Abstract
We describe a method, microarray analysis of differential splicing (MADS), for discovery of differential alternative splicing from exon-tiling microarray data. MADS incorporates a series of low-level analysis algorithms motivated by the “probe-rich” design of exon arrays, including background correction, iterative probe selection, and removal of sequence-specific cross-hybridization to off-target transcripts. We used MADS to analyze Affymetrix Exon 1.0 array data on a mouse neuroblastoma cell line after shRNA-mediated knockdown of the splicing factor polypyrimidine tract binding protein (PTB). From a list of exons with predetermined inclusion/exclusion profiles in response to PTB depletion, MADS recognized all exons known to have large changes in transcript inclusion levels and offered improvement over Affymetrix's analysis procedure. We also identified numerous novel PTB-dependent splicing events. Thirty novel events were tested by RT-PCR and 27 were confirmed. This work demonstrates that the exon-tiling microarray design is an efficient and powerful approach for global, unbiased analysis of pre-mRNA splicing.
Keywords: alternative splicing, Exon array, cross-hybridization, microarray, bioinformatics
INTRODUCTION
Alternative splicing of precursor mRNAs is a prevalent mechanism of gene regulation in higher eukaryotes. It generates enormous transcriptome diversity from a limited repertoire of protein-coding genes in the genome (Roberts and Smith 2002). Alternative splicing occurs among different tissues (Xu et al. 2002), during cellular responses to external stimuli (Ip et al. 2007), and in a wide range of human diseases (Wang and Cooper 2007). However, the full spectrum of splicing changes in a specific biological process or disease was difficult to gauge because, until recently, high-throughput platforms for profiling alternatively spliced transcripts were unavailable. This situation has changed with the recent advance in microarray technology for analysis of pre-mRNA splicing (Clark et al. 2002; Johnson et al. 2003; Pan et al. 2004; Blencowe 2006).
With the steady increase of oligonucleotide density on microarray chips, it is now possible for an expression microarray to tile its probes over all exons in a mammalian genome. A current example of a microarray with this “probe-rich” design is the Exon 1.0 array from Affymetrix (Affymetrix 2005b; Clark et al. 2007). Traditional oligonucleotide array designs are “probe-poor”; i.e., they employed a small number of probes targeting specific parts of the gene such as the 3′ end or specific splice junctions. In contrast, the Exon array employs an average of nearly 150 probes per gene distributed through the entire potential transcribed region (Affymetrix 2005b). For splicing analysis, this new design has potential advantages over earlier implementations of splice junction microarrays: The high density (more than six million probes on a single array) allows the placement of multiple probes against almost every known or predicted exon; the exon-tiling design does not depend on prior knowledge of splicing in target genes, which is attractive for discovery of new splicing patterns.
Several computational methods have been proposed for analysis of alternative splicing from Exon array data (Affymetrix 2005a; Cline et al. 2005; Clark et al. 2007; Yeo et al. 2007). Most notably, Affymetrix has developed a tool, ExACT, which compares the “splicing index” metric across different sample groups to identify differentially used exons (Gardina et al. 2006; Clark et al. 2007). It calculates an “exon expression index” to represent the abundance of transcripts containing a particular exon, and a “gene expression index” to represent the overall transcript abundance of a gene. These indices can be estimated from signal intensities of probes targeting an exon or a gene. The metric “splicing index” is defined as the ratio of exon expression index to gene expression index. A significant difference in the splicing index of an exon between samples indicates differential alternative splicing.
While this exon-tiling design has generated considerable enthusiasm (Gardina et al. 2006; Clark et al. 2007; Kwan et al. 2007; McKee et al. 2007; Yeo et al. 2007; Hung et al. 2008), it is in fact challenging to detect differential alternative splicing events. It is unclear whether the exon-tiling probes alone are sufficient for reliable analysis of splicing, without the inverse correlation between probes for exon skipping and inclusion obtained from the splice junction design (Srinivasan et al. 2005). Two studies using Affymetrix's standard analysis procedure reported low validation rates (21% and 45%) (Gardina et al. 2006; Kwan et al. 2007). Yeo and colleagues used Affymetrix's GC-based background model and a regression-based approach for detecting differential splicing (Yeo et al. 2007). They reported a 56% (9/16) validation rate. Another study by Affymetrix on tissue-specific splicing validated 84% (27/32) candidate brain-specific exons. However, the validation test in this work was done on the top 32 candidates out of 1.4 million probe sets (Clark et al. 2007). Recently, Hung and colleagues combined RNAi knockdown and Exon array profiling to identify the exon targets of a splicing regulator hnRNP L (Hung et al. 2008). Based on the analysis of the Exon array data, they selected 50 candidate genes with differential alternative splicing events after RNAi knockdown of hnRNP L. Their semiquantitative RT-PCR analysis provided strong evidence for differential splicing in 11 genes and marginal evidence in another 17 genes, yielding a validation rate of 22%–56% (Hung et al. 2008). The high degree of uncertainty in the performance of Exon 1.0 array hinders the utilization and future development of this technology. At the heart of the problem is the inherent noise in oligonucleotide probe signals due to various sources of artifacts such as background and cross-hybridization. Since most exons are targeted by no more than four probes in the current design (Affymetrix 2005b), this noise can make the estimated “splicing index” unreliable.
RESULTS
Overview of MADS
In this article, we describe microarray analysis of differential splicing (MADS), a new method for the detection of differential alternative splicing events from exon-tiling microarrays. This method exploits a series of low-level analysis algorithms to construct an efficient statistic for differential splicing. These low-level analyses take advantage of the high probe density of Exon arrays to perform (1) background correction, (2) iterative probe selection for expression index calculation, and (3) detection/removal of sequence-specific cross-hybridization to off-target transcripts (see details of these algorithms in Materials and Methods). By recognizing and correcting for the major sources of noise in Exon array probe intensities, our method can detect changes in splicing of individual exons with improved sensitivity and specificity, as demonstrated in the results below.
Evaluation of MADS using “gold-standard” alternative splicing data
To evaluate MADS, we used exons with predetermined transcript inclusion levels in response to shRNA-mediated repression of the splicing factor polypyrimidine tract binding protein (PTB), based on a previous study of PTB-dependent splicing events (Boutz et al. 2007). We compiled a list of 40 Exon array probe sets whose target exons were differentially spliced after PTB depletion (Supplemental Table 1). We also compiled a second list of 23 Exon array probe sets whose target exons had no change in splicing (Supplemental Table 2). It is important to note that the algorithm development of MADS is independent of the gold-standard set used in our evaluation. In other words, we did not use information from the gold-standard data set to over-train the MADS algorithm so that it could perform well in this data set. For each exon in this gold-standard set, we calculated its P-value for differential splicing using MADS based on Exon array profiles on three shRNA-PTB-treated samples and three mock-treated controls (Materials and Methods). We identified all exons known to have substantial differences in inclusion levels (based on reverse transcriptase-polymerase chain reaction [RT-PCR] data from Boutz et al. [2007]) between shRNA-treated and mock-treated cells. For example, an internal cassette exon in Smap was a known target of PTB. The inclusion level of this exon increased from 7% to 42% after PTB depletion (Boutz et al. 2007). Our analysis of the corresponding Exon array probe set for this exon (probe set 5521400) indicated a significant increase in exon inclusion in shRNA-treated cells (P = 1.8e-06).
To assess the overall performance of our method, we calculated the true positive fraction and false positive fraction in our gold-standard set under varying MADS P-value cutoffs. We also implemented Affymetrix's procedure ExACT (Clark et al. 2007) and calculated its true positive and false positive fractions (see Supplemental Tables 1 and 2 for MADS and ExACT P-values of gold-standard positive and negative exons). Considering the importance of controlling the false discovery rate in genome-wide analyses (Storey and Tibshirani 2003), we compared the true positive fractions of MADS and ExACT when the false positive fraction was small (<10%). We observed a substantial improvement by MADS over Affymetrix's method ExACT. For example, our method had a true positive fraction of 15/40 when no false positive was reported, compared with 8/40 from Affymetrix's method. At the false positive fraction of 2/23, our method's true positive fraction increased to 21/40, while Affymetrix's method was 11/40, a 25% difference. In fact, at this false positive fraction, our true positive fraction was comparable to the small-scale, 1342-exon splice junction array used in the initial discovery of the gold-standard positives (Boutz et al. 2007). This splice junction array had about eight probes per splicing event (Boutz et al. 2007).
We further investigated the potential cause for false negatives from our analysis. We chose the P-value cutoff of 0.049 to call differentially spliced exons in our “gold-standard” set. At this significance level, in the gold-standard exon set, our method had a false positive fraction of 2/23 and a true positive fraction of 21/40 (i.e., missing 47.5% [19/40] of the “gold-standard” positives). We divided the 40 probe sets for gold-standard positives into four distinct categories based on the estimated change in inclusion levels after PTB depletion by RT-PCR gel quantification (see Materials and Methods and Supplemental Table 3). The sensitivity of our method was positively correlated with the magnitude of the change in exon inclusion level (Fig. 1). We only detected one out of 11 exons with <5% change in inclusion level, while five out of five exons exhibiting a change >25% were detected. In addition, four false negatives were from probe sets with a single probe on the Exon 1.0 array. Overall, these results indicate that the current Exon 1.0 array is capable of detecting substantial changes in exon inclusion, but is not yet sensitive enough to identify subtle splicing changes. Future designs that increase the probe density in well-annotated exonic regions will likely improve the sensitivity of this technology.
FIGURE 1.
The sensitivity of MADS is positively correlated with the magnitude of the change in exon inclusion levels. (X-axis) Change in exon inclusion levels between shRNA-treated cells and mock-treated cells, estimated from quantification of RT-PCR gels of “gold-standard” positives (Boutz et al. 2007). (Y-axis) Number of Exon array probe sets that detect gold-standard differential splicing events (i.e., true positives) and the number of Exon array probe sets that fail to detect gold-standard differential splicing events (i.e., false negatives).
Validation of novel differential splicing events
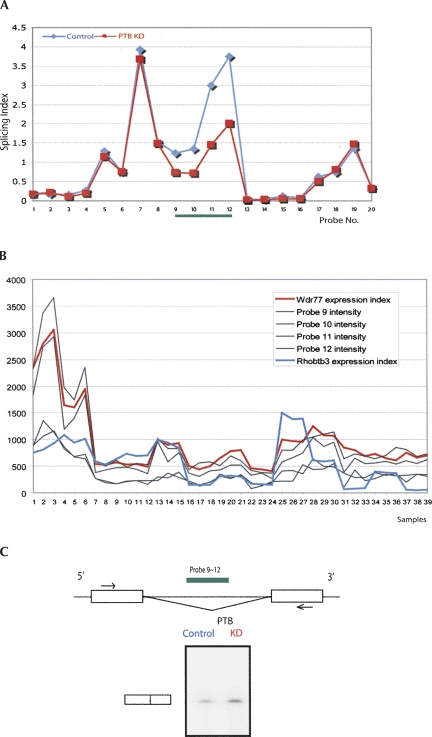
Next, we focused on the discovery of novel differential splicing events from Exon arrays. We ranked all probe sets according to their MADS P-values. We selected 30 genes from the top 1500 probe sets for validation (Supplemental Table 4). The selection of these genes did not consider prior knowledge of potential PTB binding sites or likely functional relevance of the target genes. In other words, we did not cherry-pick the 30 genes from the pool of 1500 in order to achieve a high validation rate. The primary selection criterion was to facilitate RT-PCR primer design and experiments; i.e., the selected candidate exons should be 50–100 nucleotides (nt) in length and should be flanked by constitutively spliced exons. The median MADS P-value ranking of selected candidate probe sets is 154. Of the 30 genes tested, our RT-PCR experiments confirmed differential splicing events in 27 genes, yielding a validation rate of 90% (Supplemental Fig. 1; Supplemental Table 5). We show three examples in Figure 2 (also see Supplemental Fig. 2 for UCSC Genome Browser screenshots of these exons). We found an exon in Garnl1 to be strongly up-regulated in shRNA-treated cells (Fig. 2A; also see Fig. 4 below for sequence of this exon and visualization of its probe intensities). This exon was not present in any mouse mRNAs and ESTs. Its probe design (probe set 4742986) was based on a computational exon prediction by N-SCAN (van Baren and Brent 2006). The genomic region of this exon is highly conserved, with only a single nucleotide substitution between mouse and chicken. Our RT-PCR test confirmed the expression of this novel exon as well as its differential alternative splicing after PTB depletion. In Tmem87a (Fig. 2B), we detected and validated the differential use of two mutually exclusive exons, whose upstream introns had the major-class U1/U2 splice sites (GU-AG) while the downstream introns had the minor-class U11/U12 splice sites (AU-AC) (Letunic et al. 2002). The lengths of these two exons only differed by 3 base pairs (bp). Quantification of the gel image indicated that the relative abundance of the longer Tmem87a isoform dropped from 30% to 16% after PTB depletion. We also confirmed the differential usage of alternative 3′-untranslated region (UTR) and polyadenylation sites in Ncam1, using primers specific for alternative 3′-UTRs. This differential splicing event was supported by a total of 12 probe sets (Fig. 2C). The complete list of RT-PCR confirmed novel PTB-dependent splicing events is presented in Supplemental Table 4. Of the 27 validated events, excluding the complex alternative splicing pattern in Ncam1, 22 exons were up-regulated after PTB knockdown (i.e., their splicing is repressed by PTB). Four exons in the Scarb1, Ubp1, App, and 4931406I20Rik genes were down-regulated after PTB knockdown (i.e., their splicing is enhanced by PTB) (see Supplemental Fig. 1).
FIGURE 2.
Detection and validation of novel differential alternative splicing events. (A) Garnl1: Splicing indices of Garnl1 exons suggest a differentially spliced, novel cassette exon. The splicing index was calculated as the background-corrected probe intensity divided by the estimated gene expression index (Materials and Methods). The splicing indices of probes 9–12, which target the cassette exon, indicate a substantially increased exon inclusion level after PTB depletion. RT-PCR experiments confirmed a higher ratio of the exon inclusion form over the exon skipping form in shRNA-treated cells (KD) compared with mock-treated cells (control). The RT-PCR primers are indicated (arrows) on the gene structure diagram (right). (B) Tmem87a: Splicing indices of Tmem87a suggest a pair of mutually exclusive exons. One exon (74 nt in length) was targeted by probes 5–8, whose splicing indices were lower after PTB depletion (probe set 5350538, MADS P-value 1.9e-04). Another exon (71 nt in length) was targeted by a single probe (probe 9), whose splicing index was higher after PTB depletion. RT-PCR experiments confirmed the Exon array result. (C) Ncam1: Splicing indices of 48 probes in 12 Ncam1 probe sets suggest alternative 3′-UTR and polyadenylation. Probes 1–20 target one alternative 3′-UTR whose usage is increased after PTB depletion. Probes 21–48 target three exons in another alternative 3′-UTR whose usage is reduced after PTB depletion.
FIGURE 4.
A screenshot of our stand-alone Exon array genome browser for the novel PTB-dependent exon of Garnl1. (A) Probe intensities of the novel PTB-dependent exon of Garnl1 and its flanking exons are visualized by our Exon array genome browser. (Top track) RefSeq gene structure, (middle track) probe intensities in mock-treated cells (control), (bottom track) probe intensities in shRNA-treated cells (KD). Each vertical line represents the background corrected, normalized intensity of a probe. (Box) PTB-dependent exon. The intensities of its four probes are significantly higher in KD samples compared with control samples, using the probes of the flanking constitutive exons as the control. This stand-alone Exon array genome browser is available at http://biogibbs.stanford.edu/∼jiangh/browser/. (B) The nucleotide sequence of the Garnl1 PTB-dependent exon. This exon has no mRNA/EST evidence and is supported by a computational exon prediction of N-SCAN (van Baren and Brent 2006). (Horizontal lines) Placement of four Exon array probes.
Correction for sequence-specific cross-hybridization to off-target transcripts
Our analysis also shows that cross-hybridization is a major source of false positives for exon expression and differential splicing. We developed an algorithm to detect sequence-specific cross-hybridization to off-target transcripts (see Materials and Methods). For probe set 4365403 of Rhobtb3, our initial analysis suggested a much reduced exon inclusion level in shRNA-treated cells (Fig. 3A). However, we found that all four probes of this probe set had perfect matches to the gene Wdr77 (6900236). Moreover, the intensities of these four probes had high correlation (0.62, 0.59, 0.74, 0.74, respectively) with the estimated expression levels of Wdr77 across 11 mouse tissues and our samples (Fig. 3B). Since the expression level of Wdr77 had a 1.6-fold reduction in shRNA-treated cells, we hypothesized that the apparent differential splicing of this Rhobtb3 exon was an artifact due to cross-hybridization. Indeed, RT-PCR experiments indicated no differential splicing or even the expression of this exon (Fig. 3C). We also tested probe sets in Rab31, Nsf, Smchd1, and Ndufab1. These probe sets were designed for exons supported by ESTs or computational exon predictions. The observed intensities of these probe sets were high. However, our analysis suggested that these probes cross-hybridized to highly expressed off-target transcripts (Supplemental Table 6). On the other hand, the intensities of these probe sets had poor correlation (<0.3) with their intended target genes. RT-PCR analyses indicated that these four apparently “novel” exons were not in fact expressed (Supplemental Fig. 1; Supplemental Table 6).
FIGURE 3.
Removal of false positive detection of differential splicing due to sequence-specific cross-hybridization to off-target transcripts. (A) Splicing indices of Rhobtb3 probes suggest a differentially spliced exon (targeted by probe set 4365403, probes 9–12 of this plot). The inclusion level of this exon appears to be substantially lower in shRNA-PTB-treated cells compared with mock-treated controls. (B) Detailed analyses of Rhobtb3 probe set 4,365,403 indicate cross-hybridization. (X-axis) Sample indices of 11-tissue panel and PTB samples. Samples 1–3 are mock-treated controls; samples 4–6 are shRNA-PTB-treated cells; the remaining samples are from Affymetrix's mouse 11-tissue panel, with three replicates for each tissue. (Y-axis) Probe intensity or estimated gene expression index. Each of the four black lines represents a probe in probe set 4,365,403. (Red line) Estimated gene expression indices of a potential off-target gene Wdr77. All four probes in probe set 4,365,403 are perfect matches to Wdr77. Their probe intensities are highly correlated with the estimated gene expression indices of Wdr77 across the 39 samples, with Pearson correlation coefficients of 0.62, 0.59, 0.74, and 0.74, respectively. (Blue line) Estimate gene expression indices of Rhobtb3, which are poorly correlated with the intensities of probe set 4,365,403. (C) RT-PCR using primers targeting flanking exons shows a single band corresponding to the exon-skipping form. The target exonic region of the probe set 4,365,403 (probes 9–12) is not expressed. (KD) shRNA-treated cells.
DISCUSSION
In this work, we demonstrate the efficacy of the MADS procedure for the Exon array analysis of alternative splicing. We evaluated the performance of our method, using a list of 63 probe sets targeting exons with predetermined transcript inclusion levels in response to shRNA-mediated depletion of PTB (Boutz et al. 2007). This allowed a large-scale, unbiased assessment of the Exon 1.0 array on its ability to detect differential splicing events. Although the “gold-standard” set is not sufficiently large for a robust performance analysis (such as the receiver operating characteristic analysis), we show that our method improves the true positive fraction by as much as 25%, compared with Affymetrix's method (ExACT) at the same false positive fraction. Based on prior RT-PCR gel quantification data (Boutz et al. 2007), we show that we can reliably detect exons with significant changes in their transcript inclusion levels. On the other hand, the Exon 1.0 array is less sensitive to small changes in exon inclusion than the small-scale splicing junction array (Boutz et al. 2007), which has both exon probes and splice junction probes. However, the Exon array has real advantages in the number of exons profiled. These features will be important for a number of experimental applications, such as discovering the regulatory targets of splicing regulators on a genomic scale.
Our analysis has identified a large set of novel PTB-dependent splicing events. Overall, the 27 validated differentially spliced genes cover all alternative splicing patterns: single cassette exon, skipping of multiple adjacent cassette exons, alternative donor/acceptor splice sites, mutually exclusive exon usage, intron retention, and alternative polyadenylation. This shows an advantage of the unbiased exon-tiling design, which does not require prior knowledge of the use of a particular spliced segment. The MADS P-value rankings of RT-PCR-confirmed probe sets had an interquartile range (IQR) of 45–299, with the lowest ranking being 1371 (Supplemental Table 4). Extrapolating from our validation rate (90%), hundreds of probe sets would represent bona fide novel PTB-dependent splicing events. This large exon list will allow new analyses of the sequence features needed for PTB binding and regulation. Our MADS algorithm can also be useful for Exon array analysis of other splicing regulators, or in the reanalysis of existing Exon array profiles (such as the hnRNP L data set) (Hung et al. 2008).
Our work illustrates the importance of correcting probe-level noise (such as background and cross-hybridization) for microarray analysis of alternative splicing. The inference of exon-level expression (such as alternative splicing) from the Exon 1.0 array is considerably more challenging compared with analysis of overall gene expression by conventional gene expression microarrays. The key to reliable expression estimates from short-oligonucleotide arrays is to model the performance of individual probes based on sequence information and empirical data on the probe's performance. This is the strategy behind several popular programs such as dChip (Li and Wong 2001) or GC-RMA (Wu and Irizarry 2005) for the analysis of traditional 3′ expression arrays. For Exon array analysis, since the number of probes on individual exons is small (four on average on the Exon 1.0 array), it becomes extremely important to recognize and eliminate noise in observed probe intensities due to cross-hybridization to off-target transcripts. Because of the ability of Exon 1.0 array to accurately estimate the expression levels of all well-annotated genes (Kapur et al. 2007; Xing et al. 2007), it is in principle possible for us to first conduct sequence searches for off-target transcripts that have 0, 1, 2, or 3 bp mismatches, then identify those with highly correlated signals to our probe(s) of interest. This strategy has not been feasible until the availability of the Exon array, since all previous arrays were either non-comprehensive or too probe-poor (i.e., not having enough probes to construct a robust gene-level estimate) to support such a computation. In Rhobtb3, the cross-hybridization analysis eliminated a strong false positive prediction for differential splicing. Among the initial list of the top 500 differentially spliced probe sets, the fraction of cross-hybridizing probes was 3.7%, compared with 0.8% of all probes analyzed on the array. Since our knowledge of the transcriptional landscape of mammalian genomes is far from complete (Kapranov et al. 2007b), it is possible that the true scope of cross-hybridization is even broader. The detection and filtering of cross-hybridizing probes will eliminate an important source of false positives. As demonstrated in our analysis of Rab31, Nsf, Smchd1, and Ndufab1, false discoveries of novel transcribed regions also may be caused by cross-hybridization. In the future, it will be useful to incorporate cross-hybridization correction techniques in the refinement of the global RNA transcription map inferred from extremely dense whole-genome tiling arrays (Kapranov et al. 2007a).
Our data demonstrate that the exon-tiling microarray design is an efficient and powerful approach for global analysis of alternative splicing. The current Exon 1.0 array has only four probes for most exons and no splice junction probes. Despite that, using statistical methods to recognize and correct for microarray noise, we have identified a large number of differentially spliced exons at a very low false positive rate. The limitations inherent in the design of the current Exon 1.0 arrays will be solved as newer versions of “probe-rich” Exon arrays become available in the near future. During the past decade, we have seen a dramatic increase in the density of oligonucleotide expression microarrays. There is a 40-fold increase in density for spotted/inkjet arrays in the past 11 years (DeRisi et al. 1997; NimbleGen product page: http://www.nimblegen.com/products/exp/#eukaryotic) and an 80-fold increase for photolithography in the past eight years (from Affymetrix Hu6800 to Exon 1.0 array). Since photolithography has submicron resolution, there should be considerable room for further increases in the density of these arrays. This will create even more powerful microarray platforms for exon-level profiling of eukaryotic transcriptomes.
MATERIALS AND METHODS
Statistical procedure for detecting differential alternative splicing
Briefly, our method (MADS) involves the following steps:
First, we predict the background intensities of individual probes using a sequence-specific, 80-parameter linear model that considers the composition of nucleotides at each position of a 25-mer probe (Johnson et al. 2006; Kapur et al. 2007). The advantage of this background model over the standard GC-based background model was described in detail before (Kapur et al. 2007). We train the background model using “genomic” and “anti-genomic” background probes on the Exon 1.0 array (Affymetrix 2005b). For every probe, we subtract the predicted background intensity from the observed probe intensity (Kapur et al. 2007) and use the background-corrected intensity for downstream analysis.
Second, for each gene we use a correlation-based iterative probe selection algorithm to select a subset of probes with highly correlated intensities across diverse samples (Xing et al. 2006). In this work, we use Exon array data for our own samples and the Affymetrix 11-tissue panel (Xing et al. 2006). For each gene, we apply hierarchical clustering (distance metric: 1-Pearson correlation; average linkage clustering) to cluster its core probes using their background-corrected intensities in all samples. We cut the clustering dendrogram at a pre-defined height (0.5 in this study). The intensities of core probes in the biggest sub-cluster are fitted to the Li–Wong model (Li and Wong 2001) to obtain estimated gene expression levels. After the initial Li–Wong fit, for each core probe, we calculate the Pearson correlation coefficient of its background-corrected intensities with the current gene expression estimates in all samples. If the correlation is below a pre-defined threshold (0.7 in this study), the probe will be dropped from the list of selected probes. If the correlation is above this pre-defined threshold, the probe will be retained or added to the list of selected probes. We repeat this procedure until the list of selected probes stabilizes. The final selected probes are regarded as reliable indicators of overall gene expression levels. Their background-corrected intensities are fitted to the Li–Wong model (Li and Wong 2001) to calculate gene expression indices. The idea of our probe selection procedure is similar to the iterPLIER algorithm (Affymetrix 2005c) used by Affymetrix to calculate Exon array expression indices. Lowly expressed genes whose expression indices are below a given cutoff (500 in this study) are removed from further analysis. Alternatively, we can use the background model to calculate a P-value for gene presence/absence as described before (Kapur et al. 2007) and remove genes called absent in one of the sample groups. We can also drop genes without enough selected probes (e.g., <11) to construct a robust gene-level estimate.
Third, for each probe, we calculate its splicing index as the ratio of its background-corrected probe intensity to the estimated gene expression index. We conduct two separate one-sided t-tests to assess whether the splicing indices of a probe are significantly higher or lower in one sample group over another group. After we obtain P-values for individual probes, we summarize a probe-set-level P-value using Fisher's method as follows. The P-values for individual probes are transformed via the formula x = −2log(p). Under the null hypothesis that the exon targets are not differentially spliced, the P-values follow a uniform [0,1] distribution, and the transformed P-values follow χ 2 2 distribution. The sum of the transformed P-values follows χ 2 2k distribution where k equals the number of probes. This sum of the transformed P-values is used to calculate a probe-set-level P-value, which is used to rank all probe sets. The t-test of splicing index is similar to the ANOVA-based method used by Affymetrix in the analysis of tissue-specific and cancer-specific alternative splicing (Affymetrix 2005a; Gardina et al. 2006; Clark et al. 2007). The main distinction is that MADS calculates splicing indices and P-values of individual probes separately, prior to the summarization of a probe-set-level P-value. By contrast, Affymetrix's approach first calculates an overall exon-level expression index (from no more than four probes per probe set), prior to splicing index calculation and statistical testing.
Finally, to eliminate false positives due to cross-hybridization, we use an efficient sequence search algorithm (H. Jiang and W.H. Wang, unpubl.; http://biogibbs.stanford.edu/∼jiangh/SeqMap/) to search all 25-mer oligonucleotide probes against all RefSeq-supported exon regions, allowing up to 3-bp mismatches. Once a potential off-target gene is found for a probe, we calculate the Pearson correlation coefficient between the probe's intensities and the off-target gene's expression indices (also estimated from Exon arrays) across our own samples and the Affymetrix 11-tissue panel (Xing et al. 2006). We define a probe to be cross-hybridizing if there is an off-target gene within 3-bp mismatches, and if the computed Pearson correlation coefficient is above a given threshold (0.55 in our current computation). Probe sets with cross-hybridizing probes are regarded as unreliable and are filtered from the final result. In this paper, we use cross-hybridization merely in the follow-up analysis of exons selected for validation. In the future it will be useful to incorporate the cross-hybridization analysis directly into the statistics for ranking of exons.
Compilation of a “gold-standard” exon set
To evaluate the performance of our method, it is essential to have a “gold-standard” set of exons whose splicing patterns are known in particular samples. For this purpose, we used a list of 79 exons with known inclusion/exclusion profiles in response to shRNA-mediated knockdown of the splicing factor PTB. PTB is a well-characterized splicing repressor. It binds to pyrimidine-rich elements on pre-mRNAs and suppresses the splicing of a wide range of mammalian tissue-specific exons (Black 2003). Previously, we used shRNA to deplete PTB in mouse N2A neuroblastoma cells. We confirmed 50 exons that were differentially spliced after PTB depletion and 29 exons that were unaffected by PTB, using a small-scale (1342-exon) custom splice-junction microarray followed by extensive RT-PCR tests (Boutz et al. 2007). Exons with positive or negative confirmation for differential splicing based on these earlier RT-PCR data were used to construct the gold-standard set. For each exon, RT-PCR gel images were quantified by ImageQuant TL software (GE Lifesciences). We then estimated the relative levels of the mRNA isoforms in control and shRNA-treated cells. The inclusion level of the exon in a sample was calculated as the percentage of exon-containing isoform. Since these exons were annotated on the mm5 version of the mouse genome at the time of the initial work (Boutz et al. 2007), we mapped these exons to the mm8 version using the LiftOver tool of UCSC Genome Browser (Kent et al. 2002). We then mapped Exon array probe sets to these exons, requiring that the target region of a probe set to be fully contained by an exon in our gold-standard set. After removing exons without Exon 1.0 array probe sets, we obtained 40 probe sets as our “gold-standard” positives and 23 probe sets as our “gold-standard” negatives.
Exon array hybridization and analysis
Short hairpin knockdown of PTB was performed as described before (Boutz et al. 2007). The efficiency of the PTB knockdown was monitored by Western blot using PTB-NT primary antibody and Cy5 labeled secondary antibody (GE Life Sciences). The blots were imaged using Typhoon 9410 (GE Life Sciences). The band intensities were measured using ImageQuant and normalized to GAPDH. In all cases, the efficiency of the knockdown was close to 80% (data not shown).
We conducted Exon array profiling on RNAs from three shRNA-PTB-treated samples and three mock-treated controls (using empty vectors). Probes for the Affymetrix Exon Array ST 1.0 were prepared and hybridized to the array using the GeneChip Whole Transcript Sense Target Labeling Assay (Affymetrix) according to the manufacturer's suggestions. Briefly, for each sample, 2 μg of total RNA was subjected to ribosomal RNA reduction. Following rRNA reduction, double-stranded cDNA was synthesized with random hexamers tagged with a T7 promoter sequence. The double-stranded cDNA was used as a template for amplification with T7 RNA polymerase to create antisense cRNA. Next, random hexamers were used to reverse transcribe the cRNA to produce single-stranded sense strand DNA. The DNA was fragmented and labeled with terminal deoxynucleotidyl transferase. The probes of triplicate samples of the shRNA-PTB-treated cells and mock-treated controls were hybridized to the Affymetrix mouse Exon 1.0 arrays and scanned.
We used the “gold-standard” sets of known positives and known negatives to count the numbers of true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). We calculated the true positive fraction and false positive fraction at a particular MADS P-value cutoff as:
 |
RT-PCR validation of differential alternative splicing
We conducted RT-PCR validation of novel PTB-dependent splicing events discovered from genome-wide Exon array analysis. Total RNA was collected from adherent tissue culture cells using Trizol (Invitrogen) according to the manufacturer's instructions. RNA samples were then treated with DNase I to remove residual DNA contamination and extracted with chloroform. RNA was quantified (A260) using a Nanodrop-1000 spectrophotometer (Nanodrop Technologies). Total RNA (2 μg) was reverse transcribed with SuperScript III (Invitrogen) according to the manufacturer's protocol. For each candidate differential splicing event, we used PRIMER3 (Koressaar and Remm 2007) to design forward and reverse primers against its flanking exon regions. One-fortieth volume of the RT reaction was used in a 10-μL PCR reaction containing 32P-labeled primers. PCR reactions were run in an MJ Research PTC-200 thermocycler for 25 cycles with an annealing temperature of 60°C. The reaction products were resolved on 8% denaturing polyacrylamide gel and imaged using Typhoon 9410. The images were quantified using ImageQuant TL (GE Lifesciences). The inclusion levels of the exons were calculated as the percentage of exon-containing isoforms.
Available software/datasets
Our software tools and datasets are publicly available to the research community: (1) We release the MADS source code for detection of differential splicing events from Exon 1.0 array data (http://biogibbs.stanford.edu/∼yxing/MADS/). We also provide at this URL the lists of cross-hybridizing probes on human and mouse Exon 1.0 arrays and their potential off-target genes. These lists are obtained by analyzing a diverse set of public and in-house Affymetrix Exon array profiles, using the cross-hybridization detection method described in this manuscript. (2) We develop and maintain a light-weight, stand-alone genome browser for visualization of Exon 1.0 array profiles along the genome (http://biogibbs.stanford.edu/∼jiangh/browser/; see Fig. 4 for a screen shot showing the probe intensities of the novel PTB-dependent exon of Garnl1). Using this genome browser, researchers can inspect the data in specific genes of interest to assess the exon-level expression patterns (e.g., differential alternative splicing) discovered by our computational analysis. (3) We have deposited our Exon array data set to the NCBI GEO database under the accession number GSE11344.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Elizabeth Zuo, Hong Yang, Zhengqing Ouyang, Wenxiu Ma, and Chia-Ho Lin for technical assistance, and Erik Miller, Wenzhong Xiao, Eric Chiao, and Quntian Wang for discussions. We thank David Eichmann and the University of Iowa Institute for Clinical and Translational Science (NIH grant UL1 RR024979) for computer support. This work is supported by NSF grant DMS0505732, NIH grants R01HG002341 and R01HG003903 to W.H.W., and NIH grant R24GM070857 to D.L.B. Y.X. is supported by a Hereditary Disease Foundation research grant and a research startup fund from the University of Iowa. P.S. is supported by development grant MDA4260 from the Muscular Dystrophy Association. D.L.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1070208.
REFERENCES
- Affymetrix. Alternative transcript analysis methods for exon arrays. 2005a http://www.affymetrix.com/support/technical/whitepapers/exon_alt_transcript_analysis_whitepaper.pdf.
- Affymetrix. Exon array design datasheet. 2005b http://www.affymetrix.com/support/technical/datasheets/exon_arraydesign_datasheet.pdf.
- Affymetrix. Gene signal estimates from exon arrays. 2005c http://www.affymetrix.com/support/technical/whitepapers/exon_gene_signal_estimate_whitepaper.pdf.
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Boutz P.L., Stoilov P., Li Q., Lin C.H., Chawla G., Ostrow K., Shiue L., Ares M., Jr, Black D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T.A., Sugnet C.W., Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M.S., Blume J., Cawley S., Clark T.A., Hu J.S., Lu G., Salomonis N., Wang H., Williams A. ANOSVA: A statistical method for detecting splice variation from expression data. Bioinformatics. 2005;21(Suppl. 1):i107–i115. doi: 10.1093/bioinformatics/bti1010. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Gardina P.J., Clark T.A., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L.H., Heiner M., Hui J., Schreiner S., Benes V., Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: A combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip J.Y., Tong A., Pan Q., Topp J.D., Blencowe B.J., Lynch K.W. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007a;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007b;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kapur K., Xing Y., Ouyang Z., Wong W.H. Exon arrays provide accurate assessments of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kwan T., Benovoy D., Dias C., Gurd S., Serre D., Zuzan H., Clark T.A., Schweitzer A., Staples M.K., Wang H., et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Bork P. Common exon duplication in animals and its role in alternative splicing. Hum. Mol. Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A.E., Neretti N., Carvalho L.E., Meyer C.A., Fox E.A., Brodsky A.S., Silver P.A. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome Biol. 2007;8:R159. doi: 10.1186/gb-2007-8-8-r159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Roberts G.C., Smith C.W. Alternative splicing: Combinatorial output from the genome. Curr. Opin. Chem. Biol. 2002;6:375–383. doi: 10.1016/s1367-5931(02)00320-4. [DOI] [PubMed] [Google Scholar]
- Srinivasan K., Shiue L., Hayes J.D., Centers R., Fitzwater S., Loewen R., Edmondson L.R., Bryant J., Smith M., Rommelfanger C., et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baren M.J., Brent M.R. Iterative gene prediction and pseudogene removal improves genome annotation. Genome Res. 2006;16:678–685. doi: 10.1101/gr.4766206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.S., Cooper T.A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- Xing Y., Kapur K., Wong W.H. Probe selection and expression index computation of Affymetrix exon arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Ouyang Z., Kapur K., Scott M.P., Wong W.H. Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol. Biol. Evol. 2007;24:1283–1285. doi: 10.1093/molbev/msm061. [DOI] [PubMed] [Google Scholar]
- Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Xu X., Liang T.Y., Muotri A.R., Carson C.T., Coufal N.G., Gage F.H. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput. Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]