Abstract
Aminoglycosides are one of the most widely used and clinically important classes of antibiotics that target the ribosome. Hygromycin B is an atypical aminoglycoside antibiotic with unique structural and functional properties. Here we describe the structure of the intact Escherichia coli 70S ribosome in complex with hygromycin B. The antibiotic binds to the mRNA decoding center in the small (30S) ribosomal subunit of the 70S ribosome and induces a localized conformational change, in contrast to its effects observed in the structure of the isolated 30S ribosomal subunit in complex with the drug. The conformational change in the ribosome caused by hygromycin B binding differs from that induced by other aminoglycosides. Also, in contrast to other aminoglycosides, hygromycin B potently inhibits spontaneous reverse translocation of tRNAs and mRNA on the ribosome in vitro. These structural and biochemical results help to explain the unique mode of translation inhibition by hygromycin B.
Keywords: hygromycin B, aminoglycoside, ribosome, inhibition, translation, structure
INTRODUCTION
A large number of antibiotics act by targeting the ribosome (Hermann 2005; Poehlsgaard and Douthwaite 2005; Sutcliffe 2005). Antibiotics bind to specific sites on the ribosome and selectively affect a number of steps of the ribosomal translation cycle. The mechanisms of action of some antibiotics are now well established (Hermann 2005; Sutcliffe 2005; Borovinskaya et al. 2007a,b). However, the mechanism used by hygromycin B to inhibit protein synthesis by the ribosome is not known despite years of intense study. Hygromycin B is an aminoglycoside antibiotic produced by Streptomyces hygroscopicus (Mann and Bromer 1958). Widely used in veterinary medicine (Kelley and Olsen 1960; Biehl 1986) and in cell culture selections (Gritz and Davies 1983; Rao et al. 1983), it kills bacteria (Davies et al. 1965), fungi, and higher eukaryotic cells, including mammalian cells (Gonzáles et al. 1978; Eustice and Wilhelm 1984b).
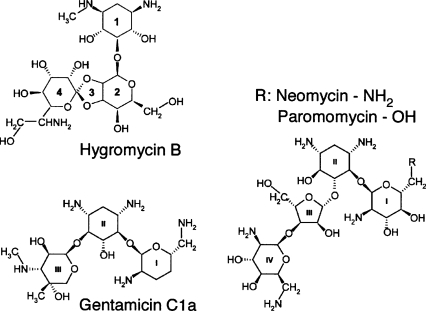
Hygromycin B possesses unique structural and functional properties when compared to other aminoglycosides. It has a dual ether linkage between its second and fourth rings, forming a third ring (Fig. 1; Neuss et al. 1970; Sutcliffe 2005). Other structurally related aminoglycosides such as neomycin, paromomycin, and gentamicin possess 4,5-substituted and 4,6-substituted 2-deoxystreptamine (2-DOS) cores which form ring II of the drugs, and a glucosamine sugar, or ring I, attached at position 4 in ring II (Fig. 1). Rings I and II, common among most of the aminoglycosides, are thought to be the primary ribosome binding determinants of the drugs and to establish the drugs' functional properties (Carter et al. 2000; Hermann 2005; Sutcliffe 2005; Borovinskaya et al. 2007a). Hygromycin B has a 2-DOS core as its first ring but does not have a glucosamine ring I, which results in distinct properties of the drug (Konno et al. 2004).
FIGURE 1.
Chemical structures of aminoglycosides hygromycin B, neomycin, paromomycin, and gentamicin. The 2-DOS ring 1 of hygromycin B is similar to ring II of the other three aminoglycosides.
Hygromycin B effectively inhibits translocation of mRNA and tRNAs on the ribosome in both bacteria (Davies et al. 1965; Cabañas et al. 1978a; Peske et al. 2004) and eukaryotes (Cabañas et al. 1978b; Gonzáles et al. 1978; Eustice and Wilhelm 1984b), and also has a subtle effect on decoding fidelity (Eustice and Wilhelm 1984a,b), as it slightly increases aminoacyl–tRNA (A-site tRNA) affinity (Cabañas et al. 1978a; Eustice and Wilhelm 1984a; Peske et al. 2004). Gentamicin, paromomycin and neomycin preferentially target bacterial (Davies et al. 1964) and mitochondrial ribosomes, and to a lesser extent, eukaryotic ribosomes (Wilhelm et al. 1978; Eustice and Wilhelm 1984b; Manuvakhova et al. 2000). These aminoglycosides exert multiple effects on protein synthesis: they cause miscoding (Davies et al. 1964; Davies et al. 1966) and also inhibit ribosomal translocation (Cabañas et al. 1978b; Misumi et al. 1978), peptide release (Youngman et al. 2007), and ribosome recycling (Hirokawa et al. 2002).
Consistent with its more specific effect on translocation, hygromycin B exhibits monophasic inhibition of polypeptide synthesis (Zierhut et al. 1979), suggesting that it binds to a single site on the ribosome. Neomycin, gentamicin, and paromomycin, in contrast, have a triphasic effect on translation that indicates the presence of multiple interacting binding sites on the ribosome for the drugs (Tai and Davis 1979; Zierhut et al. 1979; Jelenc and Kurland 1984). Two functional binding sites on the ribosome for neomycin, gentamicin, and paromomycin have been identified, one in each ribosomal subunit (Misumi et al. 1978; Borovinskaya et al. 2007a).
Structural and biochemical analysis of bacterial ribosomes revealed that hygromycin B has a binding site in RNA helix 44 (h44) of the small (30S) ribosomal subunit, next to the aminoacyl–tRNA binding site (Moazed and Noller 1987; Brodersen et al. 2000). When bound to the intact 70S ribosome, hygromycin B protects N7 of G1494 in 16S rRNA from dimethylsulfate (DMS) modification, and enhances DMS modification of N1 in A1408 (Moazed and Noller 1987). When hygromycin B binds to the isolated small ribosomal subunit, it enhances DMS reactivity of both G1494 and A1408 (Moazed and Noller 1987). In contrast, neomycin, paromomycin, and gentamicin, which bind to a slightly different binding site in h44, strongly protect A1408 and G1494 in both the intact ribosome and isolated small ribosomal subunit (Moazed and Noller 1987).
A number of mutations in the hygromycin B binding site confer resistance to the drug, including mutations U1406C, C1496U, and U1498C in 16S rRNA in Mycobacterium smegmatis (here and throughout the text an Escherichia coli numbering is used) (Pfister et al. 2003), and a U to C transition in nucleotide U1495 in 18S rRNA in the eukaryote Tetrahymena thermophila (Spangler and Blackburn 1985). In E. coli, mutation of either G1491 or C1409, which would break a base pair between these nucleotides, causes resistance to hygromycin B (De Stasio and Dahlberg 1990).
The X-ray crystal structure of the small ribosomal subunit from Thermus thermophilus in complex with hygromycin B provided the first structural model of how the drug interacts with the 30S subunit (Brodersen et al. 2000). Based on the position of the hygromycin B binding site in h44 of the ribosome it was suggested that the drug could restrict or inhibit a conformational change in h44 crucial for the movement of this helix during translocation (Frank and Agrawal 2000), although no conformational changes were directly observed in the structure (Brodersen et al. 2000). These structural data helped to explain the U1495C mutation that confers resistance to hygromycin B in eukaryotes (Spangler and Blackburn 1985) and the resistance mutations C1496U and U1498C found in M. smegmatis (Pfister et al. 2003). However, the 30S subunit crystal structure does not reveal why the U1406C mutation in M. smegmatis (Pfister et al. 2003) or the G1491 and C1409 mutations in E. coli (De Stasio and Dahlberg 1990) confer resistance to the drug. This structure also does not explain an increase of chemical reactivity of A1408 to DMS modification in the 70S ribosome and an increase of reactivity of G1494 in the 30S subunit in the presence of hygromycin B (Moazed and Noller 1987).
Hygromycin B weakly stabilizes tRNA binding to the ribosomal A site, but its potent inhibition of translation is thought to result primarily from its inhibition of mRNA and tRNA translocation, by a mechanism still to be determined (Davies et al. 1965; Cabañas et al. 1978a; Eustice and Wilhelm 1984a; Peske et al. 2004). Translocation only occurs on the intact ribosome (Spirin 1987; Horan and Noller 2007). Thus, in order to shed light on the mechanism of hygromycin B inhibition, we have determined the structure of hygromycin B in complex with the intact E. coli 70S ribosome. We have also investigated effects of hygromycin B and other aminoglycosides known to inhibit translocation on reverse tRNA and mRNA movement in the ribosome, i.e., reverse translocation (Shoji et al. 2006; Borovinskaya et al. 2007b). The observed structural interactions between the antibiotic and the ribosome, together with the biochemical data, help to explain the distinct mode of the translation inhibition by hygromycin B.
RESULTS AND DISCUSSION
Binding of hygromycin B to the 70S ribosome
In the 70S ribosome, hygromycin B binds to a single site in RNA helix h44 of the small ribosomal subunit (Fig. 2), consistent with the monophasic mode of translation inhibition by the drug (Zierhut et al. 1979). In contrast, aminoglycosides gentamicin, neomycin, and paromomycin, which exhibit a triphasic inhibition pattern (Tai and Davis 1979; Zierhut et al. 1979), bind to h44 of the small ribosomal subunit and to RNA helix 69 (H69) of the large ribosomal subunit (Borovinskaya et al. 2007a). Therefore, it is likely that helix h44 in the small subunit is targeted by all 2-DOS containing aminoglycosides, whereas the H69 binding site is involved in the triphasic effects of neomycin, gentamicin, and paromomycin on translation (Zierhut et al. 1979). The binding site of hygromycin B in h44 is located a few angstroms away from that of the other aminoglycosides. Moreover, due to the rigidity of the hygromycin B molecule, it is unable to fit into binding pockets in either h44 or H69, where other 2-DOS aminoglycosides bind (not shown).
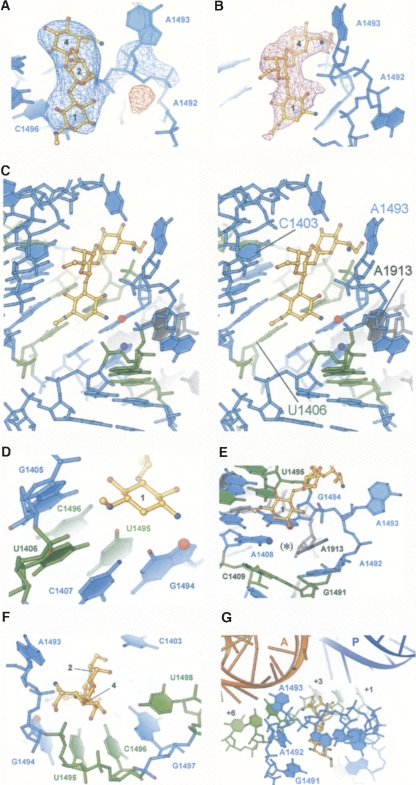
FIGURE 2.
Interactions of hygromycin B with h44 of the ribosome. (A) Two-fold averaged F obs − F obs difference Fourier electron density map covering hygromycin B bound to h44, and A1493 of 16S rRNA flipped out of h44. Positive density is shown in blue, and negative electron density is in red. 16S rRNA is in light blue. (B) Two-fold averaged and sharpened 3F obs − 2F calc difference Fourier electron density map (magenta) near hygromycin B binding site in h44. The density for rRNA is removed for clarity. (C) Stereo view of the hygromycin B binding site in h44. 16S rRNA is shown in light blue, 23S rRNA is in gray. N7 of G1494, which is protected from DMS modification in the presence of hygromycin B, is labeled with a red sphere. N1 of A1408, which shows enhanced DMS reactivity in the presence of hygromycin B, is labeled with a purple sphere. Nucleotides, mutations in which confer resistance to hygromycin B, are shown in green. (D) Interactions of ring 1 of hygromycin B with h44. Nucleotides are colored as in C. N7 of G1494, protected from DMS modification in the presence of hygromycin B, is labeled with a red sphere. (E) Hygromycin B-induced flipping of A1493 out of h44 explains increased reactivity of N1 in A1408 to DMS modification in the presence of the antibiotic. N1 of A1408 is labeled with a purple sphere. The position adopted by A1493 in the vacant 70S ribosome structure (Schuwirth et al. 2005) is marked with the black asterisk. Nucleotides and N7 of G1494 protected from DMS modification in the presence of hygromycin B are colored as in C. (F) Interactions of ring 4 of hygromycin B with h44. Nucleotides are colored as in C. Rings of the antibiotic are labeled with numbers. (G) Model of the hygromycin B binding site in h44 (light blue) with mRNA and tRNAs bound. A-site tRNA (orange), P-site tRNA (dark blue), and mRNA (green) were superimposed with the present structure based on the structures of the T. thermophilus 70S ribosome (Yusupov et al. 2001; Selmer et al. 2006). Positions in the P-site and A-site mRNA codons are numbered from +1 as the first base of the P-site codon to +6 as the third base of the A-site codon.
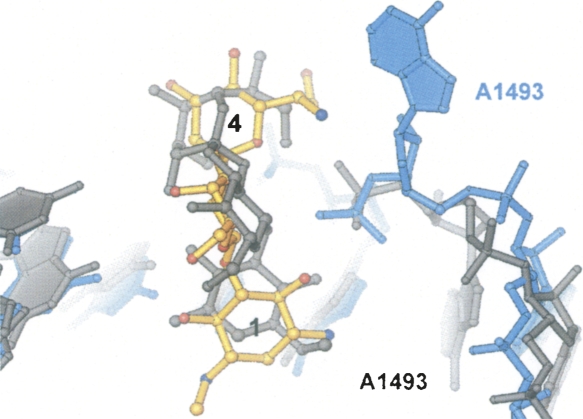
In the 70S ribosome, hygromycin B binds to the same site within h44 as seen in the structure of the 30S subunit in complex with the drug (Brodersen et al. 2000). However, the conformation of the antibiotic, its interactions with 16S ribosomal RNA, and its effects on the conformation of h44 differ substantially from those observed in the 30S subunit structure (Figs. 2, 3). The first ring of the antibiotic is rotated 180° with respect to that modeled in the 30S subunit structure (Brodersen et al. 2000), and forms a network of hydrogen bonds that spans the major groove of nucleotides C1404, G1405, U1406, and G1494–C1496, to the phosphate of G1494 (Fig. 2B–E). The second ring hydrogen bonds to C1403 (Fig. 2C,F). The fourth ring is rotated away from the P site toward the A site in the present structure compared to that in Brodersen et al. (2000), and forms hydrogen bonds with A1493 and with the phosphate of U1495 (Figs. 2F,G, 3). In the present 70S ribosome structure, hygromycin B changes conformations of three universally conserved adenosines, A1492 and A1493 in 16S rRNA and A1913 in 23S rRNA (Cannone et al. 2002). The drug induces A1493 of 16S rRNA to flip out of h44, and reorients A1492 of 16S rRNA and A1913 of 23S rRNA (Figs. 2, 3, 4). This conformational change was not observed in the structure of the 30S subunit in complex with hygromycin B (Fig. 3; Brodersen et al. 2000).
FIGURE 3.
Superposition of the hygromycin B and h44 conformations observed in the present structure of hygromycin B bound to 70S ribosome with the structure of hygromycin B bound to 30S ribosomal subunit (Brodersen et al. 2000). For the present structure, hygromycin B and 16S rRNA are shown in gold and light blue, respectively. Hygromycin B and 16S rRNA from the 30S subunit structure (Brodersen et al. 2000) are in gray.
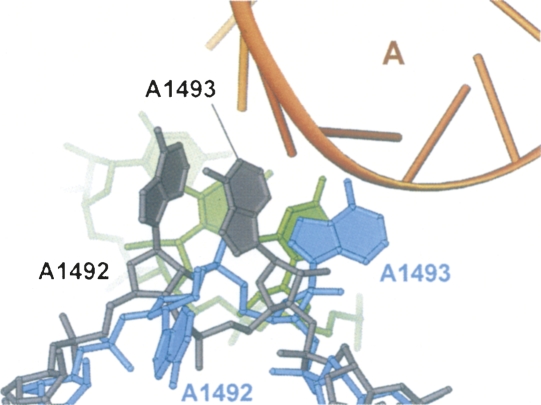
FIGURE 4.
Superposition of the h44 conformations observed in the present structure of the E. coli 70S ribosome in complex with hygromycin B (rRNA in light blue) and in the structure of the T. thermophilus 70S ribosome in complex with tRNAs, mRNA, and paromomycin (rRNA in gray) (Selmer et al. 2006). Hygromycin B, paromomycin, and the P-site codon of mRNA were removed for clarity. The A-site tRNA is shown in orange and the A-site codon of mRNA is in green. In the presence of only paromomycin (or the related aminoglycosides neomycin or gentamicin), h44 of the 70S ribosome adopts the same conformation as that observed in the presence of paromomycin and tRNAs and mRNA (Selmer et al. 2006; Borovinskaya et al. 2007a).
Binding of neomycin, gentamicin, or paromomycin to h44 of the 70S ribosome causes nucleotides A1492 and A1493 in 16S rRNA to flip out of h44 in the correct conformation to interact with the minor groove of the A-site codon–anticodon helix (Fig. 4; Selmer et al. 2006; Borovinskaya et al. 2007a). Also, upon aminoglycoside binding nucleotide A1913 in 23S rRNA flips toward the A-site anticodon binding site (Selmer et al. 2006; Borovinskaya et al. 2007a). The conformational switch of A1492 and A1493 in 16S rRNA occurs upon binding of a cognate tRNA–mRNA complex to the ribosome, and thus is thought to aid in the discrimination of cognate versus noncognate tRNA during aminoacyl–tRNA selection by an induced fit mechanism (Carter et al. 2000; Ogle et al. 2001). The fact that neomycin, gentamicin, or paromomycin themselves promote the concerted flipping of these three adenosines across the ribosomal subunit interface prior to tRNA binding (Borovinskaya et al. 2007a) indicates that these aminoglycosides likely increase A-site tRNA affinity to the ribosome (Peske et al. 2004) by stabilizing the above conformation of the three adenosines, thereby inducing miscoding and possibly inhibiting translocation (Carter et al. 2000; Ogle et al. 2001; Selmer et al. 2006).
In contrast to structures of the 30S subunit and 70S ribosome with neomycin, gentamicin, or paromomycin (Carter et al. 2000; Ogle et al. 2001; Selmer et al. 2006; Borovinskaya et al. 2007a), binding of hygromycin B orients A1493 between the positions that would be occupied by A-site and P-site tRNAs (Figs. 2A,G, 4), although the adenine base of A1493 is not very well ordered. Nucleotides A1492 in 16S rRNA and A1913 in 23S rRNA undergo a rather modest conformational change when hygromycin B binds, as opposed to the large change in the complexes with neomycin, gentamicin, and paromomycin. In the hygromycin B complex, A1492 maintains base stacking with A1913 at the tip of H69 in the 50S subunit, a contact that serves as an intersubunit bridge in the apo–70S ribosome structure (Schuwirth et al. 2005), although the conformations of both A1492 and A1913 differ from those in the vacant 70S ribosome structure.
The mode of binding of hygromycin B and conformational changes in rRNA structure observed upon binding of hygromycin B help explain its effect on the reactivity of A1408 and G1494 in 16S rRNA to chemical probes (Fig. 2C–E; Moazed and Noller 1987). In the intact 70S ribosome hygromycin B shields G1494 and forms a hydrogen bond to N7 of G1494, thus protecting it from DMS modifications (Fig. 2C–E). The rearrangement of A1492 in 16S rRNA and A1913 in 23S rRNA upon binding of hygromycin B opens up a way for the chemical probe to reach A1408, and explains enhanced reactivity of N1 in A1408 to DMS modification in the presence of the drug (Fig. 2E; Moazed and Noller 1987). The conformational changes observed in the present structure would expose both A1408 and G1494 to chemical probes in the isolated 30S subunit (Fig. 2C,E), in agreement with chemical protection data (Moazed and Noller 1987), but in contrast to the crystal structure of the 30S subunit in complex with hygromycin B (Brodersen et al. 2000) where no structural changes were observed upon binding of the drug.
The present structure also helps to explain hygromycin B resistance mutations. The methylamine group of ring 1 of hygromycin B forms hydrogen bonds with the O4 carbonyls of U1406 and U1495, and the hydroxyl adjacent to the methylamine group in ring 1 contacts the N4 exocyclic amine of C1496 (Fig. 2D). These favorable contacts between ring 1 of hygromycin B and h44 of 16S rRNA would be disrupted in the resistant mutants U1406C and C1496U in 16S rRNA in Mycobacterium smegmatis (Pfister et al. 2003) and in the U1495C mutant in 18S rRNA in Tetrahymena thermophila (Fig. 2D; Spangler and Blackburn 1985). In addition, nucleotides U1406 and C1496 base pair with U1495 and G1405, respectively, which provide crucial structural components of the hygromycin B binding pocket. Mutations in U1406 or C1496 would break these base pairs and thus distort the hygromycin B binding site (Pfister et al. 2003).
Two sets of mutations are more challenging to interpret in light of the present structure. Despite the fact that base paired nucleotides C1409–G1491 do not directly interact with hygromycin B (Fig. 2C,E), mutations in these nucleotides lead to resistance to the drug (De Stasio and Dahlberg 1990). Breaking this base pair could confer resistance by compromising stability of the helix, or by perturbing the conformation of the hygromycin B binding pocket. Hygromycin B also does not make direct contact with U1498, making it hard to explain the resistance mutation U1498C found in M. smegmatis (Fig. 2F; Pfister et al. 2003). It should be noted that the U1498C mutation had the lowest level of resistance among three mutations found in this bacterium, U1406C, C1496U, and U1498C.
Notably, the nucleotides involved in binding of hygromycin B and/or those required for hygromycin B sensitivity (Spangler and Blackburn 1985; De Stasio and Dahlberg 1990; Pfister et al. 2003) are universally conserved in all three domains of life (Cannone et al. 2002). This explains the lack of specificity and general toxicity of hygromycin B.
Effect of hygromycin B on reverse translocation
Hygromycin B is thought to inhibit translation primarily by affecting translocation (Cabañas et al. 1978a; Peske et al. 2004), the coupled movement of tRNAs and mRNA on the ribosome following the formation of each peptide bond. Hygromycin B blocks multiple-turnover translocation, and decreases the rate of single-round translocation by over 300-fold (Peske et al. 2004). Translocation, catalyzed by elongation factor G (EF-G) in a GTP-dependent reaction, is the movement of the tRNA substrates from the A (aminoacyl-) and P (peptidyl-) sites on the ribosome to the P site and E (Exit) sites, respectively (Watson 1964; Wettstein and Noll 1965; Rheinberger et al. 1981), coupled with shifting of mRNA in the 5′ direction by one codon and thus opening the A site for a new round of mRNA decoding and aminoacyl–tRNA binding. Translocation is thought to occur in several kinetically and structurally distinct steps, where the peptidyl–tRNA and deacylated tRNA move first with respect to the 50S subunit, and then move with respect to the 30S subunit (Moazed and Noller 1989; Frank and Agrawal 2000; Savelsbergh et al. 2003; Valle et al. 2003; Peske et al. 2004; Sharma et al. 2004; Schuwirth et al. 2005; Dorner et al. 2006; Wilden et al. 2006; Frank et al. 2007).
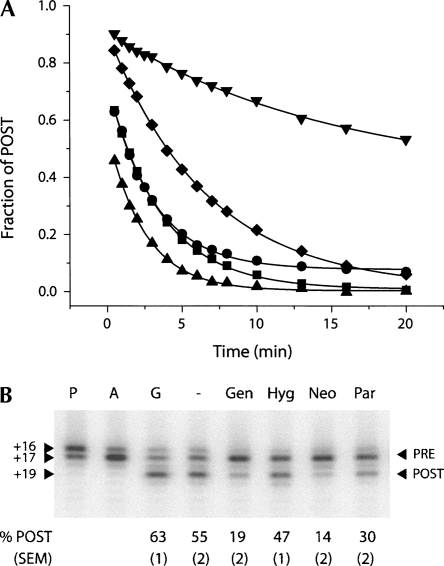
Notably, in the absence of EF-G, the ribosome is capable of performing translocation by itself, but at much slower rates (Pestka 1969; Gavrilova et al. 1976). Furthermore, the ribosome can support back-translocation, or reverse translocation, in both factor-dependent (Qin et al. 2006) and spontaneous (Shoji et al. 2006; Konevega et al. 2007) reactions. In the spontaneous reverse translocation reaction, i.e., reaction in the absence of EF-G and GTP, the pretranslocation (PRE) complex is formed by reverse translocation of peptidyl–tRNA from the P site and deacylated tRNA from the E site (the post-translocation, or POST, state) into the A and P sites, respectively (Shoji et al. 2006; Konevega et al. 2007). Movement of mRNA in the 3′ direction happens spontaneously upon E-site tRNA binding, without tRNA dissociation from the complex (Shoji et al. 2006). Interestingly, aminoglycosides affect the rate of the spontaneous reverse translocation reaction in strikingly different ways (Fig. 5A). Hygromycin B decreases the rate of reverse translocation by threefold, and also substantially affects the extent of the reaction, namely, decreases the final population of the PRE complex (Fig. 5A; Table 1). In contrast, neomycin, gentamicin, and paromomycin increase the observed rate of reverse translocation by two- to threefold and have no effect on the extent of the reaction (Fig. 5A; Table 1).
FIGURE 5.
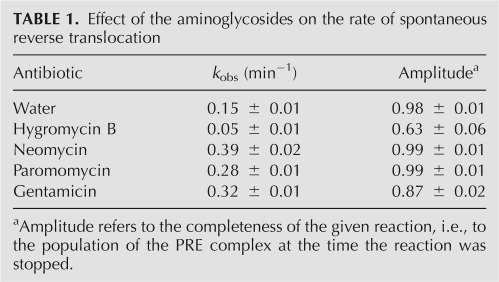
Effect of the aminoglycosides on reverse translocation. (A) Spontaneous reverse translocation upon addition of E-site tRNA to ribosomes programmed with mRNA and P-site bound N-acetyl-Val-tRNAVal in the absence of antibiotics (diamonds) and in the presence of neomycin (upward-pointing triangles), paromomycin (squares), gentamicin (circles), and hygromycin B (downward-pointing triangles). In all cases the data fit to a single exponential function (Table 1). (B) Reverse translocation promoted by antibiotics in the presence of EF-G and GTP. The P complex corresponds to deacylated tRNAfMet bound to the P-site of the mRNA programmed 70S ribosome; the A complex (PRE) forms upon binding of the N-acetyl-Val-tRNAVal to the A site of P complex; G complex (POST) forms upon addition EF-G/GTP to the A complex; the other complexes are formed upon addition of neomycin (Neo), paromomycin (Par), gentamicin (Gen), hygromycin B (Hyg), or water (−) to the G (POST) complex. The percent of POST complex remaining at the end of the incubation is indicated below each lane. Reported values represent the mean values (±SEM) derived from three independent experiments.
TABLE 1.
Effect of the aminoglycosides on the rate of spontaneous reverse translocation
Neomycin and paromomycin, along with other antibiotics that inhibit translocation, also promote reverse translocation of POST complexes into the PRE state even in the presence of EF-G and GTP (Shoji et al. 2006; Borovinskaya et al. 2007b). In the reversible reaction of translocation, the drugs were proposed to strongly inhibit the forward translocation reaction with little or no effect on reverse translocation (Shoji et al. 2006). Notably, when compared to the other 2-DOS containing aminoglycosides, hygromycin B has the weakest effect on conversion of the POST into PRE complexes in the presence of EF-G/GTP, while neomycin and gentamicin are the most efficient (Fig. 5B). This suggests that hygromycin B only weakly, if at all, stabilizes the PRE state in the presence of EF-G/GTP, when compared to the other 2-DOS aminoglycosides (Shoji et al. 2006).
Insights into the mechanism of translation inhibition by hygromycin B
There are several possible mechanisms by which antibiotics could inhibit translocation. They can stabilize tRNAs in their respective binding sites on the ribosome, thereby increasing the energy barrier of translocation. Alternatively, antibiotics can either block the conformational changes on the ribosome that are required for translocation to occur, or induce conformational changes that would interfere with translocation.
The unique behavior of hygromycin B, which strongly inhibits spontaneous reverse translocation (Fig. 5A; Table 1), can be explained by the structural results presented above. In the hygromycin-bound structure, nucleotide A1493 flips out of h44 into a position between those that would be occupied by A-site and P-site tRNAs (Fig. 2G; Yusupov et al. 2001; Selmer et al. 2006). In this position, A1493 would not interfere with A-site tRNA binding during mRNA decoding, although it might contribute to the slight increase of A-tRNA affinity caused by hygromycin B (Fig. 4; Cabañas et al. 1978a; Eustice and Wilhelm 1984a; Peske et al. 2004). However, in this position, A1493 would cause a steric block to the movement of tRNAs between the A and P sites, both in the forward and reverse directions (Figs. 2G, 5A; Table 1). Alternatively, A1493 in its conformation adopted upon hygromycin B binding could interact with the A-site codon of the mRNA (Figs. 2G, 4). Finally, hygromycin B binds to the ribosome in a position to contact the backbone of the P-site mRNA codon via its second ring (Fig. 2G). Therefore, translocation inhibition by hygromycin B (Fig. 5; Cabañas et al. 1978b; Eustice and Wilhelm 1984b; Peske et al. 2004) may result from a steric block between the A and P sites caused by A1493, and from confinement of mRNA in the A and P sites. It should be noted that hygromycin B might induce larger conformational changes in the ribosome when A-site tRNA is bound compared to what is observed here with a vacant ribosome (Jerinic and Joseph 2000).
The structural results also help to explain the observation that hygromycin B only weakly promotes POST to PRE conversion in the presence of EF-G and GTP (Fig. 5B). By slightly increasing the affinity of the A site for tRNA and restricting mRNA movement, hygromycin B would lead to a small increase in PRE complex formation by completely abolishing EF-G/GTP-dependent forward translocation, while promoting slow reformation of the PRE complex via tRNA dissociation and rebinding to the A site (Shoji et al. 2006). A similar mechanism of restricted mRNA and tRNA movement coupled with tRNA dissociation and rebinding would also explain hygromycin B inhibition of spontaneous reverse translocation (Fig. 5A; Table 1). Notably, the inhibitory action of hygromycin B does not require the presence of elongation factors (Cabañas et al. 1978a). Unlike neomycin, gentamicin, and paromomycin, hygromycin B has no effect on binding of EF-G to the 70S ribosome in the presence of nonhydrolyzable GTP (Campuzano et al. 1979). Indeed, based on the data we present here, hygromycin B would interfere with translocation primarily by affecting mRNA movement on the ribosome and increasing A–tRNA affinity, both in the presence and in the absence of EF-G. Future experiments will be required to confirm this model.
In contrast, neomycin, paromomycin, and gentamicin strongly promote reverse translocation both in the presence of EF-G/GTP and in the spontaneous reverse translocation reactions. Presumably, they increase A-tRNA affinity and stabilize the pretranslocation state of the ribosome, thus inhibiting the forward translocation reaction predominantly with little or no effect on the reverse reaction (Shoji et al. 2006; Borovinskaya et al. 2007a).
The results presented here provide evidence that different 2-DOS aminoglycoside antibiotics inhibit translation through structurally and functionally distinct mechanisms. All four of the 2-DOS aminoglycosides examined here target the decoding site of the ribosome by binding to h44 of the small ribosomal subunit. Binding of the antibiotics to this site is sufficient to cause substantial inhibition of translation through two different modes of drug binding. Whereas neomycin, paromomycin, and gentamicin induce a local conformational rearrangement that traps A-site tRNA on the ribosome (Carter et al. 2000; Selmer et al. 2006; Borovinskaya et al. 2007a), hygromycin B may act by confining the movement of mRNA and by blocking the tRNA path between the A and P sites. Potent inhibition of translocation and ribosome recycling by neomycin, paromomycin, and gentamicin may also depend on a second aminoglycoside binding site in H69 of the large ribosomal subunit (Borovinskaya et al. 2007a). The distinct modes of translation inhibition seen with different classes of aminoglycosides could have implications for the design of new antibiotics, and for improving their veterinary use and clinical use in aminoglycoside antibiotic therapy (Senior 1999; Manuvakhova et al. 2000; Kimura et al. 2005; Croft et al. 2006).
MATERIALS AND METHODS
Crystallization, data collection, and structure refinement
Ribosomes from E. coli strain MRE600 depleted of protein S1 were crystallized as described (Schuwirth et al. 2005). Large single crystals were cryo-protected as described (Borovinskaya et al. 2007b) prior to flash-freezing in liquid nitrogen. Hygromycin B (Sigma) at a saturating concentration of 0.08 mM (Cabañas et al. 1978a; Zierhut et al. 1979; Moazed and Noller 1987; Jerinic and Joseph 2000; Peske et al. 2004) was soaked into ribosome crystals for 24 h during cryo-stabilization.
Diffraction data were measured from one single crystal (Table 2) cooled to 93 K using 0.2°–0.3° oscillations at the SIBYLS (12.3.1) beam line at the Advanced Light Source, which is equipped with an ADSC Q315 area detector. Data were reduced and scaled using Denzo/Scalepack (Otwinowski and Minor 1997) and Truncate (Table 2; Collaborative Computational Project No. 4 1994). The crystal diffracted X-rays anisotropically, as indicated by the completeness of the data set (Table 2) and signal-to-noise level as a function of resolution (not shown).
TABLE 2.
Data collection and refinement statistics
The 3.5 Å structure of the E. coli 70S ribosome (Schuwirth et al. 2005) was used as the starting model for further refinement in CNS (Brünger et al. 1998). The model was first subjected to rigid body refinement against the antibiotic-bound data. Modeling of hygromycin B into difference electron density maps was carried out as follows. An initial model of hygromycin B (Brodersen et al. 2000) was subjected to energy minimization and to a conformational search using Monte Carlo algorithms with Maestro (Schrödinger, LLC), then a drug conformation of minimum energy was chosen. The topology and parameter files describing the antibiotic model were generated with HIC-Up (Kleywegt and Jones 1998). The model was manually docked into (F obs − F obs) difference electron density maps, with apo-70S ribosomes as the reference and phases derived from Pirate density modification (Cowtan 2000). The position of bases A1492, A1493 in 16S rRNA, A1913 in 23S rRNA, and other nucleotides in the antibiotic binding pocket were adjusted to fit the difference electron density. The model was then refined using rounds of manual rebuilding with O (Jones et al. 1991) and torsional dynamics (Brünger et al. 1998). After torsional dynamics refinement, new electron density maps were generated using sharpened (3F obs − 2F calc) coefficients (Brünger et al. 1998) and Pirate-derived phases (Cowtan 2000), followed by twofold crystallographic averaging. The new map revealed asymmetric density for the first ring of hygromycin B that was best explained by rotating the ring 180° from its position in the model described by Brodersen et al. (2000). This alternative model was also consistent with a minimum energy conformation of hygromycin B calculated with Maestro (Schrödinger, LLC). The refinement statistics are presented in Table 2.
Toeprinting assays
The position of mRNA on the ribosome in translocation and reverse translocation experiments was determined by toeprinting, essentially as described (Fig. 5; Shoji et al. 2006; Borovinskaya et al. 2007b). All experiments were carried out in polymix buffer (Ehrenberg et al. 1990). Briefly, mRNA 292 (Shoji et al. 2006; Borovinskaya et al. 2007b) (0.5 μM) annealed to a [32P]-labeled primer was added to ribosomes depleted of protein S1 (0.7 μM), along with deacylated tRNAfMet (1 μM), and incubated at 37°C for 20 min to allow tRNAfMet to bind the ribosomal P site. The PRE complex was then formed by adding N-acetyl-Val-tRNAVal (to 1 μM) and incubating at 37°C for 10 min to allow this tRNA to bind the ribosomal A site. The resulting complex was then diluted by 20% into polymix buffer containing GTP (0.3 mM) and EF-G (1 μM), and incubated at 37°C for 10 min to form a POST complex. Finally, the POST complex was diluted by 10% in the presence of either water, or saturating (Cabañas et al. 1978a,b; Misumi et al. 1978; Zierhut et al. 1979; Moazed and Noller 1987; Jerinic and Joseph 2000; Peske et al. 2004) concentrations of either hygromycin B (0.08 mM), neomycin (0.1 mM, Sigma), gentamicin (0.36 mM, Fluka), or paromomycin (0.1 mM, Sigma), and incubated for 10 min at 37°C. Aliquots were removed for primer extension analysis at each stage of complex formation.
For primer extension reactions, each aliquot above was treated with AMV reverse transcriptase, and extension products were resolved by 6% (v/v) polyacrylamide denaturing gel electrophoresis, as described (Shoji et al. 2006). Toeprints were quantified using a phosphorimager (Molecular Dynamics) as follows. The percentage of POST complex was calculated as %POST=100× (post)/(pre+post), where pre and post are intensities of toeprints corresponding to PRE and POST complexes, respectively. The PRE complex corresponds to the sum of toeprints at positions +16 and +17 on the mRNA, whereas the POST complex corresponds to the toeprint at +19 on the mRNA (Hartz et al. 1989).
To measure the rate of spontaneous reverse translocation (Shoji et al. 2006), the P-site binding complex was formed as described above, using N-acetyl-Val-tRNAVal (1 μM). Reverse translocation was initiated by adding deacylated tRNAfMet (8 μM) to bind the E site at time t = 0, and the reaction was incubated at 37°C. The antibiotics were also added at time t = 0 at the concentrations noted above. A 2-μL aliquot was removed at each time point and immediately added to 10 μL of prewarmed extension mix and further incubated at 37°C for 2 min. The extension products were resolved by 6% (v/v) polyacrylamide denaturing gel electrophoresis as described above. The fraction of POST complex at time t, F(t), was determined by calculating F(t) = [(post)/(pre + post)]/F p , where F p is the equivalent value derived from the P lane. The data were then plotted as a function of time and fit to the single exponential function: F(t) = [F 0 + A × exp(−k obs × t)], where A and k obs are an amplitude and an observed rate constant, respectively (Table 1; Shoji et al. 2006). The validity of fitting the reaction kinetics to a single exponential was deciphered based on the residuals, i.e., the differences in observed and calculated values (not shown).
Figure preparation
The figures were made using programs Ribbons (Carson 1997), ISIS Draw (MDL Information Systems, Inc.), Origin (OriginLab) and Adobe Photoshop (Adobe Systems, Inc.).
Accession codes
The coordinates and structure factors for the 70S ribosome in complex with hygromycin B have been deposited in the Protein Data Bank with accession codes 3DF1, 3DF2, 3DF3, and 3DF4.
ACKNOWLEDGMENTS
We thank K. Frankel and S. Classen for help with data measurement at the SIBYLS (12.3.1) beam line at the Advanced Light Source, A. Borovinskiy for help with figure preparation and overall support, K. Durkin for the help with energy minimization calculations, and J. Doudna for helpful comments on the manuscript. This work was supported by NIH Grants GM65050 (J.H.D.C.) and GM072528 (K.F.), National Cancer Institute Grant CA92584 (for the SIBYLS beam line), and U.S. Department of Energy Grants DE-AC03-76SF00098, KP110201, and LBNL LDRD 366851 (J.H.D.C.), and DE-AC03 76SF00098 (for the SIBYLS beam line).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1076908.
REFERENCES
- Biehl L.G. Anthelmintics for swine. Vet. Clin. North Am. Food Anim. Pract. 1986;2:481–487. doi: 10.1016/s0749-0720(15)31258-5. [DOI] [PubMed] [Google Scholar]
- Borovinskaya M.A., Pai R.D., Zhang W., Schuwirth B.S., Holton J.M., Hirokawa G., Kaji H., Kaji A., Cate J.H.D. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007a;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Borovinskaya M.A., Shoji S., Holton J.M., Fredrick K., Cate J.H. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem. Biol. 2007b;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen D.E., Clemons W.M., Jr, Carter A.P., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cabañas M.J., Vázquez D., Modolell J. Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur. J. Biochem. 1978a;87:21–27. doi: 10.1111/j.1432-1033.1978.tb12347.x. [DOI] [PubMed] [Google Scholar]
- Cabañas M.J., Vázquez D., Modolell J. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun. 1978b;83:991–997. doi: 10.1016/0006-291x(78)91493-6. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Vázquez D., Modolell J. Dissociation of guanosine nucleotide-elongation factor G-ribosome complexes. Biochemistry. 1979;18:1570–1574. doi: 10.1021/bi00575a029. [DOI] [PubMed] [Google Scholar]
- Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Müller K.M., et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. Ribbons. In: Sweet R.M., Carter C.W. Jr, editors. Methods in enzymology. Vol. 277. Academic Press; New York: 1997. pp. 493–502. [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cowtan K. General quadratic functions in real and reciprocal space and their application to likelihood phasing. Acta Crystallogr. D Biol Crystallogr. 2000;D56:1612–1621. doi: 10.1107/s0907444900013263. [DOI] [PubMed] [Google Scholar]
- Croft S.L., Seifert K., Yardley V. Current scenario of drug development for leishmaniasis. Indian J. Med. Res. 2006;123:399–410. [PubMed] [Google Scholar]
- Davies J., Gilbert W., Gorini L. Streptomycin, suppression and the code. Proc. Natl. Acad. Sci. 1964;51:883–890. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Gorini L., Davis B.D. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- Davies J., Jones D.S., Khorana H.G. A further study of misreading of codons induced by streptomycin and neomycin using ribopolynucleotides containing two nucleotides in alternating sequence as templates. J. Mol. Biol. 1966;18:48–57. doi: 10.1016/s0022-2836(66)80075-x. [DOI] [PubMed] [Google Scholar]
- De Stasio E.A., Dahlberg A.E. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J. Mol. Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- Dorner S., Brunelle J.L., Sharma D., Green R. The hybrid state of tRNA is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M., Rojas A.M., Weiser J., Kurland C.G. How many EF-Tu molecules participate in aminoacyl-tRNA binding and peptide bond formation in Escherichia coli translation? J. Mol. Biol. 1990;211:739–749. doi: 10.1016/0022-2836(90)90074-V. [DOI] [PubMed] [Google Scholar]
- Eustice D.C., Wilhelm J.M. Fidelity of the eukaryotic codon–anticodon interaction: Interference by aminoglycoside antibiotics. Biochemistry. 1984a;23:1462–1467. doi: 10.1021/bi00302a019. [DOI] [PubMed] [Google Scholar]
- Eustice D.C., Wilhelm J.M. Mechanisms of action of aminoglycoside antibiotics in eukaryotic protein synthesis. Antimicrob. Agents Chemother. 1984b;26:53–60. doi: 10.1128/aac.26.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Agrawal R.K. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J., Gao H., Sengupta J., Gao N., Taylor D.J. The process of mRNA–tRNA translocation. Proc. Natl. Acad. Sci. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova L.P., Kostiashkina O.E., Koteliansky V.E., Rutkevich N.M., Spirin A.S. Factor-free (“nonenzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- Gonzáles A., Jiménez A., Vázquez D., Davies J.E., Schindler D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim. Biophys. Acta. 1978;521:459–469. doi: 10.1016/0005-2787(78)90287-3. [DOI] [PubMed] [Google Scholar]
- Gritz L., Davies J. Plasmid-encoded hygromycin B resistance: The sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae . Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D.S., Gold L. Selection of the initiator tRNA by Escherichia coli initiator factors. Genes & Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hermann T. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Hirokawa G., Kiel M.C., Muto A., Selmer M., Raj V.S., Liljas A., Igarashi K., Kaji H., Kaji A. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan L.H., Noller H.F. Intersubunit movement is required for ribosomal translocation. Proc. Natl. Acad. Sci. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc P.C., Kurland C.G. Multiple effects of kanamycin on translational accuracy. Mol. Gen. Genet. 1984;194:195–199. doi: 10.1007/BF00383516. [DOI] [PubMed] [Google Scholar]
- Jerinic O., Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kelley G.W., Olsen L.S. Critical tests of hygromycin B as an ascaricide of swine. Cornell Vet. 1960;50:60–65. [PubMed] [Google Scholar]
- Kimura S., Ito K., Miyagi T., Hiranuma T., Yoshioka K., Ozasa S., Matsukura M., Ikezawa M., Matsuo M., Takeshima Y., et al. A novel approach to identify Duchenne muscular dystrophy patients for aminoglycoside antibiotics therapy. Brain Dev. 2005;27:400–405. doi: 10.1016/j.braindev.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J., Jones T.A. Databases in protein crystallography. Acta Crystallogr. 1998;D54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- Konevega A.L., Fischer N., Semenkov Y.P., Stark H., Wintermeyer W., Rodnina M.V. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat. Struct. Mol. Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- Konno T., Takahashi T., Kurita D., Muto A., Himeno H. A minimum structure of aminoglycosides that causes an initiation shift of trans-translation. Nucleic Acids Res. 2004;32:4119–4126. doi: 10.1093/nar/gkh750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R.L., Bromer W.W. The isolation of a second antibiotic from Streptomyces hygroscopicus . J. Am. Chem. Soc. 1958;80:2714–2716. [Google Scholar]
- Manuvakhova M., Keeling K., Bedwell D.M. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6:1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi M., Nishimura T., Komai T., Tanaka N. Interaction of kanamycin and related antibiotics with the large subunit of ribosomes and the inhibition of translocation. Biochem. Biophys. Res. Commun. 1978;84:358–365. doi: 10.1016/0006-291x(78)90178-x. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Neuss N., Koch K.F., Molloy B.B., Day W., Huckstep L.L., Dorman D.E., Roberts J.D. Structure of hygromycin B, an antibiotic from Streptomyces hygroscopicus; The use of CMR. spectra in structure determination, I. Helv. Chim. Acta. 1970;53:2314–2319. doi: 10.1002/hlca.19700530846. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Sweet RM., Carter C.W. Jr, editors. Methods in enzymology. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Peske F., Savelsbergh A., Katunin V.I., Rodnina M.V., Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA–mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Pestka S. Studies on the formation of transfer ribonucleic acid–ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- Pfister P., Risch M., Brodersen D.E., Böttger E.C. Role of 16S rRNA helix 44 in ribosomal resistance to hygromycin B. Antimicrob. Agents Chemother. 2003;47:1496–1502. doi: 10.1128/AAC.47.5.1496-1502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J., Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- Qin Y., Polacek N., Vesper O., Staub E., Einfeldt E., Wilson D.N., Nierhaus K.H. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Rao R., Allen N.E., Hobbs J.N., Jr, Alborn W.E., Jr, Kirst H.A., Paschal J.W. Genetic and enzymatic basis of hygromycin B resistance in Escherichia coli . Antimicrob. Agents Chemother. 1983;24:689–695. doi: 10.1128/aac.24.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinberger H.J., Sternbach H., Nierhaus K.H. Three tRNA binding sites on Escherichia coli ribosomes. Proc. Natl. Acad. Sci. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Katunin V.I., Mohr D., Peske F., Rodnina M.V., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H.D. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M., Dunham C.M., Murphy IV F.V., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Senior K. Duchenne muscular dystrophy improved by gentamicin. Mol. Med. Today. 1999;5:461. doi: 10.1016/s1357-4310(99)01588-9. [DOI] [PubMed] [Google Scholar]
- Sharma D., Southworth D.R., Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S., Walker S.E., Fredrick K. Reverse translocation of tRNA in the ribosome. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler E.A., Blackburn E.H. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics paromomycin and hygromycin. J. Biol. Chem. 1985;260:6334–6340. [PubMed] [Google Scholar]
- Spirin A.S. Structural dynamic aspects of protein synthesis on ribosomes. Biochimie. 1987;69:949–956. doi: 10.1016/0300-9084(87)90228-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J.A. Improving on nature: Antibiotics that target the ribosome. Curr. Opin. Microbiol. 2005;8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Tai P.-C., Davis B.D. Triphasic concentration effects of gentamicin on activity and misreading in protein synthesis. Biochemistry. 1979;18:193–198. doi: 10.1021/bi00568a029. [DOI] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Watson J.D. The synthesis of proteins upon ribosomes. Bull. Soc. Chim. Biol. (Paris) 1964;46:1399–1425. [PubMed] [Google Scholar]
- Wettstein F.O., Noll H. Binding of transfer ribonucleic acid to ribosomes engaged in protein synthesis: Number and properties of ribosomal binding sites. J. Mol. Biol. 1965;11:35–53. doi: 10.1016/s0022-2836(65)80169-3. [DOI] [PubMed] [Google Scholar]
- Wilden B., Savelsbergh A., Rodnina M.V., Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc. Natl. Acad. Sci. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J.M., Pettitt S.E., Jessop J.J. Aminoglycoside antibiotics and eukaryotic protein synthesis: Structure–function relationships in the stimulation of misreading with a wheat embryo system. Biochemistry. 1978;17:1143–1149. doi: 10.1021/bi00600a001. [DOI] [PubMed] [Google Scholar]
- Youngman E.M., He S.L., Nikstad L.J., Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol. Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H.D., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zierhut G., Piepersberg W., Böck A. Comparative analysis of the effect of aminoglycosides on bacterial protein synthesis in vitro. Eur. J. Biochem. 1979;98:577–583. doi: 10.1111/j.1432-1033.1979.tb13219.x. [DOI] [PubMed] [Google Scholar]