothetical effects of miRNAs, we also plot the amount of protein production relative to the protein production from the mRNA without any miRNA-mediated inhibition. In each figure, we compare the A-capped mRNA (blue line) with the m7G-capped mRNA (green line), and include an mRNA of intermediate k1 value (yellow line), to indicate the trend of reducing k1. Theses analyses make the following points.
FIGURE 2.
Effects of translational inhibition on individual steps of translation. These graphs depict the percentage of unrepressed protein production by altered rate constants and their effects for m7G-capped (m7G, green, k1 = 2), A-capped (A, blue, k1 = 0.09), and an intermediately translated mRNA (yellow, k1 = 0.5) for comparison. These modeled mRNA are identical except for the initiation complex formation rates (k1). Protein production refers to the percentage of unrepressed production for a respective k1 value. The region of rates inhibited by miRNA is from 0% to 100% repression of the respective values, where 100% repression results in no protein production. The circles indicate the observed in vitro miRNA-mediated inhibition in Mathonnet et al. (2007) with m7G-capped mRNA to allow comparisons to the amount of translation that will occur with mRNA having reduced k1 values. (A–B) Protein production from increasing repression of cap-dependent k1 shown as total and relative amounts, respectively. (C–D) Protein production from increasing repression of cap-independent k1 shown as total and relative amounts, respectively. (E–F) Protein production from increasing repression of k2 shown as total and relative amounts, respectively. (G–H) Protein production from increasing repression of k3 shown as total and relative amounts, respectively.
First, if miRNAs directly repress recognition of the cap structure, miRNAs would no longer be able to repress translation of A-capped mRNAs since they would only repress the k1 rate of m7G-capped mRNA. Thus, in this possible model, although low, the protein production from the A-capped mRNAs would not be affected by miRNAs (Fig. 2A,B). This result is consistent with the proposed interpretation that miRNAs directly affect cap recognition per se.
Second, if miRNAs inhibit formation of the initiation complex in a manner independent of direct cap recognition, the model predicts increased sensitivity of A-capped mRNA to miRNA-mediated repression relative to the m7G-capped mRNA. This result is most easily seen in Figure 2D, where relative amounts of protein from each mRNA show that the A-capped mRNA (blue line) is repressed more than the m7G-capped mRNA (green line). Since this predicted result is the opposite of the effect observed experimentally, this result argues that miRNAs do not affect a cap-independent step in assembly of the translation initiation complex.
Third, this modeling indicates that if miRNAs repress a later stage in translation initiation such as “AUG recognition” (k2 in our model), then they will also fail to repress protein production from mRNAs with A-caps (Fig. 2E,F). This result is most easily seen in Figure 2F, where relative amounts of protein from each mRNA show that the A-capped mRNA (blue line) is much more resistant to repression than the m7G-capped mRNA (green line). This is simply due to the fact that k1 is the predominant rate-determining step for A-capped mRNAs, and therefore alterations in k2 have little effect. Thus, the failure of miRNAs to repress protein production from A-capped mRNAs could be explained by miRNAs repressing a late stage in translation initiation.
Similarly, if miRNAs repress k3, then they will also fail to repress protein production from mRNAs with A-caps, again because k1 is the predominant rate-determining step under most conditions unless k3 is extremely slow (Fig. 2G,H). Thus, the failure of miRNAs to repress protein production from A-capped mRNAs could be explained by miRNAs repressing a late stage in translation initiation such as the subunit joining step.
Based on these observations, we suggest that miRNAs either directly affect cap recognition, or affect a late stage in the translation initiation process such as AUG recognition or 60S joining. Our analysis is inconsistent with miRNAs repressing a cap-independent step in translation initiation complex assembly. An important general principle from this analysis is that if miRNA-mediated repression affects a step in translation initiation that is a relatively fast step in initiation for that particular mRNA, then little effect on overall protein production will be observed. For example, if miRNAs reduce the rate of AUG recognition (k2 in this model), then mRNAs with a slow k1 will be resistant to miRNA-mediated repression.
Other alterations to mRNAs that alter the process of cap recognition in vivo and thereby prevent miRNA-mediated repression are subject to the same caveat. For example, when cap recognition is bypassed by eIF4F being delivered to mRNAs by tethering with an RNA binding protein, the mRNA is resistant to miRNA-mediated repression (Pillai et al. 2005). However, at least in other experiments, the absolute amount of protein produced from these tethered constructs is very low, ∼5% that of cap-dependent translation, suggesting that tethering is inefficient, and therefore may have also caused a significant decrease in k1 (De Gregorio et al. 1999). Therefore, the failure to repress translation driven by tethering eIF4F with miRNAs may be subject to the same caveats as the A-capped mRNAs.
Implications for miRNA-mediated repression of IRES-driven translation
A second line of argument for miRNAs affecting cap-dependent translation is that when translation initiation is driven by an IRES in some cases mRNAs are unable to be repressed by miRNA action in vitro (Mathonnet et al. 2007). The logic here is that the IRES-mediated translation bypasses the recognition of the cap structure, so if miRNAs repress cap recognition, then IRES-dependent translation should be resistant to miRNA-mediated repression.
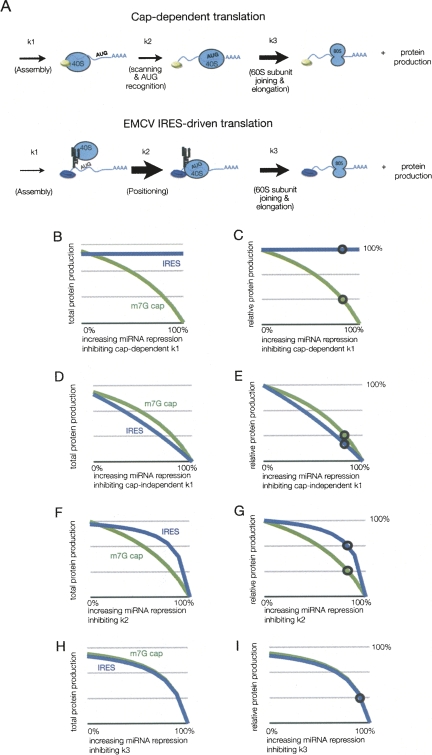
In order to consider how miRNAs might affect translation initiation from mRNAs with IRES elements, it was first necessary to consider how IRES-mediated translation occurs and what the relationship between k1 and k2 might be for IRES-driven translation. For the EMCV IRES, which is the only IRES examined in vitro for miRNA-mediated repression because of its relatively high translation rate compared to other IRES elements, the translation initiation process again proceeds in three steps, although they are notably different from translation initiation on capped mRNAs (Fig. 3A). First, the IRES element in EMCV recruits the translation initiation complex directly, through interactions with eIF4G, and that process can be stimulated by the poly(A) tail (Humphreys et al. 2005) and PABP, the poly(A) binding protein (Bergamini et al. 2000; Svitkin et al. 2007). Second, in contrast to capped mRNAs, the AUG is positioned at the site of ribosome recruitment, so there is no scanning and AUG recognition occurs in a spatially restricted manner (Kaminski et al. 1994). Due to the absence of scanning and the direct proximity of the AUG to the site of ribosome recruitment, we hypothesize that for translation initiation driven by the EMCV IRES, k1 is relatively slow compared to k2. (Kaminski et al. 1990; Pilipenko et al. 1994). This is consistent with the observation that at a moderate concentration of the eIF4A inhibitor hippuristanol (0.4 μM), EMCV IRES-driven translation is reduced more significantly than the cap-dependent translation (Bordeleau et al. 2006). Note that for the EMCV IRES to give a high rate of translation initiation and still have a slow k1, k2 must be quite efficient. Thus, in order for IRES containing mRNA to produce a roughly equal amount of protein as do capped mRNAs (Mathonnet et al. 2007), we modeled translation initiation of EMCV IRES as a situation wherein k2 is quite high (approximately fourfold that of scanning-dependent translation) and k1 is low (50% of cap-dependent value). We then examined how possible modes of miRNA-mediated repression would be predicted to affect this process using rates estimated from in vitro experimental results from Mathonnet et al. (2007).
FIGURE 3.
Effect of miRNA inhibition on EMCV IRES-driven translation. (A) Depiction of the relative rates used in modeling cap-dependent and -independent (EMCV IRES) driven translation. The size of the arrow corresponds to the magnitude of the rate indicated. Panels B–I display the results of modeling potential sites of miRNA action on IRES and cap-dependent translation depicting protein production. The region of rates inhibited by miRNA is from 0% to 100% repression of the respective values, where 100% repression results in no protein production. The circles on m7G cap lines indicate the observed in vitro miRNA-mediated inhibition in Mathonnet et al. (2007). The circles on the IRES lines indicate the amount of translation if repressed equivalently to the m7G mRNA. (B–C) Protein production from increasing repression of cap-dependent k1 shown as total and relative amounts, respectively. (D–E) Protein production from increasing repression of cap-independent k1 shown as total and relative amounts, respectively. (F–G) Protein production from increasing repression of k2 shown as total and relative amounts, respectively. (H–I) Protein production from increasing repression of k3 shown as total and relative amounts, respectively.
This modeling demonstrates the following points. First, the model predicts that IRES-driven translation would be resistant to any changes in k1 that were directly dependent on the cap structure (Fig. 3B,C). Thus, these results are consistent with the possibility that miRNAs inhibit cap dependent translation.
Second, the model predicts that translation from IRES elements will be more sensitive than cap-driven translation to perturbations of translation complex assembly (k1) that are cap independent (Fig. 3D,E). We interpret this specific result to argue that miRNAs do not affect a cap-independent step in assembly of the translation initiation complex.
A third, and important, point is that the model predicts that protein production from the EMCV IRES would be not significantly repressed over a wide range of k2 values (Fig. 3F,G). Thus, assuming k1 is limiting for IRES-driven translation, if miRNAs reduce the rate of downstream events such as “AUG recognition” (k2) then the EMCV IRES would be expected to be insensitive to miRNA-mediated repression (Fig. 3F,G). The actual k1 and k2 values are not important for this conclusion; merely that an elevated k2 and reduced k1 produces the results shown below for a wide range of values (data not shown). It should also be noted that if miRNAs inhibit k2 our modeling predicts that translation from the EMCV IRES should be repressed ∼20%, which is similar to what was observed by Mathonnet et al. (2007), who observed 17% repression. A fourth, and final point, is that translation from the IRES element is equally sensitive to repression of k3 as cap-dependent translation (Fig. 3H,I).
Taken together, we interpret this modeling to suggest that the failure of miRNAs to repress translation from IRES elements is consistent with miRNAs affecting cap recognition, but could also be explained by other mechanisms of translation inhibition. Considering only effects on initiation, the results involving miRNA-mediated repression of IRES-driven translation can be interpreted in two manners. First, if miRNAs repress recognition of the cap structure, miRNAs would no longer be able to repress translation from the EMCV IRES. Thus, this result is consistent with the proposed interpretation that miRNAs directly affect cap recognition per se, but does not provide an explanation for the small repression observed with miRNAs. Alternatively, since k1 is likely to be the predominant rate-determining step in IRES driven translation initiation, if miRNAs repress a later step in the process, then miRNAs will only repress protein production from the EMCV IRES to a small degree, as has been observed (Mathonnet et al. 2007; Wakiyama et al. 2007).
Implications for miRNA repression with elevated levels of eIF4F
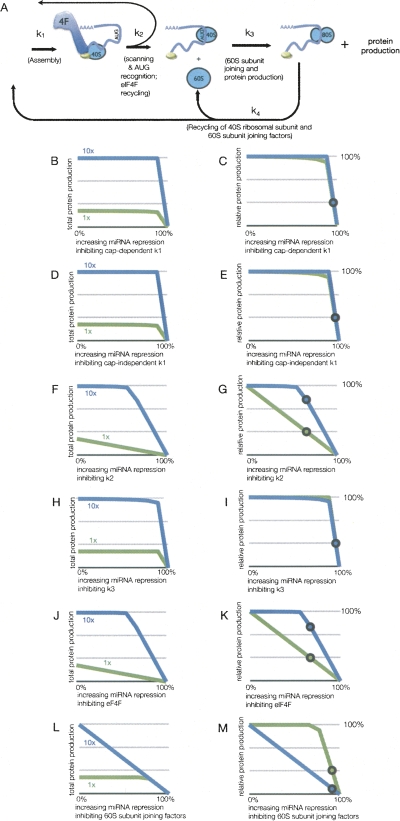
Another observation used to argue for miRNAs targeting a cap-dependent step in translation is that addition of extra eIF4F complex to in vitro translation systems prevents miRNAs from repressing translation (Mathonnet et al. 2007). The reasonable interpretation put forth is that if a miRNA-mediated repression complex competes with eIF4F for binding the cap structure, then enhancing cap recognition using high levels of eIF4F would block miRNA-mediated repression. Increasing concentrations of eIF4F would be expected to increase the overall rate of the first step (k1) by mass action, without changing any of the actual rate constants. To model the effects of changing eIF4F concentration, we created a modified model of translation initiation incorporating the concentrations of eIF4F and subunit joining factors. In this model, we added two species: eIF4F and the 60S subunit joining factors (depicted in Fig. 4A). Additionally, k3 in the first model is split into two steps: subunit joining (k3) and factor recycling rate (k4).
FIGURE 4.
Resistance of eIF4F enhanced translation to miRNA-mediated translational repression. (A) Depiction of the rates and species used for modeling translation incorporating eIF4F and 60S subunit joining factors. For panels B–M, the results of modeling potential sites of miRNA action on mRNA with standard (1×) or elevated (10×) amounts of eIF4F depicting protein production, with the percentage representing that versus unrepressed translation for each condition. The region of rates inhibited by miRNA is from 0% to 100% repression of the respective values, where 100% repression results in no protein production. The amount of eIF4F modeled is either the standard extract concentration (green, 1×) or 10-fold increased eIF4F (blue, 10×), which is indicated in the respective graphs. The circles on 1× lines indicate the observed in vitro miRNA-mediated inhibition of m7G mRNA with no additional eIF4F in Mathonnet et al. (2007). The circles on the 10× lines indicate the amount of translation if repressed equivalently to the mRNA with no additional eIF4F. (B–C) Protein production from increasing repression of cap-dependent k1 shown as total and relative amounts, respectively. (D–E) Protein production from increasing repression of cap-independent k1 shown as total and relative amounts, respectively. (F–G) Protein production from increasing repression of k2 shown as total and relative amounts, respectively. (H–I) Protein production from increasing repression of k3 shown as total and relative amounts, respectively. (J–K) Protein production from increasing repression of eIF4F shown as total and relative amounts, respectively. (L–M) Protein production from increasing repression of 60S subunit joining factors shown as total and relative amounts, respectively.
The eIF4F/subunit joining model contains three conserved species, whose amounts cannot change, for which only alterations in association with different complexes can occur. They are the 40S small ribosomal subunit; translation initiation factors, which are considered as a combined factor eIF4F; and the subunit joining factors for the 60S large ribosomal subunit. These were estimated as follows: The amount 40S ribosomal subunit was set arbitrarily high (see Materials and Methods for specific values), as it is thought to generally not be a limiting factor for translation initiation. In contrast, the level of eIF4F, as the canonical limiting factor, was set significantly lower so translation would be dependent on its concentration as observed experimentally (e.g., Mathonnet et al. 2007). Finally, the amount of subunit joining factors for the 60S large ribosomal subunit were estimated to be more abundant than eIF4F but still substoichiometric when compared to 40S levels, consistent with in vivo levels (Singh et al. 2007).
We then utilized this model to calculate how changes in the different rate constants or concentrations of species (as hypothetically caused by miRNA-mediated repression) would affect the amount of protein production at either normal levels of eIF4F (1×) or at 10-fold elevated levels (10×). These analyses revealed the following points.
First, under normal concentrations of eIF4F, protein production is primarily dependent on the amount of eIF4F levels and rate of scanning (k2), as variations in both these values have linear effects on protein output (1× line in Fig. 4G,K). In this model, moderate reductions in the rates of k1 and k3 did not have a strong effect on protein output for both normal and extra eIF4F, which is different from the simple model (cf. Figs. 2A,4B and Figs. 2E,4H). This is due to the consideration of the concentrations of eIF4F and the 60S subunit joining factors in the latter model, which are limiting for these steps. However, this eIF4F/subunit joining model provided substantially similar results for the analysis of the A-capped and IRES mRNAs to the simpler model used earlier (data not shown). One minor difference is that the degree of inhibition of k2 required to observe a similar decrease in protein output is lower than in the simple model (cf. Figs. 2F and 4G).
A second observation was that while translation with normal levels of eIF4F is very susceptible to repression at k2, addition of exogenous eIF4F renders it largely resistant to k2 repression. This result is most easily seen in Figure 4G, where relative amounts of protein from each mRNA show that translation with high levels of eIF4F (blue line) is much more resistant to repression at k2 than translation in the presence of normal levels of eIF4F (green line). Notably, in this model under these conditions, translation becomes limited by the concentration of factors at the 60S joining step (k3; Fig. 4M). This suggests that under conditions of high eIF4F a different step in translation initiation becomes rate limiting, which is supported by the observation that addition of eIF4F above a certain level fails to further enhance protein production (Mathonnet et al. 2007). Moreover, if the new rate-limiting step in translation initiation is not the step inhibited by miRNA function, then miRNAs will be unable to repress the translation of mRNAs in the presence of high concentrations of eIF4F.
DISCUSSION
Our modeling analyses have revealed three issues that should be considered in the analyses of experiments used to determine how miRNAs affect specific steps in translation initiation. First, kinetic analyses demonstrate that a miRNA will have little effect on the overall rate of protein production from a specific mRNA if the step it affects is not the predominant rate-determining step under the conditions examined. Second, increases in the rate of a substep may lead to a different step in the pathway becoming the predominant rate-determining step. If such a new predominant rate-determining step is not the site of miRNA-mediated repression, then the overall production will show no repression by miRNAs. Third, the use of IRES to map steps of action in translation repression is complicated by the fact that the predominant rate-determining step may be different between cap-dependent translation and IRES mediated translation, which could then kinetically mask any effect by miRNAs. Note that these issues can be overcome by either directly measuring the rate constants of specific substeps in initiation with and without miRNA repression, or by determining the concentrations of intermediates in the translation initiation pathway of the repressed and unrepressed mRNAs. For example, if miRNAs repress 60S joining, then in the presence of miRNAs, a 48S complex should accumulate at the AUG with miRNA repression, which is exactly what has been recently reported (Wang et al. 2008).
Although our results do not rule out that miRNAs can directly affect the function of the cap structure in enhancing translation, our analyses do demonstrate that many of the arguments used to argue that miRNAs repress cap-dependent translation are subject to other interpretations. First, the failure of miRNAs to repress translation from A-capped mRNAs would also be expected if miRNAs repress at a later stage of translation initiation and A-capped mRNAs assemble the translation complex at a slower rate. Similarly, the failure of miRNAs to repress mRNAs tethered to eIF4F would be expected if tethering is inefficient and therefore repression of any other step in translation initiation would have no effect on overall protein production (De Gregorio et al. 1999; Pillai et al. 2005). Second, the failure of miRNAs to repress translation when extra eIF4F is provided (Mathonnet et al. 2007) could be easily explained if the new predominant rate-determining step under these conditions is not the step affected by miRNA-mediated repression. Finally, the analysis of mRNAs containing IRES elements is complicated since they (1) may also be most limited for assembly and thereby unaffected by alterations in downstream steps, and (2) include the absence of other steps in translation initiation (besides cap recognition) that could also be targets of miRNA function (such as scanning). This raises the possibility that miRNAs might repress translation at a different stage of the initiation process, independent of cap recognition per se.
Other observations in the literature are consistent with miRNAs repressing translation initiation by acting at a late step in the process, independent of the cap structure. For example, A-capped mRNAs can be repressed by miRNAs if they are provided with a poly(A) tail. Similarly, mRNAs with IRES elements can also be repressed by miRNAs in vivo, either with an m7G- or an A-cap (Humphreys et al. 2005; Petersen et al. 2006; Lytle et al. 2007). More directly, the observation that miRNAs lead to the accumulation of 48S complexes on repressed mRNAs including those stalled at the AUG codon, supports the model that miRNAs alter some step in AUG recognition of 60S joining (Wang et al. 2008). Such an effect of miRNAs on 60S joining is also supported by observations suggesting that eIF6 interacts with Argonaute proteins and is required for miRNA repression in Caenorhabditis elegans (Chendrimada et al. 2007), although recent results suggest this is not true in Drosophila cells (Eulalio et al. 2008b).
The specific hypothesis that miRNAs affect a step in AUG recognition and/or 60S joining is consistent with the observation of pseudo-polysomes by Hentze and coworkers (Thermann and Hentze 2007). They found that when repressed by miRNAs, mRNA were found in large complexes, termed pseudo-polysomes that sedimented in the polysome region on a sucrose gradient but were not translating. This may be analogous to similar rapidly sedimenting complexes observed by Kozak and Shatkin (1978) upon inhibition of translation by edeine in extracts. The mechanism of miRNA-mediated repression could work similarly to edeine inhibition by affecting AUG start site selection causing multiple 40S subunits scanning beyond the authentic AUG.
Aside from the translation initiation steps covered by our model, there are no doubt additional, direct or indirect, effects of miRNAs on translation initiation. For example, since poly(A) tails can stimulate translation, and miRNAs can promote deadenylation, some repression by miRNAs can be attributed to deadenylation of the substrate mRNAs (Wakiyama et al. 2007). However, since poly(A) tails are not required for miRNA-mediated repression either in vivo or in vitro (Humphreys et al. 2005; Pillai et al. 2005; Wu et al. 2006; Wakiyama et al. 2007), miRNAs must be able to repress translation in a manner independent of the poly(A) tail. Another issue affecting miRNA repression in vivo is that sequestration of mRNAs complexed with the miRNA repression machinery into P-bodies, which could enhance the effect of an initial translation inhibition, perhaps by reducing the local concentration of translation factors (Jakymiw et al. 2005; Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005). These complexities highlight how it will be necessary, even once we understand the primary effects of miRNAs on translation, to integrate those effects into the cytoplasmic physiology of the mRNAs.
MATERIALS AND METHODS
Computational modeling of mRNA translation and miRNA-mediated repression
In establishing an initial model for translation, we established two key variables. The first is the small ribosomal subunit, which is conserved (it is neither created nor destroyed), and alters only in the state of the protein complexes with which it is associated. The second is protein, which is produced during the simulation, but not degraded. A more complex model was also constructed (the eIF4F/subunit joining model), which was made with two additional parameters, the factors required for 60S joining and the initiation factor eIF4F, both of which are conserved. However, they can be in a variety of states: bound to mRNA, scanning, translating, and unbound. Our approach is to describe each species as a function of mass action rate constants between each state using a series of ordinary differential equations.
The individual rates and rate equations used in the model are indicated below and diagrammed in Figure 1A for the simple model and Figure 4A for the eIF4F/subunit joining model. The first rate is the assembly of a translation initiation complex leading to the small ribosomal subunit binding to the mRNA. This rate is k1 for the simple model and incorporates both the k1 rate plus the amount of eIF4F for the eIF4F/subunit joining model. The second rate is k2, comprising scanning and the recognition of the AUG start codon. The third is subunit joining, which includes 40S recycling for the simple model and includes a 60S subunit joining factor for the more complex version. Finally, the eIF4F/subunit joining model has an additional step of recycling, which is k4.
The individual rates and rate equations used in the model are indicated below. These equations were modeled in the SimBiology 2.2 package in Matlab version 7.5.0 (2007b+). Data for the figures were obtained by parameter scanning and examination of the protein output. The simulated model time was 100 sec, and was sufficient to reach stabilization in the various species concentrations (i.e., free 40S, scanning 40S, 40S at the AUG initiation site, etc.), which was achieved in a maximum of 10 sec of simulation.
Equations used in the models
Parameters:
40S: Unbound 40S subunit
40S–mRNA: 40S ribosome bound to the mRNA
AUG: 40S ribosome at the AUG start site
60S: 60S subunit joining factors
80S: 80S ribosome
eIF4F: eIF4F
Protein: Protein
Simple model:
d[40S](t) = k3[AUG]dt − k1[40S]dt
d[40S–mRNA](t) = k1[40S] dt – k2[40S–mRNA]dt
d[AUG](t) = k2[40S–mRNA]dt – k3[AUG] dt
dProtein(t) = k3[AUG] dt
eIF4F/subunit joining model:
d[40S](t) = k4[80S]dt − k1[40S] [eIF4F]dt
d[40S-mRNA](t) = k1[40S] [eIF4F]dt – k2[40S–mRNA]dt
d[48S](t) = k2[40S–mRNA]dt – k3[AUG] [60S]dt
d[80S](t) = k3[AUG] [60S]dt − k4[80S]dt
d[60S](t) = k4[80S]dt − k3[AUG] [60S]dt
d[eIF4F](t) = k2[40S–mRNA]dt − k1[40S] [eIF4F]dt
dProtein(t) = k3[AUG] [60S]dt
Rate constants used in the models
Simple model:
k1 (2/second): small subunit binding to mRNA
k2 (2/second): AUG start site recognition
k3 (5/second): 60 subunit joining, factor recycling
eIF4F/subunit joining model:
k1 (2/molecule • second): small subunit binding to mRNA
k2 (2/second): AUG start site recognition
k3 (5/molecule • second): 60S subunit joining
k4 (1/second): 40 and 60S subunit joining factor recycling
Conserved quantities used in the eIF4F/subunit joining model
The rationale for choosing the values for the species in the more complex eIF4F/subunit joining model are as follows. The small ribosomal subunit was set to be arbitrarily high, consistent with it not being rate limiting for initiation. eIF4F was conversely set very low, so that the protein output would depend on it, as it is the rate-limiting factor in initiation. Finally, the subunit joining factors are substoichiometric to the small ribosomal subunit, consistent with their in vivo abundance (Singh et al. 2007).
Small ribosomal subunit: 100
Subunit joining factors for the 60S large ribosomal subunit: 25
eIF4F: 6
Estimations of rate constants
The estimations of the rate constants used in this study are relative rate constants. These correspond to data from the literature and educated assumptions regarding the steps modeled in this translation. The logic behind assignment of the steps is as follows. First, the binding step k1 is faster in m7G-capped mRNA, which have eIF4E bound than in A-capped mRNA, which are compromised in its binding. The important aspect for this study is that the relative rate is slower in A capped, and we have modeled a variety of k1 rates to simulate the range of possible values for m7G-capped and A-capped mRNA as correlated with protein output. Second, the second-rate k2 was set to be equivalent to k1 to provide the model with a minimum of assumptions. For the IRES rate estimations, a reduced k1 rate was combined with a k2 rate that was sufficiently high to allow 90% of the translation as compared to an essentially unlimited k2 value. The repressed value of k2 was based on the amount of protein produced for repressed m7G cap/poly(A)+ mRNA compared to unrepressed. Note that data for m7G cap/poly(A)+ from Wakiyama et al. (2007) likely incorporates deadenylation reduced initiation; so to estimate k2 repression in this case, we modeled this using data from the m7G-capped unadenylated mRNA to eliminate the effects of deadenylation on repression. Third, k3 is modeled as being fast relative to the other two steps. The justification for this is that complexes bound to AUG with the 60S subunit are rarely observed in translation. For the simple model, k3 represents subunit joining, start of translation, and recycling. For the eIF4F/subunit joining model, this step is divided into two steps. Subunit joining and start of translation are designated by k3. The recycling of 40S and subunit joining factors for the 60S large ribosomal subunits is represented by the k4 rate constant. The k4 rate is relatively slower than the other rates in the model; nevertheless, the simulation's overall protein production was not altered by changes of several orders of magnitude around its value.
ACKNOWLEDGMENTS
We thank Drs. Debrah Thompson and Carolyn Decker for critical reading of the manuscript. T.N. was supported in part by the NIH (T32 CA09213). An NIH grant (R37 GM45443) and funds from the Howard Hughes Medical Institute supported this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1072808.
REFERENCES
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini G., Preiss T., Hentze M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Preiss T., Hentze M.W. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Gen. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Kaminski A., Howell M.T., Jackson R.J. Initiation of encephalomyocarditis virus RNA translation: The authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A., Belsham G.J., Jackson R.J. Translation of encephalomyocarditis virus RNA: Parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Kozak M., Shatkin A.J. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., Sharp P.A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Nottrott S., Simard M.J., Richter J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pilipenko E.V., Gmyl A.P., Maslova S.V., Belov G.A., Sinyakov A.N., Huang M., Brown T.D., Agol V.I. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J. Mol. Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Singh C.R., Udagawa T., Lee B., Wassink S., He H., Yamamoto Y., Anderson J.T., Pavitt G.D., Asano K. Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J. Mol. Biol. 2007;370:315–330. doi: 10.1016/j.jmb.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standart N., Jackson R.J. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes & Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Costa-Mattioli M., Herdy B., Perreault S., Sonenberg N. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP) RNA. 2007;13:2330–2340. doi: 10.1261/rna.606407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Hentze M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Takimoto K., Ohara O., Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Love T.M., Call M.E., Doench J.G., Novina C.D. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wang B., Yanez A., Novina C.D. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl. Acad. Sci. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]