Abstract
PUF proteins comprise a highly conserved family of sequence-specific RNA binding proteins that regulate target mRNAs via binding directly to their 3′UTRs. The Caenorhabditis elegans genome encodes several PUF proteins, which cluster into four groups based on sequence similarity; all share amino acids that interact with the RNA in the cocrystal of human Pumilio with RNA. Members of the FBF and the PUF-8/9 groups bind different but related RNA sequences. We focus here on the binding specificity of representatives of a third cluster, comprising PUF-5, -6, and -7. We performed in vivo selection experiments using the yeast three-hybrid system to identify RNA sequences that bind PUF-5 and PUF-6, and we confirmed binding to optimal sites in vitro. The consensus sequences derived from the screens are similar for PUF-5 and PUF-6 but differ from those of the FBF or PUF-8/-9 groups. Similarly, neither PUF-5 nor PUF-6 bind the recognition sites preferred by the other clusters. Mutagenesis studies confirmed the unique RNA specificity of PUF-5/-6. Using the PUF-5 consensus derived from our experiments, we searched a database of C. elegans 3′UTRs to identify potential targets of PUF-5, several of which indeed bind PUF-5. Therefore the consensus has predictive value and provides a route to finding genuine targets of these proteins.
Keywords: PUF protein, C. elegans, RNA binding protein, RNA–protein interactions, development, mRNA control
INTRODUCTION
Regulation of mRNA stability and translation determines how much protein is produced from an mRNA and often is governed by elements in the 3′ untranslated region (UTR). These elements bind regulatory proteins and microRNAs that then mediate their effects on mRNA stability, translation, and localization. Indeed, 3′UTR-borne complexes are critical in a wide range of biological contexts (Kuersten and Goodwin 2003; Mazumder et al. 2003; Colegrove-Otero et al. 2005; de Moor et al. 2005; Vardy and Orr-Weaver 2007). Not surprisingly, dysfunctional controls can lead to disease (Fan et al. 2005; Esquela-Kerscher and Slack 2006; Osborne and Thornton 2006; Hamilton et al. 2007). The specificity of the complexes—which mRNAs they control—is determined by the RNA sequence selectivity of the RNA binding proteins within them.
PUF proteins embody many general features of 3′UTR regulatory proteins. PUF proteins bind specific RNA sequences in 3′UTRs. The resulting RNA–protein complexes trigger translational repression, deadenylation, and/or mRNA decay (Ahringer and Kimble 1991; Wreden et al. 1997; Zhang et al. 1997; Chagnovich and Lehmann 2001; Goldstrohm et al. 2006, 2007; Kadyrova et al. 2007). The exact outcome varies with the PUF protein and mRNA (e.g., Hook et al. 2007). PUF proteins are critical in developmental processes, including the maintenance of stem cells in Caenorhabditis elegans, Drosophila, and planaria (Lehmann and Nusslein-Volhard 1987; Lin and Spradling 1997; Forbes and Lehmann 1998; Crittenden et al. 2002; Salvetti et al. 2005), and are implicated in stem cell control in vertebrates as well (Moore et al. 2003). PUF proteins also are critical in the nervous system: They are required for long-term memory, and control neuron excitability, and morphogenesis (Schweers et al. 2002; Dubnau et al. 2003; Mee et al. 2004; Menon et al. 2004; Ye et al. 2004).
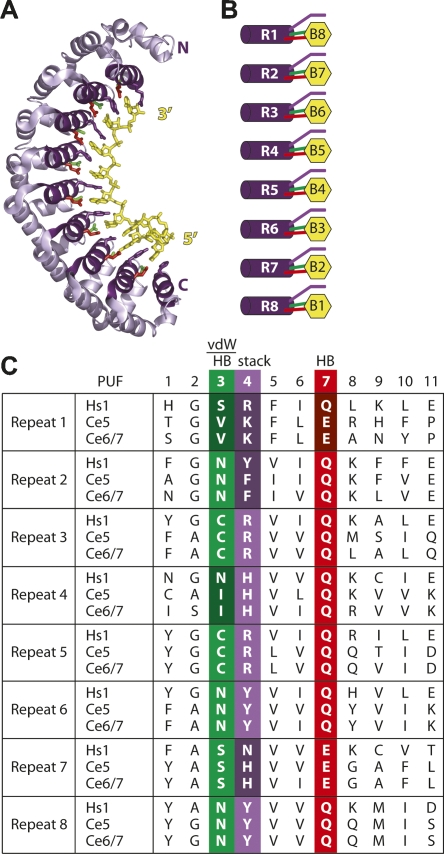
PUF proteins are defined by the presence of eight triple-helical repeats termed PUF repeats (Edwards et al. 2001; Wang et al. 2001). The repeats are arranged in an elongated crescent shape with one helix from each repeat presenting the amino acids required for RNA binding to the concave face of the protein (Fig. 1A). In the complex of human Pumilio with a target RNA, each repeat contacts a single nucleotide using hydrogen bonding, van der Waals, and stacking interactions (Fig. 1A–C; Wang et al. 2002).
FIGURE 1.
PUF proteins share similar structure. (A) Model of the human Pumilio–RNA interaction, based on the human Pumilio–RNA cocrystal in Wang et al. (2002). (B) Contacts made between PUF repeats and RNA. Coloring corresponds with that in panel C and to positions in the RNA recognition α-helix. (C) Alignment of the RNA recognition helices of C. elegans PUF-5, PUF-6, and PUF-7 with human Pumilio. Amino acid residues differing between PUF-5/-6/-7 and Pumilio are shaded more darkly. Residues involved in van der Waals contacts, stacking interactions, and hydrogen bonding are identified above the alignment.
The amino acids involved in binding RNA are well conserved, yet different PUF proteins discriminate different sequences. This paradoxical difference is understood in a single instance involving two C. elegans proteins, FBF and PUF-8. In this case, FBF requires the insertion of a single nucleotide in the middle of the PUF-8 RNA binding site; PUF-8 binds only in the absence of that nucleotide. The identity of the “extra” nucleotide is unimportant for FBF binding (Opperman et al. 2005). The inference is that for FBF and PUF-8, most of the interactions that mediate contacts between the proteins and the bases in their cognate RNAs are the same, but the two proteins bind distinct RNAs depending on the presence or absence of a single nucleotide. Other PUF proteins may gain specificity by using novel amino acid–nucleotide contacts in divergent PUF repeats.
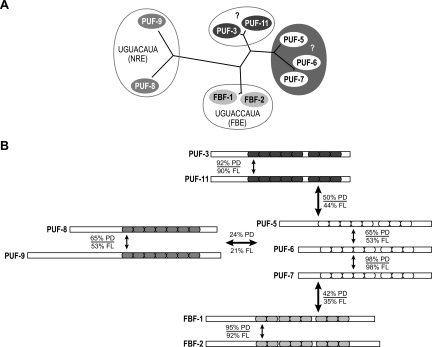
C. elegans possesses multiple canonical PUF proteins that we have grouped into four clusters based on the similarities of their amino acid sequences (Fig. 2). Of these, the FBF group, consisting of FBF-1 and FBF-2, has been characterized in the greatest detail, both in terms of its binding specificity and biological roles. FBF-1 and FBF-2 recognize identical sequences and control stem cell proliferation and sex determination in the germline (Zhang et al. 1997; Crittenden et al. 2002; Lamont et al. 2004; Bachorik and Kimble 2005; Bernstein et al. 2005; Opperman et al. 2005; Thompson et al. 2005; Cho et al. 2007). PUF-8 is less well understood but participates in sex determination and spermatogenesis (Subramaniam and Seydoux 2003; Bachorik and Kimble 2005). Its close relative, PUF-9, controls differentiation of epidermal stem cells and progression through the heterochronic pathway, possibly through a conserved interaction with a hunchback-like mRNA (Nolde et al. 2007). The two remaining clusters of C. elegans PUF proteins have not been analyzed in great detail. The PUF-5/-6/-7 proteins function redundantly in the late stages of oogenesis (Lublin and Evans 2007), while silencing PUF-3/-11 via RNAi yields an embryonic lethal phenotype (Sonnichsen et al. 2005). The simplest interpretation of the data is that each group recognizes and controls distinct mRNAs.
FIGURE 2.
C. elegans PUF proteins cluster into four groups. (A) Four distinct clusters of C. elegans PUF proteins based on amino acid sequence similarity. (B) Proteins within a group share a higher degree of identity than those from other groups. Percentage of identity is reported for the PUF domain (PD) and full-length proteins (FL). Colored boxes represent the PUF repeats within each protein. Colors correspond to those in panel A.
We sought to characterize the binding specificity of the C. elegans PUF-5/-6/-7 cluster. We identified a consensus binding element with high affinity for PUF-5. This element is recognized by PUF-6, and distinct from elements recognized by FBF and PUF-8 proteins. The site is longer than those of FBF or PUF-8 and possesses an additional and essential UGU trinucleotide not found in other PUF binding sites. Our knowledge of PUF-5 sequence specificity was used to identify potential mRNA targets of PUF-5 and PUF-6 in a database of C. elegans 3′UTR sequences.
RESULTS
Four clusters of C. elegans PUF proteins
C. elegans expresses nine canonical PUF proteins. Using the relatedness of their amino acid sequences, we grouped these into four clusters (Fig. 2). Two other PUF-related sequences, PUF-4 and PUF-10, are present in the genome but appear to be pseudogenes, as no ESTs have been reported. The four clusters can be distinguished by comparing the amino acid sequences of their RNA binding domains (Fig. 2). Outside their RNA binding domains, the clusters shown in Figure 2 are even more apparent: Proteins within a cluster are related, but distinct from other groups (not shown).
Proteins in the FBF and PUF-8/-9 groups bind distinct RNA sequences (Opperman et al. 2005). The FBF group comprises FBF-1 and FBF-2, which both specifically bind the FBF binding element (FBE) (Fig. 2A; Bernstein et al. 2005; Opperman et al. 2005). PUF-8 and PUF-9 both bind to a sequence first identified as the binding site for the Drosophila Pumilio protein, to which they are the closest C. elegans relatives (Fig. 2A; Opperman et al. 2005; Nolde et al. 2007). This binding element is termed the Nanos response element (NRE) or Pumilio binding element (PBE); here, we use the term NRE.
Of the remaining C. elegans PUF proteins, PUF-5, PUF-6, and PUF-7 form one group, while PUF-3 and PUF-11 form another (Fig. 2). PUF-6 and PUF-7 are 98% identical, while PUF-5 is 55% identical to both PUF-6 and PUF-7 (Fig. 2B). Of the amino acids that directly contact RNA in the human Pumilio–NRE cocrystal, all are the same in PUF-5, PUF-6, and PUF-7 (Fig. 1C). The RNA binding specificities of the PUF-5/-6/-7 and PUF-3/-11 groups are unknown and their mRNA targets unidentified. We hypothesized that proteins within these groups bind similar RNA sequences, distinct from those bound by proteins in other groups.
One cannot predict the RNA binding specificities of PUF proteins ab initio. The C. elegans PUF proteins are very similar to one another in terms of the amino acids that interact with RNA in the human Pumilio–RNA structure. In the particular case of the PUF-5/-6/-7 family, 18 of the 24 amino acids involved in human Pumilio's base-specific contacts are identical in PUF-5/-6/-7 (Fig. 1C). Five of the six amino acids that differ are located in the amino-terminal half of the PUF domain, suggesting that the 3′ end of the RNA binding site of PUF-5 and PUF-6 might differ from that of Pumilio.
PUF-5 binds a unique RNA sequence
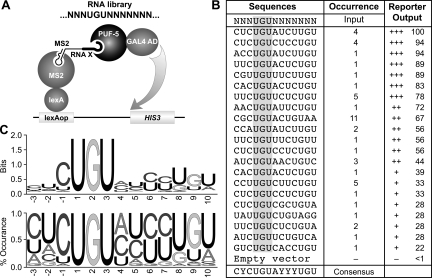
To determine the preferred RNA binding sequence for PUF-5, we performed a selection experiment using the yeast three-hybrid assay. We constructed a library in a three-hybrid RNA expression vector, p3HR2. This vector possesses a stem-loop structure that segregates the RNA of interest from the rest of the RNA molecule. Additionally, the ADE2 selectable marker has been removed to facilitate DNA manipulations. The library consisted of a fixed UGU trinucleotide, conserved in all known PUF protein binding sites, flanked by three randomized nucleotide positions upstream and seven randomized nucleotides downstream (Fig. 3A). Sequencing of independent clones from the library yielded random representation at the intended positions (not shown). The library was introduced into the yeast strain YBZ-1 along with a PUF-5/AD fusion protein (where “AD” means activation domain). Interaction between that protein and any RNA sequence in the library would activate transcription of the HIS3 reporter gene and thus could be identified by growth on selective media. Using this strategy, we obtained 21 unique RNA sequences (Fig. 3B). Each RNA sequence activated transcription only in the presence of the PUF-5/AD fusion protein (not shown). These sequences were used to derive a consensus PUF-5 binding element using WebLogo software, depicted in two formats in Figure 3C (Crooks et al. 2004).
FIGURE 3.
Derivation of the PUF-5 binding element (5BE). (A) The three-hybrid screen used to identify the 5BE. The sequence of the randomized region of the RNA is illustrated centrally. (B) Sequences isolated from the positive clones from the three-hybrid screen. The isolated sequences and consensus sequence are depicted in column one. The number of times a sequence was isolated, and the relative strength of the interaction in the three-hybrid system are also depicted. (C) The 5BE consensus sequence as derived using the WebLogo software is represented as information content and nucleotide frequency at each position.
An analogous screen was performed using a PUF-6/AD protein. This screen was designed to detect only high affinity interactions. Each of the 34 positive clones isolated from the screen with PUF-6 contained the same single sequence, which was identical to the sequence exhibiting the highest affinity for PUF-5 (Fig. 3B). The lack of other sequences, such as those recovered by PUF-5/AD, is probably due to the stringent selection, since other PUF-5 binding RNAs bind PUF-6 in directed tests.
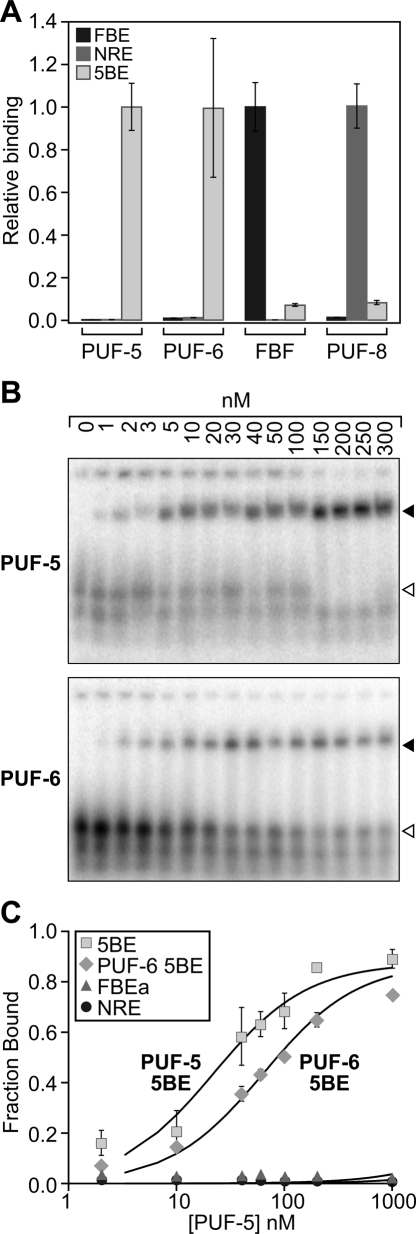
To compare the specificity of PUF-5 binding to that of other C. elegans PUF proteins, we evaluated the binding of PUF-5 to known PUF binding sites, and of the PUF-5 binding element to other C. elegans PUF proteins. For this purpose, we used the sequence obtained in the screen that gave the highest signal output from the reporter gene, CUCUGUAUCUUGU (consequently named the PUF-5 binding element, or 5BE). In the three-hybrid assay, PUF-5 bound to its cognate binding site (5BE) but not to the sites for FBF (FBE) or PUF-8 (NRE) (Fig. 4A). PUF-6 also bound only to the 5BE (Fig. 4A). Similarly FBF and PUF-8 preferred their cognate sites (Fig. 4A). These results are consistent with the hypothesis that the PUF-5/-6/-7 proteins bind similar RNA elements.
FIGURE 4.
PUF proteins bind unique sequences. (A) Three-hybrid analysis of C. elegans PUF proteins. The relative affinity of each RNA–protein interaction as measured by quantitative β-galactosidase assay has been normalized to the cognate RNA for each protein. (B) Representative EMSA gels illustrating PUF-5 and PUF-6 binding the 5BE. The filled arrowhead depicts the bound complex, while the open arrowhead identifies the free RNA species. (C) Quantitation of the fraction RNA bound vs. PUF protein concentration for four different RNA–protein complexes: PUF-5:5BE (■); PUF-5:FBE (▲); PUF-5:NRE (●); and PUF-6:5BE (♦).
To further quantify the affinity and selectivity of PUF-5 for its binding site, we conducted electrophoretic mobility shift assays (EMSAs) using purified recombinant protein. GST-PUF-5 and GST-PUF-6 bound the 5BE with similar affinity (Fig. 4B,C). PUF-5 binds to the 5BE with an apparent dissociation constant of ∼35 nM, while PUF-6 binds at ∼55 nM (Fig. 4B,C). The data are consistent with the PUF protein binding a single 5BE in a noncooperative manner. PUF-5 does not bind detectably to other known PUF binding elements, the FBE or the NRE (Fig. 4C).
5BE is the optimal PUF-5 binding element
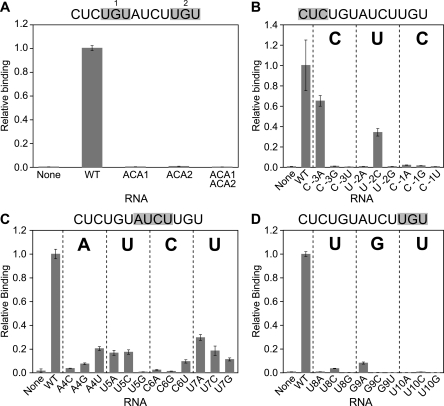
To characterize PUF-5 binding specificity in greater detail, we carried out point mutagenesis across the entire 5BE. We analyzed mutations from positions −3 to +10 (relative to the 5′-UGU element), spanning the entire sequence that had been randomized in the selection. Substitution of any nucleotide within this region was detrimental to binding (Fig. 5).
FIGURE 5.
Perturbation of the 5BE is detrimental to binding. Quantitative three-hybrid assays were performed; the β-galactosidase activity per cell has been normalized to that of the wild-type 5BE. Data are grouped by region in the binding site; mutations are in (A) the two UGU trinucleotides; (B) the region upstream of the 5′-UGU trinucleotide; (C) the central region of the binding site; (D) the 3′-UGU trinucleotide.
The 5BE contains two UGU trinucleotide elements (+1 to +3 and +8 to +10). PUF-5 binding was abolished by changing either element to ACA, demonstrating that both UGU trinucleotides are required (Fig. 5A). Upstream of the first UGU, mutations C(–3)A (changing C to A at position –3) and U(–2)C reduced binding, while all other mutations abolished it (Fig. 5B). Mutations between the two UGU trinucleotides also were deleterious; most substitutions resulted in a fivefold to 10-fold decrease in binding activity (Fig. 5C). Finally, any single mutations in the second UGU trinucleotide decreased binding greater than 10-fold (Fig. 5D). Substitution of the region downstream from the second UGU with unrelated sequence did not disrupt binding (Fig. 6; not shown). These results suggest that the sequence obtained from the three-hybrid screen is the optimal PUF-5 binding element.
FIGURE 6.
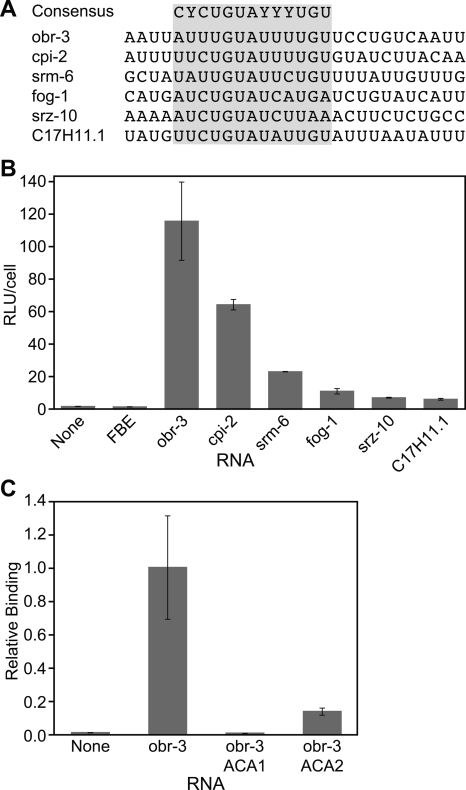
Potential candidate target sequences bind PUF-5 in the three-hybrid system. (A) Putative PUF-5 target sequences bound in the three-hybrid system. (B) Reporter gene expression for sequences that bound PUF-5. (C) The importance of both UGU trinucleotides in the PUF-5 binding element from the 3′UTR of obr-3.
Identification of candidate PUF-5 target mRNAs
To test the validity of our mutagenesis studies and to begin identifying mRNA targets of PUF-5, we searched a database of C. elegans 3′UTR sequences using the optimum PUF-5 binding sequence we had derived from our screen and mutagenesis assays (Figs. 3, 5).By use of a pattern search algorithm with a weight matrix based on the specificity data obtained in the three-hybrid experiments, we found 172 3′UTRs with a potential PUF-5 binding site (Supplemental Table 1).
We tested a selection of candidate 3′UTR sequences for binding to PUF-5 using the three-hybrid system (Fig. 6). Of 12 candidate genes analyzed, six bound to PUF-5 in the three-hybrid assay (Fig. 6A,B). These include 3′UTR sequences from obr-3, cpi-2, srm-6, fog-1, srz-10, and C17H11 (Fig. 6A). To confirm that these RNAs interacted in a manner similar to the 5BE, we mutated both UGU elements in the obr-3 3′UTR. As with the 5BE, mutation of either trinucleotide decreased binding to PUF-5 (Fig. 6C). Therefore, the 5BE accurately predicts PUF-5 binding sites in endogenous mRNAs. However, additional studies will be required to determine whether the mRNAs we identified are genuine targets in vivo.
DISCUSSION
Our work expands the repertoire of sequences bound by C. elegans PUF proteins. PUF-5 and PUF-6 recognize similar RNA sequences, and these are distinct from those seen by members of the FBF and PUF-8/-9 groups. The difference in binding specificity may underlie regulation of different target mRNAs.
Structural analysis of a human PUF protein bound to RNA suggested that amino acid side chains in one α-helix contacted one nucleotide in the RNA in direct succession (Wang et al. 2002). PUF-5 binds an extended RNA sequence: 10 nucleotides are present from the beginning of the first UGU to the end of the second. This compares to eight with PUF-8 (and human Pumilio), and nine with FBF. This clearly implies that recognition by PUF-5 differs from the one-to-one mode seen with human Pumilio. For FBF, a nucleotide in the central region of the binding element is critical for binding, but its identity is inconsequential (Opperman et al. 2005). This crucial determinant of FBF specificity is referred to as a “spacer” nucleotide, as it spaces the recognition elements seen in the human Pumilio–RNA cocrystal. Recent structural studies showed that longer binding sites can be formed by extrusion of the nucleotide away from the protein, exposed to solvent (Gupta et al. 2008; Miller et al. 2008). Our studies (Figs. 3, 5) suggest flexibility in positions +4 to +7, though pyrimidines are strongly favored at +5, +6, and +7; this region may serve as a spacer separating the two UGU elements, which make sequence specific contacts with the protein.
The precise sequence requirements in the intervening region (between the two UGUs) are context-dependent. In our randomization screen (Fig. 3), we identified several sequences with fewer than 3 nucleotides between the two UGU elements, suggesting that the intervening nucleotides may be “spacers” rather than sites of protein contacts. We propose that one or more of those nucleotides “flip out” away from the protein and are stacked on one another. This could explain differences between targeted mutagenesis studies (Fig. 5) and the sequences isolated from the randomization screen (Fig. 3). In particular, several single nucleotide substitutions in the 5BE severely decrease binding affinity, yet the same substitution in a different RNA (containing multiple substitutions) results in a more modest decrease. For example, substitution of any single nucleotide at position +5, +6, or +7 (UCU) significantly decreased binding, yet a sequence containing CUC rather than UCU exhibited only a slight effect. In one simple model, the identities of the flipped bases are important for the efficacy of their own base-stacking interactions, while allowing interactions with the protein elsewhere. Alternatively, bases could contact the protein in a context-dependent manner. Regardless, our results demonstrate new features of PUF–RNA interactions, including flexibility in spacing and recognition of UGU at the 3′ end of the site.
The presence of two UGU trinucleotides in the PUF-5 binding site is novel among known PUF protein sites, and raises several questions. First, how is the second UGU recognized? If repeats 6–8 of PUF-5 recognize the first UGU (as in human Pumilio), then how is the second UGU recognized? Repeats 1–3 of PUF-5 (which are predicted to recognize the 3′ end of the RNA sequence) are not similar in sequence to repeats 6–8 ; thus their mode of interaction probably differs. New amino acid–nucleotide contacts that recognize UGU are likely present in PUF-5. Indeed, it is striking that the amino acids involved in base recognition differ in repeats 1–4 of PUF-5/-6/-7 and Pumilio (Fig. 1C): Four of the six differences in recognition amino acids are located in repeats 1 and 2. We predict that these differences underlie PUF-5/-6 specificity for UGU rather than AUA. However, given the extended length of the 5BE, RNA contacts with UGU could be made outside the recognition helices, or even outside the PUF repeat region. Recent work has identified a structure of Pumilio bound to “noncognate” RNA sequences with sevenfold reduced affinity (Gupta et al. 2008). Our work has focused on the optimal binding site, but deviations from that sequence may nonetheless be important in vivo.
In principle, the presence of two UGU elements in the binding site could mean that two separate protein molecules recognize the site, each interacting with a UGU sequence. While this is a tenable hypothesis, mutation of the nucleotides downstream from the second UGU has little effect on PUF-5 binding (not shown); similarly, the PUF binding sites identified in Figure 6 all differ in sequence downstream from the second UGU. Thus, if a second PUF-5 molecule binds the second UGU, it must not discriminate the flanking nucleotides. Biochemical experiments do not suggest cooperativity (e.g., Fig. 4). PUF-5 could bind as an obligate dimer, though our preliminary analyses do not support this view.
PUF-5, PUF-6, and PUF-7 are required for the late stages of oogenesis in C. elegans, consistent with expression of PUF-5 in the germline (Lublin and Evans, 2007). RNAi against all three genes, but not PUF-5 or PUF-6/-7 separately, yields a defect in oogenesis. This implies redundancy, consistent with the overlapping RNA binding specificity of PUF-5 and PUF-6/-7. We propose that PUF-5/-6/-7 regulate overlapping (or identical) sets of target mRNAs.
The binding element determined by mutagenesis studies was used to predict potential mRNA targets for PUF-5 and PUF-6; indeed, PUF-5 binds to several of these RNA sequences (Fig. 6). In addition, there is a direct correlation between the similarity of the predicted binding elements to the 5BE and the affinity with which PUF-5 is able to bind the site. We identified a potential target (fog-1), which has known roles in regulating cell fate decisions in the germline (Barton and Kimble 1990; Luitjens et al. 2000; Thompson et al. 2005; Lamont and Kimble 2007), consistent with roles for PUF-5/-6/-7 in the germline. From the candidate mRNA targets we identified, other roles for PUF-5/-6/-7 can be inferred. For example, several predicted targets, obr-3 and cpi-2 among them, encode proteins involved in modulating signaling cascades. A large number of G-protein–coupled receptor mRNAs, including srm-6, srz-10, and C17H11.1, contain PUF-5/-6/-7 binding sites in their 3′UTRs, suggesting a possible role in cell signaling and chemotaxis. In addition, a group of ion channel mRNAs were predicted to be targets of PUF-5/-6/-7, similar to recent reports describing Drosophila Pumilio binding to para mRNA, which encodes a voltage-gated sodium channel (Muraro et al. 2008). Our preliminary data suggest that PUF-6 is also expressed in neurons (D. Byrd, C.R. Stumpf, J. Kimble, and M. Wickens, unpubl.).
Analysis of C. elegans PUF proteins reveals, at the RNA level, that different PUF proteins discriminate different RNA sequences. With FBF and PUF-8, the same set of atomic interactions may be used, punctuated by a single spacer nucleotide. With PUF-5, some of these contacts (PUF repeats 6–8 and the 5′-UGU) are likely to be preserved, and two, not one, spacer nucleotides relative to Pumilio may be essential for binding. Novel modes of recognition are required to accommodate the preference for the second UGU. Structural analysis is needed to reveal the underlying mechanisms of discrimination and distinguish those features that are general from those that are idiosyncratic.
MATERIALS AND METHODS
Strains, plasmids, and sequences
The yeast strain YBZ-1 has been described previously (Hook et al. 2005). Plasmid p3HR2 was created by removing the ADE2 gene from pIIIA/MS2-2 and then inserting a stem-loop structure into the multiple cloning site. To create the random library, an oligonucleotide (CCGGCTAGCNNNTGTNNNNNNNAATTTAATAAAGCATG) was inserted such that it would be expressed in the loop of the stem upon transcription of the RNA. All additional RNA expression constructs were created in the same manner. Full-length PUF-5 and PUF-6 were amplified from cDNA and cloned into pGADT7 for three-hybrid assays. For studies using recombinant protein, full-length PUF-6 and PUF-5 (amino acid 77-553) were cloned into pGEX-6P1 (GE Healthcare). Synthetic RNAs of the following sequences were ordered from Dharmacon:
5BE: 5′-UAGCCUCUGUAUCUUGUAAUUUAAUAAA-3′;
FBE: 5′-UUACCAUAGAAUCAUGUGCCAUACAUCA-3′; and
NRE: 5′-UUGUUGUCGAAAAUUGUACAUAAGCCAA-3′.
Three-hybrid assays
Three-hybrid assays were performed as previously described (Bernstein et al. 2002; Hook et al. 2005). The initial selection was carried out in the presence of 2 mM 3-AT. PUF-5 is expressed to a higher level than PUF-6. β-Galactosidase assays were performed in quadruplicate and normalized to cell number.
Electrophoretic mobility shift assay
PUF proteins were purified as described (Bernstein et al. 2005). EMSAs were performed as described (Cheong and Hall 2006). Briefly, reactions were incubated at room temperature in EMSA binding buffer (10 mM HEPES, 1 mM EDTA, 50 mM KCl, 1 mM DTT, 0.1 mg/mL BSA, 0.01% Tween 20) for 2 h. Samples were mixed with loading buffer (2% Ficoll, 1%DMSO, 0.02% bromophenol blue), and half the reaction loaded onto a 5% native gel (Bio-Rad Inc.). Gels were run at 100 V for 45 min at 4°C, exposed to a storage phosphor screen for 30 min, and scanned on a Typhoon 9410 scanner (GE Healthcare Biosciences). Data were analyzed using Prism 4 (GraphPad Software).
Target identification
Potential targets for PUF-5 were selected by searching the C. elegans 3′UTR database using the ScanForMatches algorithm and a reference sequence derived from the mutational analysis of the 5BE (Dsouza et al. 1997; Hajarnavis et al. 2004).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dana Byrd for comments on the manuscript and members of the Wickens and Kimble laboratories for advice and discussion. We thank Laura Vanderploeg and the Biochemistry Media Laboratory for help with figure preparation. M.W. is supported by grants from the NIH (GM31892, GM50942), and J.K. is a member of the Howard Hughes Medical Institute. C.R.S. was supported by the NIH Molecular Biosciences Training Grant and GM31892 during this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1095908.
REFERENCES
- Ahringer J., Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- Bachorik J.L., Kimble J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. 2005;102:10893–10897. doi: 10.1073/pnas.0504593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.K., Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans . Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D.S., Buter N., Stumpf C., Wickens M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. doi: 10.1016/S1046-2023(02)00015-4. [DOI] [PubMed] [Google Scholar]
- Bernstein D., Hook B., Hajarnavis A., Opperman L., Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnovich D., Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc. Natl. Acad. Sci. 2001;98:11359–11364. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C.G., Hall T.M. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Rogers K.W., Fay D.S. The C. elegans glycopeptide hormone receptor ortholog, FSHR-1, regulates germline differentiation and survival. Curr. Biol. 2007;17:203–212. doi: 10.1016/j.cub.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Colegrove-Otero L.J., Minshall N., Standart N. RNA-binding proteins in early development. Crit. Rev. Biochem. Mol. Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Crittenden S.L., Bernstein D.S., Bachorik J.L., Thompson B.E., Gallegos M., Petcherski A.G., Moulder G., Barstead R., Wickens M., Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans . Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor C.H., Meijer H., Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Dsouza M., Larsen N., Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Dubnau J., Chiang A.S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U., et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fan J., Heller N.M., Gorospe M., Atasoy U., Stellato C. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur. Respir. J. 2005;26:933–947. doi: 10.1183/09031936.05.00120204. [DOI] [PubMed] [Google Scholar]
- Forbes A., Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Seay D.J., Hook B.A., Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Gupta Y.K., Nair D.T., Wharton R.P., Aggarwal A.K. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008;16:549–557. doi: 10.1016/j.str.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Hajarnavis A., Korf I., Durbin R. A probabilistic model of 3′ end formation in Caenorhabditis elegans . Nucleic Acids Res. 2004;32:3392–3399. doi: 10.1093/nar/gkh656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton T.A., Novotny M., Datta S., Mandal P., Hartupee J., Tebo J., Li X. Chemokine and chemoattractant receptor expression: Post-transcriptional regulation. J. Leukoc. Biol. 2007;82:213–219. doi: 10.1189/jlb.1206754. [DOI] [PubMed] [Google Scholar]
- Hook B., Bernstein D., Zhang B., Wickens M. RNA-protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook B.A., Goldstrohm A.C., Seay D.J., Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- Kadyrova L.Y., Habara Y., Lee T.H., Wharton R.P. Translational control of maternal cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Goodwin E.B. The power of the 3′ UTR: Translational control and development. Nat. Rev. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Lamont L.B., Kimble J. Developmental expression of FOG-1/CPEB protein and its control in the Caenorhabditis elegans hermaphrodite germ line. Dev. Dyn. 2007;236:871–879. doi: 10.1002/dvdy.21081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont L.B., Crittenden S.L., Bernstein D., Wickens M., Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lehmann R., Nusslein-Volhard C. Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature. 1987;329:167–170. [Google Scholar]
- Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lublin A.L., Evans T.C. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev. Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Luitjens C., Gallegos M., Kraemer B., Kimble J., Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans . Genes & Dev. 2000;14:2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B., Seshadri V., Fox P.L. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Mee C.J., Pym E.C., Moffat K.G., Baines R.A. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J. Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K.P., Sanyal S., Habara Y., Sanchez R., Wharton R.P., Ramaswami M., Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Miller M.T., Higgin J.J., Tanaka Hall T.M. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore F.L., Jaruzelska J., Fox M.S., Urano J., Firpo M.T., Turek P.J., Dorfman D.M., Pera R.A. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro N.I., Weston A.J., Gerber A.P., Luschnig S., Moffat K.G., Baines R.A. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolde M.J., Saka N., Reinert K.L., Slack F.J. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev. Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman L., Hook B., DeFino M., Bernstein D.S., Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Osborne R.J., Thornton C.A. RNA-dominant diseases. Hum. Mol. Genet. 2006;15(Spec No 2):R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Lena A., Batistoni R., Deri P., Rainaldi G., Locci M.T., Evangelista M., Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Schweers B.A., Walters K.J., Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E., et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans . Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 2003;13:134–139. doi: 10.1016/s0960-9822(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Thompson B.E., Bernstein D.S., Bachorik J.L., Petcherski A.G., Wickens M., Kimble J. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development. 2005;132:3471–3481. doi: 10.1242/dev.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L., Orr-Weaver T.L. Regulating translation of maternal messages: Multiple repression mechanisms. Trends Cell Biol. 2007;17:547–554. doi: 10.1016/j.tcb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Wang X., Zamore P.D., Hall T.M. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wang X., McLachlan J., Zamore P.D., Hall T.M. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wreden C., Verrotti A.C., Schisa J.A., Lieberfarb M.E., Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Ye B., Petritsch C., Clark I.E., Gavis E.R., Jan L.Y., Jan Y.N. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M.P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]