Abstract
Replication of picornaviruses is dependent on VPg uridylylation, which is linked to the presence of the internal cis-acting replication element (cre). Cre are located within the sequence encoding polyprotein, yet at distinct positions as demonstrated for poliovirus and coxsackievirus-B3, cardiovirus, and human rhinovirus (HRV-A and HRV-B), overlapping proteins 2C, VP2, 2A, and VP1, respectively. Here we report a novel distinct cre element located in the VP2 region of the recently reported HRV-A2 species and provide evolutionary evidence of its functionality. We also experimentally interrogated functionality of recently identified HRV-B cre in the 2C region that is orthologous to the human enterovirus (HEV) cre and show that it is dispensable for replication and appears to be a nonfunctional evolutionary relic. In addition, our mutational analysis highlights two amino acids in the 2C protein that are crucial for replication. Remarkably, we conclude that each genetic clade of HRV and HEV is characterized by a unique functional cre element, where evolutionary success of a new genetic lineage seems to be associated with an invention of a novel cre motif and decay of the ancestral one. Therefore, we propose that cre element could be considered as an additional criterion for human rhinovirus and enterovirus classification.
Keywords: picornavirus, rhinovirus, cis-acting replication element, replication, classification
INTRODUCTION
Human rhinoviruses (HRV), the most frequent cause of respiratory infection (Denny 1995), represent the largest genus of non-enveloped, single positive stranded RNA viruses in the Picornaviridae family. Using seroneutralization assays, HRV have been classified into 99 serotypes (Kapikian 1967) and further divided in two different species, HRV-A (74 serotypes) and HRV-B (25 serotypes), based on capsid protein sequences (Ledford et al. 2004; Savolainen et al. 2002). In addition, recent studies have reported new HRV strains clustering into a new HRV species designated as HRV-A2, HRV-C, or HRV-X (Arden et al. 2006; Lamson et al. 2006; Kistler et al. 2007; Lau et al. 2007; Lee et al. 2007; McErlean et al. 2007; Renwick et al. 2007). Similar to all Picornaviridae members, HRV genomes of ∼7200 base pairs are organized in four different regions: a long 5′-untranslated region (UTR), a single open reading frame (ORF), a short 3′-UTR, and a poly(A) tail (Kitamura et al. 1981).
The 5′- and 3′-UTRs are known to contain important structural motifs. The 5′-UTR contains two highly conserved elements, the 5′-terminal cloverleaf and the internal ribosomal entry site (IRES). The 5′-terminal cloverleaf interacts with the viral protease 3CDpro (Andino et al. 1990, 1993; Gamarnik and Andino 1997; Parsley et al. 1997; Rieder et al. 2003) and cellular proteins such as PCBP2 (Andino et al. 1990, 1993; Gamarnik and Andino 1997; Parsley et al. 1997; Perera et al. 2007) to form a ribonucleoprotein complex that is implicated in the switch from viral translation to replication (Huang et al. 2001; Sharma et al. 2005; Perera et al. 2007). This domain is also required both in cis and in trans for negative (Gamarnik and Andino 1998; Barton et al. 2001) and positive strand initiation (Andino et al. 1990), respectively. In contrast, the IRES motif promotes initiation of translation from the uncapped viral genome. The 3′-UTR of piconaviruses is required for efficient viral RNA replication (Jacobson et al. 1993; Pilipenko et al. 1996; Melchers et al. 1997; Todd et al. 1997; Duque and Palmenberg 2001; Brown et al. 2004, 2005), although the exact mechanism is not yet understood.
In addition to the structural elements described above, cis-acting replication elements (cre) have been identified within the coding region of several picornavirus genomes (Fig. 1). The first cre motif was originally identified in the VP1 encoding region of HRV14 (a HRV-B member) (McKnight and Lemon 1996, 1998). Similar cre elements have been described for different picornaviruses in diverse genome regions such as in 2C for poliovirus (Goodfellow et al. 2000) and coxsackievirus-B3 (van Ooij et al. 2006), 2A for HRV2 (Gerber et al. 2001) and VP2 for Theiler's and Mengo viruses (two cardioviruses) (Lobert et al. 1999). Moreover, the presence of cre motif sequences for HRV3 and HRV72 has been identified in the VP1 coding sequence, similar to HRV14 VP1 cre, and in 2A for HRV1a and HRV16 serotypes, similar to HRV2 cre (McKnight 2003). In contrast to all these previous cre elements identified in the polyprotein-encoding region, the foot-and-mouth disease virus (FMDV) presents a cre element in the 5′-UTR adjacent to the IRES (Mason et al. 2002). These cre exhibit different nucleotide sequences, but are all comparable in size and share a similar stem–loop structure (Yang et al. 2002; Thiviyanathan et al. 2004). Based on sequence comparisons and mutational analysis of entero- and rhinovirus cre elements, a common motif, R1NNNA1A2R2NNNNNNR3, has been proposed for the loop segment of these two genus (Yang et al. 2002; Yin et al. 2003). Extensive studies have been performed to understand the function of these cre motifs in virus replication. Both in vivo and in vitro experiments clearly demonstrated the involvement of these cre in VPg uridylylation (Paul et al. 1998; Rieder et al. 2000; Gerber et al. 2001; Yang et al. 2002; Yin et al. 2003; Richards et al. 2006). VPgpUpU is assumed to serve as primer for the viral polymerase 3Dpol in RNA synthesis. Not only does the loop appear to be important for the function of cre motif, the stem also seems to play a crucial role (Yin et al. 2003; Pathak et al. 2007). Since VPg is linked to the 5′-end of the positive and negative strand genomes, it is tempting to postulate that cre motifs are involved in the initiation of synthesis of both strands. However, this remains a subject of controversy, since some studies have argued in favor of a role of cre motif only in positive strand RNA synthesis (Goodfellow et al. 2003; Morasco et al. 2003), while others established an involvement of cre in negative sense synthesis (McKnight and Lemon 1998; Yang et al. 2002). More surprising were the observations that heterologous exchanges of cre elements were permissive in some cases (Gerber et al. 2001; Yin et al. 2003; Yang et al. 2004; Shen et al. 2007), thus suggesting that their roles in RNA replication are therefore independent of their location along the genome.
FIGURE 1.
Schematic diagrams of human rhinovirus (HRV) and enterovirus (HEV) genomic organization showing the location of known cre elements in HEV, HRV-A, and HRV-B as well as newly predicted HRV-A2 cre and putative HRV-B cre (underlined) that are characterized in the text.
In a previous study, we observed that HRV-B and human enterovirus (HEV) were more closely related to each other than either is to HRV-A for 2A, 2B, 2C, and 3D proteins (Tapparel et al. 2007). This was explained as the result of hypothetical recombination events between HRV-A and the HEV ancestor of HRV-B soon after the divergence of HEV and HRV-A (Supplemental Fig. 1). Moreover, a putative 2C cre motif absent in all HRV-A genomes was identified in the 25 HRV-B serotypes that conserved significantly essential nucleotides within the consensus loop (R1NNNA1A2R2NNNNNNR3). As a functional cre motif was already known in HRV14 VP1 protein (HRV-B), the first goal of this study was to determine whether this putative cre located within the HRV14 2C coding region was required for virus replication. By mutational analysis and creation of 2C cre disrupted-mutant (DM), we provide strong evidence that this putative 2C cre motif is nonfunctional for virus replication and appears to be an evolutionary relic. Interestingly, we pointed out two amino acids within the cre region of the 2C protein that are absolutely necessary for HRV14 replication. Moreover, by analyzing sequences of newly reported HRV strains, we were able to identify a unique and conserved VP2 cre motif present in all new HRV-A2 members, but absent in both HRV-A and HRV-B species.
RESULTS
Here we discuss internal cre of human enterovirus and rhinovirus that are essential for virus replication (Fig. 1) and provide experimental evidence of nonfunctionality of an additional putative HRV14 cre in the 2C region and evolutionary evidence of a novel predicted cre element located in the VP2 region of the recently reported HRV-A2 strains.
Mutations within the HRV14 2C cre loop affectvirus growth
We previously identified a putative cre element in the HRV-B 2C coding sequence in addition to the already characterized HRV14 VP1 cre motif by full-length genome comparison of HRV-A, HRV-B, and HEV (Tapparel et al. 2007). In an attempt to determine the functionality of this second cre motif, we performed a mutational analysis in the corresponding region of the HRV14 infectious clone. Since nucleotides A1, A2, R2, and R3, known as essential for cre motif function, were conserved in HRV14 2C cre, as in all HRV-B (unlike R1) (Tapparel et al. 2007), these positions were mutated as described (Fig. 2A,B). The effect of these mutations on HRV14 replication was then measured by immunofluorescence 12 h post-infection and confirmed by in situ hybridization with antisense RNA probe (data not shown).
FIGURE 2.
Effects of nucleotide changes within the putative HRV14 2C cre motif on viral growth. (A) Quantification of mutant virus growth in HeLa cells. Each HRV14 2C cre mutant is named according to the substituted nucleotide. The amino acid changes that follow the nucleotide mutations are indicated in parentheses after the mutation and those conserving the amino acid properties are marked by an asterisk. The frequency plot of amino acid residue conservation in 25 HRV-B serotypes is shown at the foot of Panel A (http://weblogo.berkeley.edu/). Quantification of virus growth was measured by immunofluorescence 12 h post-infection and expressed as the mean of positive cells per total cells and expressed relative to that of HRV14-WT. (B) Schematic representation of the putative HRV14 2C cre structure as predicted by MFOLD from nucleotides 4256 to 4303. Nucleotide substitutions at position 4277, 4278, 4279, and 4286 (corresponding, respectively, to A1, A2, R2, and R3 positions within the consensus R1NNNA1A2R2NNNNNNR3 sequence) are represented in bold. (C) Quantification of virus growth for HRV14-DM versus HRV14-WT. Measurements were conducted by immunofluoresence as described in A. (D) Schematic representation of 2C cre motif for HRV14-DM (nucleotides 4256–4303) following the introduction of 12 silent mutations (surrounded nucleotides) as predicted by MFOLD. None of the crucial positions within the consensus cre loop sequence were mutated, except one at position 4279 (A → G) known to be permissive for efficient HRV14 replication (A). (E) HeLa cells infected by HRV14-WT, HRV14-DM, or Mock. Infected positive cells were detected by immunofluorescence (IF).
As shown in Figure 2, A4277C (Lys → Gln), A4278C (Lys → Thr), A4279C (Lys → Asn), and A4286T (Thr → Ser) caused a decrease of six- to sevenfold in viral replication. The replication is further reduced (30- to 50-fold) with the following mutations: A4277G (Lys → Glu), A4278T (Lys → Ile), AA4277/8GG (Lys → Gly), A4286C (Thr → Pro), and A4286G (Thr → Ala). In contrast, A4279G does not change viral replication efficiency (Fig. 2A). Of note, none of these mutations disrupts the classical hairpin structure (data not shown). Taking into consideration the critical positions within the loop, the results were as expected apart from A4278G (Lys → Arg) and A4286G (Thr → Ala). Indeed, A4278G (Lys → Arg) changes the critical A2 residue but replication is as efficient as wild type, whereas A4286G (Thr → Ala) respects the R3 position but abrogates viral replication. The explanation for this can be found when observing the 2C amino acid sequence. All mutations changing the amino acid encoded by nucleotides A1A2R2 (originally a Lys) in a nonconservative way decrease replication, whereas the only conservative change (A4278G [Lys → Arg]) has no effect. Similarly, R3 is the first nucleotide of a codon encoding for a threonine in HRV14, and replacement of this threonine with any other amino acid disrupts replication such as mutations A4286G (Thr → Ala), A4286C (Thr → Pro), and A4286T (Thr → Ser). As all except one mutation (A4279G) changed the amino acid sequence within the 2C protein, the interpretation of results was complicated since any variations in virus growth could be caused by changes either in the 2C protein or in the putative 2C cre motif. Of note, sequencing of mutants after growth allowed us to exclude any reversion.
HRV14 replication is not affected by disrupting the putative 2C cre stem–loop
To firmly conclude that the putative HRV14 2C cre motif is not necessary for viral replication, we introduced 12 silent mutations disrupting HRV14 2C cre classical stem–loop (Fig. 2D, HRV14-DM). This disruption renders unlikely any use of this motif for VPg uridylylation by 3Dpol without altering the resulting coded viral protein product. Creation of disrupted cre mutants was successfully applied previously, notably in HRV14 VP1 cre (Yang et al. 2004) and coxsackievirus-B3 2C cre functional studies (van Ooij et al. 2006). After quantification of virus growth by immunofluorescence, we observed similar replication levels for HRV14-DM and HRV14-WT (Fig. 2C-E). Again, the absence of reversion was confirmed by sequencing.
In conclusion, the mutageneses approach together with the disruption mutant show that the putative HRV14 2C cre element is dispensable for viral replication, but the amino acid sequence is critical. In particular, we identified two amino acids (corresponding to 118th and 123rd amino acids in 2C coding sequence) essential for replication, since their individual changes resulted in a strong decrease in HRV14 replication efficiency.
Comparison of 2C cre structure between HEVand HRV-B
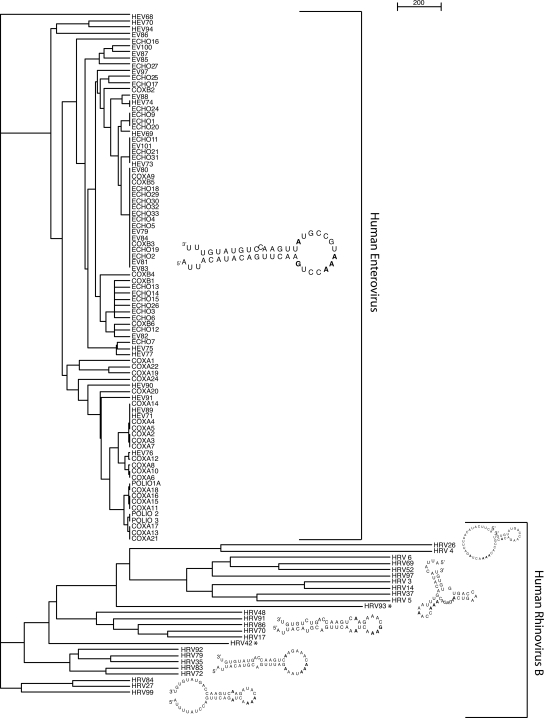
The absence of the putative HRV14 2C cre function suggests that the ancestral HRV-B 2C cre motif, probably originating from HEV 2C cre (Tapparel et al. 2007), has evolved into a nonfunctional motif and was replaced by a functional VP1 cre motif. To confirm this hypothesis and to determine if “intermediate” less conserved cre motifs could also exist within the enterovirus genus, we compared the evolution rate and conservation of the 2C cre motif among HEV and HRV-B. For this purpose, we calculated a structure-based phylogenetic tree of all HEV and HRV-B 2C cre elements. Our phylogenic analysis includes 86 publicly available full-length HEV (http://www.picornaviridae.com/sequences/sequences.htm) and the 25 HRV-B serotype sequences and could elucidate the evolutionary pathways differentiating these two groups. The tree depicted in Figure 3 represents a structural clustering tree based on the conserved secondary structures of 2C cre for each group and the measurement of structural distance between these groups. The tree topology clearly shows a highly conserved cluster for HEV 2C cre structures and five loosely conserved smaller clusters for the putative HRV-B 2C cre structures without any overlap between HEV and HRV-B members. Apparently, evolutionary pressure on the conservation of the HEV 2C cre stem–loop is much higher than for the HRV-B 2C cre. The length of terminal branches also presents important differences between the HEV and the HRV-B clusters. The mean structural distance between any of the HEV structures is smaller than for the HRV-B sequences. In addition, no intermediate strain exists that would result in a clustering of a HRV-B strain within the HEV cluster. From these data, we conclude that there is a close link between a functional cre element and the short mean structural distance of that element within a group. This is further confirmed when looking at the structure-based phylogenetic trees of the two functional cre of HRV-A 2A and HRV-B VP1 (Supplemental Figs. 2, 4).
FIGURE 3.
WPGMA structure-based cluster tree for 86 HEV and 25 HRV-B 2C cre elements. A consensus 2C cre structure is shown for each cluster except for HRV42 and HRV93, which do not belong to any specific cluster (marked *). The crucial positions within the consensus cre structures are marked by bold letters for each cluster.
Taken together, these results support those obtained with HRV14-DM and the conclusion that the putative HRV14 2C cre is nonfunctional. In the absence of functional pressure, this motif structure evolves more rapidly within HRV-B. In addition, the tight structural cluster for 2C cre among HEV members, VP1 and 2A cre among HRV-B and HRV-A, respectively, suggests that a selective pressure, most likely related to the function of this motif, is present across all HEV, HRV-A, and HRV-B members.
Identification of a conserved predicted cre motifin the HRV-A2 VP2 coding region
New HRV strains diverging significantly from HRV-A and HRV-B species were identified recently. Different nomenclature names (HRV-A2, HRV-X, HRV-QPM, or HRV-C) have been assigned to these strains and we will use the HRV-A2 denomination for consistency. Indeed, whole genome comparison shows that HRV-A2 species is distinct but close to HRV-A (McErlean et al. 2008). Using the RNAz program, we scanned the six full-length genomes available for these new strains in order to localize their cre elements. In addition to the 5′-terminal cloverleaf, the IRES, and the 3′-UTR hairpin elements constantly found in picornaviruses (data not shown), we identified the presence of a unique and conserved predicted cre structure within the VP2 coding region for the six new HRV genomes analyzed (Fig. 4), and not within 2A or VP1 as is the case for HRV-A and HRV-B, respectively. The presence of this new predicted cre structure was then confirmed in the VP2 coding region for additional HRV-A2 partial sequences recently published (Kistler et al. 2007; McErlean et al. 2007). Although not yet investigated due to the lack of infectious clones and the inability of these new viruses to grow in standard cell culture, the probability that this predicted VP2 cre motif acts as a nondispensable functional element is extremely high. This assumption is based on four main observations: first, this element is only found in VP2 and matches perfectly the classical cre hairpin structure with a loop region of exactly 14 nucleotides (nt); second, the critical R1, A1, A2, R2, and R3 nucleotides within the consensus cre sequence are conserved for all HRV-A2 analyzed; third, similar to the observations made for the HEV 2C cre (Fig. 3), HRV-A 2A cre, and HRV-B VP1 cre (Supplemental Figs. 2, 4), the structural tree of HRV-A2 VP2 cre presents very short structural distances between the individual strains elements (Supplemental Fig. 3). Fourth, the mean base pair distance, defined as the number of base pairs to change to transform one structure into another, for HRV-A2 VP2 cre, is as low as for the other functional cre elements (HEV 2C cre: 2.97, HRV-A 2A cre: 2.39, putative HRV-B 2C cre: 16.27, HRV-B VP1 cre: 5.15, predicted HRV-A2 VP2 cre: 1.81). According to the structure trees, this also shows that the cre structures of all HRV-A2 strains are very similar.
FIGURE 4.
Conservation of a predicted VP2 cre secondary structure for HRV-A2. Multiple sequence alignment across all available full-length HRV-A2 (NAT001, NAT045, EF186077, and EF582385-7) and VP2 partial sequences (Nat059, NAT069, Nat083, EU131891-2, and EF512649-65) showing a consensus secondary cre structure in VP2. The structure is shown in the dot-bracket format above alignment. Each corresponding bracket represents a consensus base pair of the alignment columns beneath. Sequences are color-coded according to consistent and compensatory mutations in the aligned sequences regarding the conserved structure (see figure text box). The sequence conservation profile is shown in gray bars below the alignment. The secondary structure of the conserved predicted cre VP2, color-coded according to the different types of base pairs in the corresponding alignment columns, is shown on the left side. The conserved cre motif nucleotides are marked in bold. Note that the amino acid sequence corresponding to the loop region is almost 100% conserved in all species (C-G-F-S-D-R-L-K-Q-I-T-I-G/N-S-T). Mutations supporting the structure (consistent mutations) occur almost exclusively at the third codon position, which leads to synonymous codons and the amino acid conservation.
In summary, each HRV species identified to date, and from whom a full-length genome is available, possesses a functional cre element located at different, but specific, positions (Fig. 1).
Boxplot analysis
The analysis of the distances between the terminal nodes in the WPGMA structural cluster trees of HRV-A 2A cre, putative HRV-B 2C cre, HRV-B VP1 cre, predicted HRV-A2 VP2 cre, and HEV 2C cre shows striking differences between these elements (Fig. 5). The size of the distance distribution as well as its mean describe how “tight” the individual cre are clustered. In other words the more similar the elements are to each other within one species the more conserved are the secondary structures between the strains. The experimental evidence for the nonfunctionality of the putative HRV-B 2C cre element is strengthened by the data of the distance distributions in the boxplot for this element. There is a large variety in distances as well as a high mean distance between single strain elements, which means that these structures do not form a species-specific evolutionary conserved secondary structure. On the other hand, the newly predicted cre element for HRV-A2 VP2 shows a very narrow distance distribution for its members as well as the lowest mean structural distance compared to all the other functional cre elements. It is therefore very likely that this predicted HRV-A2 VP2 cre is functional, although further genetic and biochemical studies will be necessary to finally confirm this assumption.
FIGURE 5.
Boxplots of structural distances within the respective WPGMA trees of HRV-A 2A cre, putative HRV-B 2C cre, HRV-B VP1 cre, predicted HRV-A2 VP2 cre, and HEV 2C cre. The mean distances are 400, 1229, 654, 282, and 475, respectively. Note that the nonfunctional putative HRV-B 2C cre shows the largest distances, thus showing that the structures of the individual cre sequences of these species do not form a tight cluster of mostly similar structures as is the case for the other cre motifs. The distance distributions of all the other elements are below that of the nonfunctional cre element. The distances are directly related to the scores for the clusters from the LocARNA package.
DISCUSSION
The presence of internal cre motif was first described for HRV14 (McKnight and Lemon 1996), and the requirement of this motif for viral RNA replication came as a surprising observation. Indeed, for many years, it was considered that the different structures previously identified within the 5′- and 3′-UTR of rhino- and enteroviruses were sufficient for this function. Since then, cre motifs have been identified within the protein-coding sequence of poliovirus, coxsackievirus-B3, HRV2, Theiler's, and Mengo viruses (Lobert et al. 1999; Goodfellow et al. 2000; Gerber et al. 2001; van Ooij et al. 2006). Although these cre elements were positioned in different regions according to the different viruses (Fig. 1), their classical hairpin structure of ∼60 nt was demonstrated as essential for the VPg uridylylation process. Uridylylated VPg then serves as primer at the genome 3′ extremities for RNA replication. Whether it is necessary for replication of both strands or for only one strand remains an open question.
In a recent study, we identified the presence of a new putative cre motif within the HRV14 2C coding region in addition to the one already described in VP1 (Tapparel et al. 2007). Although it would be surprising that HRV14 required the presence of two functional cre motifs for its genome replication, we cannot rule out that these two elements may have complementary roles in RNA replication, one being committed to the synthesis of the positive strain and the other to the negative synthesis. Both the approaches of point mutation of the critical putative 2C cre motif residues and the total disruption of the cre structure give a clear indication that the putative HRV14 2C cre motif is nonfunctional. Indeed, derivatives with disrupted motif but conserved amino acid sequences replicate at levels equivalent to wild-type virus. We recently showed that HEV and HRV-B were more closely related to each other than either is to HRV-A for 2A, 2B, 2C, and 3Dpol coding regions (Tapparel et al. 2007). Thus, it can be postulated that the presence of the putative 2C cre motif in all HRV-B strains may only result from a leftover from HEV and HRV-A early recombination events. The topology of the HRV-B and HEV 2C cre structural trees further corroborates this hypothesis. Whereas the evolution of the HEV 2C cre is very slow, the putative HRV-B 2C cre tree shows rapid evolution. Of note, some HRV serotypes (HRV92, HRV79, HRV35, HRV83, and HRV72) exhibit a better classical 2C cre stem–loop structure than the one displayed by HRV14, and we cannot totally exclude that the putative HRV-B 2C cre is absolutely nonfunctional for these specific serotypes. Nevertheless, this hypothesis seems highly unlikely based on the high conservation of the HRV-B VP1 structural tree throughout all HRV-B members. Finally, our mutational analysis revealed the presence of two essential amino acids located between positions 4277–9 and 4286–8, corresponding to the 118th and 123rd amino acids within the HRV14 2C coding sequence, respectively. The presence of these two amino acids lying exactly within the putative HRV14 2C cre loop sequence may explain in part the conservation of such motifs, as their mutation may strongly affect one of the different 2C protein functions and, consequently, HRV14 replication.
Protein 2C (also known as 2CATPase) is highly conserved among all picornaviruses and has been shown to be required for several steps during virus infection. Indeed, 2C is known to be involved in viral RNA replication, encapsidation, and intracellular membrane remodeling (Cho et al. 1994; Pfister et al. 2000; Suhy et al. 2000; Teterina et al. 2006). This protein consists of three different parts. The central domain, where the putative HRV14 2C cre is located, contains the nucleoside triphosphate (NTP) binding motif, including motifs A and B known to contribute to binding the phosphate moiety of NTP (Gorbalenya et al. 1990; Mirzayan and Wimmer 1992) and to NTP hydrolysis (Gorbalenya et al. 1990; Rodriguez and Carrasco 1993; Mirzayan and Wimmer 1994; Samuilova et al. 2006), respectively. HRV14 2C cre is not situated within a functionally known motif, but only 9 nt upstream of the A motif. Our results suggest that amino acids 118 and 123 of the cre motif are critical for one of the 2C protein functions, but further experiments are required to define their precise roles.
The whole-genome scan of new HRV strains resulted in the discovery of a new predicted cre motif located within the VP2 protein-coding region. Based on the fact that (1) this cre motif is not found elsewhere along the genome; (2) it perfectly respects the stem–loop structure including the R1, A1, A2, R2, and R3 positions; and (3) the structural tree of the predicted HRV-A2 VP2 cre shows no important discrepancies in branch length, similarly to those of HRV-B VP1 cre, HRV-A 2A cre, and HEV 2C cre structures, we assume that this classical hairpin structure is essential for HRV-A2 replication. The validity of our newly predicted cre in HRV-A2 is strengthened by the following points: This study used more sequences than the previous one (Tapparel et al. 2007) to predict the putative new HRV-A2 VP2 cre. Furthermore, the mean base pair distance between all the individual strain structures is as low as for the other cre motifs known to be functional. This reflects the high evolutionary constraints on this locus in keeping with its stable stem–loop structure and is additionally supported by consistent nucleotide mutations in the stem region. This point is also strengthened by the fact that all HRV-A2 strains form one common structure cluster in the tree (Supplemental Fig. 3), which was not the case for the putative HRV-B 2C cre (Fig. 3). This type of analysis was not used in our previous paper, but points out that a limited number of species and an exclusive view on the consensus structure can lead to a prediction of a cre that is shown here to be nonfunctional in HRV14. However, the most important difference in the putative HRV-B 2C cre and the predicted HRV-A2 VP2 cre stems from the fact that in HRV-B an experimentally verified cre was already known, whereas no cre element has been described in HRV-A2 until now.
This new rhinovirus species was previously identified based on VP4-VP2 and VP1 phylogenetic analysis. In addition to their differences in capsid sequences, HRV-A, HRV-B, and HRV-A2 are thus distinguishable from each other by the localization of their respective cre motifs. This signifies that the classification of HRV genomes based on their capsid sequence matches the classification based on cre location. Following this observation, it will be interesting to scan the genome of the other newly identified rhinoviruses whose full-length genome sequences are not yet available (Lee et al. 2007) to find the location of their cre elements. Using cre as an additional classification criterion, rhinovirus A, A2, and B could be considered as three independent species, whereas the four enterovirus species could be reclassified into one unique species. Interestingly, this observation correlates with the homology comparison made between each HEV and HRV species. Indeed, the percentages of homologies between each HEV species are significantly higher than those between each HRV species at both nucleotide (full-length genome) and amino acid levels (Supplemental Fig. 5). Beyond the classification per se, our findings suggest that secondary functional structures are key elements in shaping the evolutionary pathways of human picornaviruses and that each human genogroup evolves independently within the borders determined by these nondispensable constraints. When we extended our computational analysis with structural constraints based on HEV and HRV cre elements to the whole full-length picornavirus sequences available, no classical cre motif could be observed for the other Picornaviridae (data not shown). This suggests that either the structure of this element strongly diverges among other picornavirus genera, as already observed for cardiovirus (Lobert et al. 1999) and FMDV cre (Mason et al. 2002) or that cre is only present in some Picornaviridae. However, the latter argument is unlikely due to the presence of the VPg protein in all Picornaviridae members. Finally, our data confirm that cre are functionally independent of their position along the genome. However, the driving force that directs cre location for each species remains an open question, but, clearly, the localization of this structure on the genome results from specific constraints.
In conclusion, our analysis demonstrated that the conserved putative HRV14 2C cre found also in all HRV-B serotypes is nonfunctional and likely represents an evolutionary residue from the HEV and HRV-B common ancestor. This study clearly shows the importance of the structural constraints for entero- and rhinovirus cre functionality. Moreover, we were able to identify a highly conserved cre in the VP2 protein-coding region of newly identified HRV-A2 strains. Beyond the potential usefulness for picornavirus classification, our observation highlights cre motifs as key determinants in shaping human picornavirus evolutionary ability. Finally, we propose to use variations in cre locations as an additional criterion facilitating human rhinovirus and enterovirus classification.
MATERIALS AND METHODS
Cell and media
HeLa-OH cells were grown in Eagle's Minimum Essential Medium (EMEM; Lonza) supplemented with 2 mM L-glutamine, 1 μg/mL amphotericin, 100 μg/mL gentamicin, 20 μg/mL vancomycin, 10% fetal calf serum (FCS) at 37°C in a 5% CO2-containing atmosphere.
Construction of 2C cre mutants
A PCR fragment of 1602 nt containing HRV14 2C coding sequence amplified with the forward primer 5′-GGCATTCAGAATAGTAAATGAACATG-3′ and the reverse primer 5′-GTTGGGGGGCTTAGTGTGTT-3′ from the plasmid pWR3.26-HRV14 (HRV14 full-length sequence under the T7 promoter control, kindly provided by Wai-Ming Lee [University of Wisconsin]) was subcloned into the plasmid pCR2.1-TOPO (Invitrogen). The resulting plasmid was named pCR2.1-TOPO-HRV14 and used to perform the mutagenesis with the Quickchange site-directed mutagenesis kit (Stratagene). The primers used for mutagenesis are listed in Supplemental Fig. 6. The mutated fragments were then excised at AvrII and XcmI unique restriction sites and ligated back into pWR3.26-HRV14. The mutation was confirmed by sequencing.
In vitro transcription and transfection
Twenty micrograms of plasmid pWR3.26-HRV14 harboring wt or putative HRV14 2C cre mutants were linearized at a unique MluI restriction site downstream of the 3′-viral poly(A) genome. Using T7 RNA polymerase, RNA transcripts were synthesized from the linear templates with the MEGAscript T7 kit (Ambion) 3 h at 37°C and purified with the RNeasy Mini Kit (Qiagen). Transcript RNA was quantified and checked by 0.1% sodium dodecyl sulfate–1% agarose gel analysis. HeLa-OH cells were seeded at 6 × 105 cells in 35-mm wells of a six-well plate. The following day, cells were transfected with 2 μg of RNA transcripts containing wt or mutant HRV14 2C cre sequence using the TransMessenger Transfection Reagent kit (Qiagen). After 3 h at 37°C, 2 mL of McCoy's 5A Medium (Invitrogen)–2% FCS was used to replace the transfection mix. Cells were then incubated at 33°C for 48 h.
Quantification of virus growth
Virus growth was measured by immunofluorescence. Two days post-transfection, cell supernatant was recovered and clarified 5 min at 1000 rpm in a Multifuge 4 KR. Two hundred microliters of clarified supernatant were mixed with 2 mL of McCoy's 5A Medium–2% FCS to infect HeLa cells grown overnight on 22 mm × 22 mm coverslips in 33-mm wells. Virus growth of wt and mutant HRV14 was finally quantified by immunofluorescence 12 h post-infection. The cells were washed twice with PBS lacking Ca2+ and Mg2+ (PBS−) and fixed 1.5 h in acetone at −20°C. Cells were air-dried for a few minutes at room temperature before incubation with the primary antibody, a rabbit anti-HRV14 serum (ATCC number: VR-284AS/Rb, diluted 1/1000 in PBS−–1% BSA), for 45 min at 37°C in a humidity chamber. After intensive washing with PBS−, Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen) was added and the cells were incubated for 45 min at 37°C in the dark. After final rinsing with PBS−, coverslips were mounted in fluorotec embedding medium (Bio-Science products AG). Quantification of virus growth was calculated as the percentage of positive cells in three independent experiments.
Comparative sequence analysis
The virus sequences were aligned using MAFFT version 6.240 (Katoh et al. 2002). All alignments were then analyzed with RNAz version 1.0 (Washietl et al. 2005). The conserved secondary structure elements found by the program were manually investigated for the presence of the typical 14-nt cre hairpin–loop as well as the conserved sequence motif. Subregions of the alignments were extracted using the EMBOSS package version 5.0.0 (Rice et al. 2000) and a consensus sequence and structure was calculated with RNAalifold, which is part of the Vienna Package version 1.7. The trees for Figure 3 and Supplemental Figures 2, 3, and 4 were calculated using multiple sequence alignments of the respective cre motifs and the mlocARNA pipeline version 1.0 (Will et al. 2007) for structural based cluster trees. Secondary structures and energies for individual cre sequences were computed using the RNAfold program (part of the Vienna Package) and the program MFOLD version 3.2 (Zuker 2003). The boxplots in Figure 5 were created using the pairwise distances between every terminal node in every single weighted pair group method with averaging (WPGMA) tree of the species-specific cre elements from mlocARNA. The length distributions were plotted group wise using Matlab 7.2.0.283 (Mathworks, http://www.mathworks.com).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We would like to thank Lara Turin, Sandra Frayard Van Bell, and Chantal Gaille for technical assistance and Luc Perrin for support and comments on the manuscript. We also thank Rosemary Sudan for editorial assistance. This study was supported by the Swiss National Science Foundation (No 3200B0-101670 to L.K. and 3100A0112588/I to E.M.Z.), the Canton of Geneva, and the University of Geneva Dean's programme for the promotion of women in science (C.T.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1031408.
REFERENCES
- Andino R., Rieckhof G.E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′-end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.J., O'Donnell B.J., Flanegan J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Kauder S.E., Cornell C.T., Jang G.M., Racaniello V.R., Semler B.L. Cell-dependent role for the poliovirus 3′-noncoding region in positive-strand RNA synthesis. J. Virol. 2004;78:1344–1351. doi: 10.1128/JVI.78.3.1344-1351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Cornell C.T., Tran G.P., Nguyen J.H., Semler B.L. An authentic 3′-noncoding region is necessary for efficient poliovirus replication. J. Virol. 2005;79:11962–11973. doi: 10.1128/JVI.79.18.11962-11973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M.W., Teterina N., Egger D., Bienz K., Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- Denny F.W., Jr The clinical impact of human respiratory virus infections. Am. J. Respir. Crit. Care Med. 1995;152:S4–12. doi: 10.1164/ajrccm/152.4_Pt_2.S4. [DOI] [PubMed] [Google Scholar]
- Duque H., Palmenberg A.C. Phenotypic characterization of three phylogenetically conserved stem-loop motifs in the mengovirus 3′-untranslated region. J. Virol. 2001;75:3111–3120. doi: 10.1128/JVI.75.7.3111-3120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Two functional complexes formed by KH domain containing proteins with the 5′-noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes & Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber K., Wimmer E., Paul A.V. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: Identification of a cis-replicating element in the coding sequence of 2Apro . J. Virol. 2001;75:10979–10990. doi: 10.1128/JVI.75.22.10979-10990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry Y., Richardson A., Meredith J., Almond J.W., Barclay W., Evans D.J. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I.G., Polacek C., Andino R., Evans D.J. The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J. Gen. Virol. 2003;84:2359–2363. doi: 10.1099/vir.0.19132-0. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Wolf Y.I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- Huang H., Alexandrov A., Chen X., Barnes T.W., III, Zhang H., Dutta K., Pascal S.M. Structure of an RNA hairpin from HRV-14. Biochemistry. 2001;40:8055–8064. doi: 10.1021/bi010572b. [DOI] [PubMed] [Google Scholar]
- Jacobson S.J., Konings D.A., Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′-terminus of the poliovirus RNA genome and its role in viral RNA amplification. J. Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian A.Z. Rhinoviruses: A numbering system. Nature. 1967;213:761–762. doi: 10.1038/213761a0. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A., Avila P.C., Rouskin S., Magrini V., Credle J.J., Schnurr D.P., Boushey H.A., Mardis E.R., Li H., DeRisi J.L. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J. Infect. Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B.L., Rothberg P.G., Larsen G.R., Adler C.J., Dorner A.J., Emini E.A., Hanecak R., Lee J.J., Van der Werf S., et al. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J., Dean A., St George K., Briese T., Lipkin W.I. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J. Infect. Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Yip C.C., Tsoi H.W., Lee R.A., So L.Y., Lau Y.L., Chan K.H., Woo P.C., Yuen K.Y. Clinical features and complete genome characterization of a distinct human rhinovirus genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 2007;196:986–993. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford R.M., Patel N.R., Demenczuk T.M., Watanyar A., Herbertz T., Collett M.S., Pevear D.C. VP1 sequencing of all human rhinovirus serotypes: Insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.M., Kiesner C., Pappas T., Lee I., Grindle K., Jartti T., Jakiela B., Lemanske R.F., Shult P.A., Gern J.E. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert P.E., Escriou N., Ruelle J., Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.W., Bezborodova S.V., Henry T.M. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 2002;76:9686–9694. doi: 10.1128/JVI.76.19.9686-9694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McErlean P., Shackelton L.A., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J. Clin. Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McErlean P., Shackelton L.A., Andrews E., Webster D.R., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS ONE. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K.L. The human rhinovirus internal cis-acting replication element (cre) exhibits disparate properties among serotypes. Arch. Virol. 2003;148:2397–2418. doi: 10.1007/s00705-003-0177-7. [DOI] [PubMed] [Google Scholar]
- McKnight K.L., Lemon S.M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K.L., Lemon S.M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers W.J., Hoenderop J.G., Bruins Slot H.J., Pleij C.W., Pilipenko E.V., Agol V.I., Galama J.M. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′-untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayan C., Wimmer E. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology. 1992;189:547–555. doi: 10.1016/0042-6822(92)90578-d. [DOI] [PubMed] [Google Scholar]
- Mirzayan C., Wimmer E. Biochemical studies on poliovirus polypeptide 2C: Evidence for ATPase activity. Virology. 1994;199:176–187. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- Morasco B.J., Sharma N., Parilla J., Flanegan J.B. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 2003;77:5136–5144. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsley T.B., Towner J.S., Blyn L.B., Ehrenfeld E., Semler B.L. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Pathak H.B., Arnold J.J., Wiegand P.N., Hargittai M.R., Cameron C.E. Picornavirus genome replication: Assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J. Biol. Chem. 2007;282:16202–16213. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.V., van Boom J.H., Filippov D., Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- Perera R., Daijogo S., Walter B.L., Nguyen J.H., Semler B.L. Cellular protein modification by poliovirus: The two faces of poly(rC)-binding protein. J. Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister T., Jones K.W., Wimmer E. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 2000;74:334–343. doi: 10.1128/jvi.74.1.334-343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Poperechny K.V., Maslova S.V., Melchers W.J., Slot H.J., Agol V.I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: A quasi-globular multi-domain RNA structure maintained by tertiary (“kissing”) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- Renwick N., Schweiger B., Kapoor V., Liu Z., Villari J., Bullmann R., Miething R., Briese T., Lipkin W.I. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J. Infect. Dis. 2007;196:1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richards O.C., Spagnolo J.F., Lyle J.M., Vleck S.E., Kuchta R.D., Kirkegaard K. Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase. J. Virol. 2006;80:7405–7415. doi: 10.1128/JVI.02533-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder E., Paul A.V., Kim D.W., van Boom J.H., Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder E., Xiang W., Paul A., Wimmer E. Analysis of the cloverleaf element in a human rhinovirus type 14/poliovirus chimera: Correlation of subdomain D structure, ternary protein complex formation and virus replication. J. Gen. Virol. 2003;84:2203–2216. doi: 10.1099/vir.0.19013-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez P.L., Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- Samuilova O., Krogerus C., Fabrichniy I., Hyypia T. ATP hydrolysis and AMP kinase activities of nonstructural protein 2C of human parechovirus 1. J. Virol. 2006;80:1053–1058. doi: 10.1128/JVI.80.2.1053-1058.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- Sharma N., O'Donnell B.J., Flanegan J.B. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 2005;79:3565–3577. doi: 10.1128/JVI.79.6.3565-3577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Wang Q., Yang Y., Pathak H.B., Arnold J.J., Castro C., Lemon S.M., Cameron C.E. Human rhinovirus type 14 gain-of-function mutants for oriI utilization define residues of 3C(D) and 3Dpol that contribute to assembly and stability of the picornavirus VPg uridylylation complex. J. Virol. 2007;80:12485–12495. doi: 10.1128/JVI.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy D.A., Giddings T.H., Jr, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: An autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C., Junier T., Gerlach D., Cordey S., Van Belle S., Perrin L., Zdobnov E.M., Kaiser L. New complete genome sequences of human rhinoviruses shed light on their phylogeny and genomic features. BMC Genomics. 2007;8:224. doi: 10.1186/1471-2164-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina N.L., Levenson E., Rinaudo M.S., Egger D., Bienz K., Gorbalenya A.E., Ehrenfeld E. Evidence for functional protein interactions required for poliovirus RNA replication. J. Virol. 2006;80:5327–5337. doi: 10.1128/JVI.02684-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiviyanathan V., Yang Y., Kaluarachchi K., Rijnbrand R., Gorenstein D.G., Lemon S.M. High-resolution structure of a picornaviral internal cis-acting RNA replication element (cre) Proc. Natl. Acad. Sci. 2004;101:12688–12693. doi: 10.1073/pnas.0403079101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd S., Towner J.S., Brown D.M., Semler B.L. Replication-competent picornaviruses with complete genomic RNA 3′-noncoding region deletions. J. Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij M.J., Vogt D.A., Paul A., Castro C., Kuijpers J., van Kuppeveld F.J., Cameron C.E., Wimmer E., Andino R., Melchers W.J. Structural and functional characterization of the coxsackievirus B3 CRE(2C): Role of CRE(2C) in negative- and positive-strand RNA synthesis. J. Gen. Virol. 2006;87:103–113. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will S., Reiche K., Hofacker I.L., Stadler P.F., Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Rijnbrand R., McKnight K.L., Wimmer E., Paul A., Martin A., Lemon S.M. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 2002;76:7485–7494. doi: 10.1128/JVI.76.15.7485-7494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Rijnbrand R., Watowich S., Lemon S.M. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J. Biol. Chem. 2004;279:12659–12667. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- Yin J., Paul A.V., Wimmer E., Rieder E. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: Analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J. Virol. 2003;77:5152–5166. doi: 10.1128/JVI.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]