Abstract
Pre-mRNA splicing is carried out by the spliceosome, which identifies exons and removes intervening introns. Alternative splicing in higher eukaryotes results in the generation of multiple protein isoforms from gene transcripts. The extensive alternative splicing observed implies a flexibility of the spliceosome to identify exons within a given pre-mRNA. To reach this flexibility, splice-site selection in higher eukaryotes has evolved to depend on multiple parameters such as splice-site strength, splicing regulators, the exon/intron architecture, and the process of pre-mRNA synthesis itself. RNA secondary structures have also been proposed to influence alternative splicing as stable RNA secondary structures that mask splice sites are expected to interfere with splice-site recognition. Using structural and functional conservation, we identified RNA structure elements within the human genome that associate with alternative splice-site selection. Their frequent involvement with alternative splicing demonstrates that RNA structure formation is an important mechanism regulating gene expression and disease.
Keywords: RNA secondary structure, alternative splicing, genome analysis, phylogenetic conservation
INTRODUCTION
Alternative splicing, the process by which multiple mRNA isoforms are made from a single pre-mRNA, occurs in >70% of human genes, thereby significantly enriching the proteomic diversity of higher eukaryotic organisms (Johnson et al. 2003). Many different parameters have been shown to influence the splicing decision. These include the strength of splice sites (Yeo and Burge 2004), the number of enhancers and silencers associated with the splicing unit (Black 2003), the intron/exon architecture (Fox-Walsh et al. 2005), and the process of transcription by RNA polymerase II (Kornblihtt 2005). As demonstrated in select cases, an additional factor that influences splice-site selection is local RNA secondary structure (Eperon et al. 1988; Clouet d'Orval et al. 1991; Graveley 2005; Hiller et al. 2007). Prompted by these observations, we set out to determine to what degree RNA secondary structure effected alternative splicing on a global level.
Single-stranded RNA is likely to adopt local secondary folds and tertiary interactions that may involve up to hundreds of nucleotides. Although pre-mRNAs typically are depicted in a linear fashion, we have to assume that higher-order structures exist that maintain a good portion of the double-stranded RNA. Depending on the thermodynamic stability, these structures may persist long enough to interfere or modulate splice-site recognition. In principle, RNA secondary structures can inhibit or activate spliceosome assembly because the recognition of splice sites, enhancers, and silencers typically depends upon interactions between protein factors and a single-stranded portion of the pre-mRNA (Maris et al. 2005; Hiller et al. 2007). Thus, local RNA structures can interfere with spliceosomal assembly if they conceal splice sites or enhancer binding sites within stable helices. On the other hand, local RNA structures also can promote spliceosomal assembly by masking splicing repressor binding sites (Hertel 2008).
The importance of RNA secondary structure in modulating splice-site selection has been documented in some cases. For example, two classes of conserved RNA elements have been identified in the Dscam exon 6 cluster, which contains 48 alternative exons; a common docking site and selector sequences unique to each exon 6 variant. Each selector sequence can base pair with the docking site to form a secondary structure, thereby activating and directing mutually exclusive exon pairing (Graveley 2005). An inhibitory role of RNA secondary structure was demonstrated for splice-site recognition of SMN2 exon 7. The formation of an RNA hairpin close to the 5′ splice site of SMN2 exon 7 interfered with its interaction with U1 snRNP, resulting in reduced exon inclusion levels (Singh et al. 2007). These examples support the idea that local RNA secondary structures play a more significant role in modulating splice-site recognition than perhaps currently appreciated.
To test the hypothesis that stable RNA structures that mask splice sites promote alternative splicing, we performed a genome-wide analysis to identify highly conserved RNA secondary structures that overlap the exon/intron junctions of internal exons. Within the human genome, we demonstrate that the potential to form stable RNA secondary structures highly correlates with alternative splicing. The structural analysis was extended further by evaluating the phylogenetic conservation of RNA secondary structures and the associated alternative splicing. Remarkably, up to 4% of conserved alternative splicing events were shown to associate with conserved RNA secondary structures encompassing at least one of the competing splice sites. We conclude that the presence of stable RNA secondary structures in human frequently mediates alternative splicing.
RESULTS
Stable RNA secondary structures associate with alternative splicing
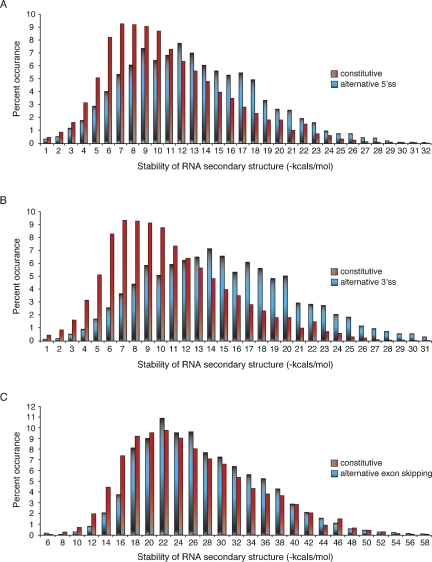
Using the UCSC Genome Browser, human sequence information was extracted for four different types of splicing events, constitutive splicing (13,426 exons), alternative 5′ splice-site selection (2728 exons), 3′ alternative splice-site selection (4179 exons), and exon inclusion/exclusion events (8385 exons) (Karolchik et al. 2008). Using MFOLD (Zuker 2003), all splice junctions then were analyzed for their ability to form RNA secondary structures within a 60-nucleotide (nt) window centered on splice sites. The distribution of RNA secondary structure stability within each category of alternatively spliced exons was then compared to the distribution of constitutively spliced exons (Fig. 1). Each category of splicing events contains a considerable fraction of exon/intron junctions that are unlikely to form RNA secondary structures (high free-energy values) and exon/intron junctions that may form very stable RNA secondary structures (low free-energy values). Remarkably, compared to constitutive exons, the RNA stability distribution of exon/intron junctions that undergo alternative 5′- or 3′-splice-site selection is significantly shifted toward more stable RNA structures (P values ≤ 2 × 10−94 and ≤ 4 × 10−131, respectively) (Fig. 1A,B). As expected, comparing two randomly selected pools of constitutively spliced exons show no difference between the two populations (Supplemental Fig. 1, P value ≤ 0.59). Given the enrichment of stable RNA secondary structures within human exons subject to alternative 5′- or 3′-splice-site selection (Supplemental Fig. 2A,B), we estimate that ∼15% of these EST-verified alternative splicing events strongly correlate with the potential to form stable RNA secondary structures. Using a similar discrimination argument, alternatively spliced exons are underrepresented at splice sites that do not have the ability to form stable RNA secondary structures (Fig. 1A,B; Supplemental Fig. 2A,B).
FIGURE 1.
Stability distributions of RNA secondary structures overlapping splice sites. The energetics of MFOLD-generated RNA secondary structures were determined for constitutive and alternative splice sites using a 60-nt widow. (A) The graph displays the relative stability distribution of RNA secondary structures overlapping alternative 5′ splice sites (blue bars) and constitutive splice sites (red bars). Free energies of RNA structure formation were binned in stability categories that increase by 1 kcal/mol. The x-axis displays increasing stability from left to right; the y-axis represents the number of splice sites within each stability bin. A two-tailed t-test was performed to show that the distribution profiles are significantly different (t = 20.7, df = 17446, P value ≤2 × 10−94). (B) Comparison of alternative 3′ splice sites (blue bars) and constitutive splice sites (red bars) (t = 24.6, df = 18,628, P value ≤4 × 10−131). (C) Comparison of alternative exon inclusion/exclusion events (blue bars) and constitutive exons (red bars). The folding energy sum of RNA structures formed around the 5′ and 3′ splice sites is shown here (t = 7.0, df = 21,782, P value ≤2 × 10−12). Analysis of either the 5′ or 3′ splice site alone results in essentially the same profile overlap (data not shown).
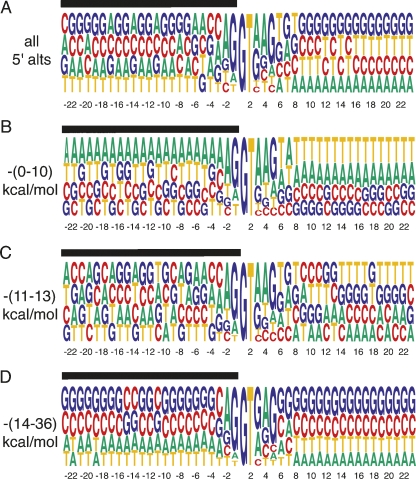
Based on these observations it is expected that splice sites capable of forming strong secondary structures are preferentially flanked by nucleotides that allow high energy base pairing. To test this prediction, the nucleotide representation around all alternative 5′ splice sites was investigated. No significant sequence bias was observed when all alternative 5′ splice sites were queried (Fig. 2A). However, when alternative 5′ splice sites associated with a high potential for secondary structure formation (ΔG between −14 and −34 kcal/mol) were analyzed a noticeable bias for GC base pairing could be discerned from the sequence alignments (Fig. 2D). Similarly, alternative splice sites with low secondary structure potential display reduced base pairing potential as illustrated by the preference for A and T (Fig. 2B). Together, these observations suggest that a significant fraction of alternative splice-site choice is mediated through RNA secondary structures.
FIGURE 2.
Alignment of alternative 5′ splice sites +23 to −23 nt from the splice site. (A) A nucleotide frequency matrix constructed from all 5′ alternative splice sites. (B) Alternative 5′ splice sites with free energies between (0–10)-kcal/mol favor low stability RNA structures as indicated by the preference for A and T. (C) 5′ splice sites with folding free energies between (11–13)-kcal/mol shows no discernable sequence preference. (D) Alternative 5′ splice sites with free energies between (14–36)-kcal/mol favor the formation of strong RNA secondary structures as indicated by the preference for G and C. The x-axis denotes the distance from the splice site, the black bar the exon.
The RNA structure analysis of all exon inclusion/exclusion events did not result in visual differences as striking as those observed for alternative 5′- and 3′-splice-site events (Fig. 1C). While a statistically significant variation was determined between alternative exons and constitutive exons (P value ≤ × 10−12), the magnitude of this difference does not permit strong discrimination between alternative and constitutive exon inclusion events (Supplemental Fig. 2C). Interestingly, exons exceeding the most common exon length (exons longer than 200 nt) display a much more pronounced bias for stable RNA secondary structures (Supplemental Fig. 3), indicating that the recognition of longer exons may be more frequently regulated through RNA structure formation. These results suggest that the mechanisms of alternative exon inclusion/exclusion in human vary with exon size.
Identification of phylogenetically conserved RNA structures associated with alternative splicing
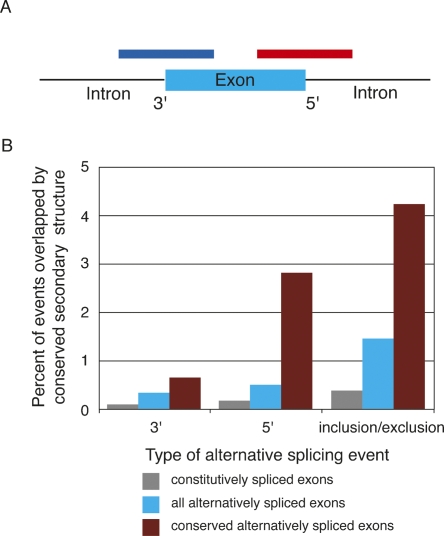
One limitation of the MFOLD minimization approach is that calculated RNA secondary structures are hypothetical and derived at simplified conditions. While invaluable for determining the potential of RNA to form a secondary structure, MFOLD structures do not always represent in vivo structures where RNAs are heavily associated with RNA binding proteins and RNA processing factors. To extend our correlation between alternative splicing and RNA secondary structure formation, we investigated whether the secondary structures identified are conserved phylogenetically, thus implying functionality. To do so, we took advantage of the RNA secondary structure prediction program Evofold, a multiple-genome alignment program that uses the characteristic differences of the substitution process in stem-pairing and unpaired regions to identify conserved RNA structures (Pedersen et al. 2006). From the sequence alignment of eight species ranging from human to zebrafish, Evofold generated 47,511 conserved secondary structures. The human coordinates for these structures were downloaded from the UCSC genome table browser (http://genome.ucsc.edu/cgi-bin/hgTables) and annotated. For each conserved structure, we then asked how often the coordinates span the splice sites of constitutive exons, alternative exons, alternative 5′ splice sites, or alternative 3′ splice sites (Fig. 3A).
FIGURE 3.
Conserved RNA secondary structure correlation with alternative splicing. (A) Location of Evofold secondary structures spanning the 5′ (red) or 3′ (blue) splice sites of an exon. (B) Bar graph displaying the relative number of splice sites overlapped by Evofold-predicted secondary structures for constitutively spliced exons (gray), alternatively spliced exons (blue), and conserved alternatively spliced exons (maroon). The observed differences in Evofold structure representation in each alternative splicing category is highly significant with P values ranging from < 0.004 (conserved 3′ alt events) to < 2.2 × 10−16 (conserved cassette events). A list of Evofold coordinates and the genes they overlap can be found in Supplemental Tables 1–3.
Strikingly, when compared to constitutive exons, alternatively spliced exons display a fourfold enrichment of Evofold structures overlaying splice sites (Fig. 3B). These results demonstrate that up to 1.5% of all analyzed alternative splicing events correlate with the presence of conserved RNA secondary structures. The high level of structure conservation further suggests that the presence of the stable RNA secondary structure is functionally important, possibly permitting regulation of the alternative splicing event. This interpretation predicts that the alternative splicing event associated with a conserved RNA secondary structure may also be conserved between species. To test this hypothesis, we compiled a list of alternative splicing events conserved between humans and zebrafish from the Alternative Splicing Annotation Project database (http://www.bioinformatics.ucla.edu/ASAP2/), resulting in 230 conserved 5′ alternative splicing events, 306 conserved constitutive events, and 992 conserved exon inclusion/exclusion events (Kim et al. 2007). Consistent with the interpretation that conserved RNA secondary structures are mediating alternative splicing, the incidence of Evofold structures that mask splice sites of conserved alternative splicing events is increased 15-fold when compared to constitutive exons (Fig. 3B). Lists of all Evofold structures that overlap splice sites of conserved alternative splicing events are given in Supplemental Tables 1–3. We conclude that a significant fraction of phylogenetically conserved alternative splicing events are mediated through local RNA secondary structures that mask at least one of the competing splice sites.
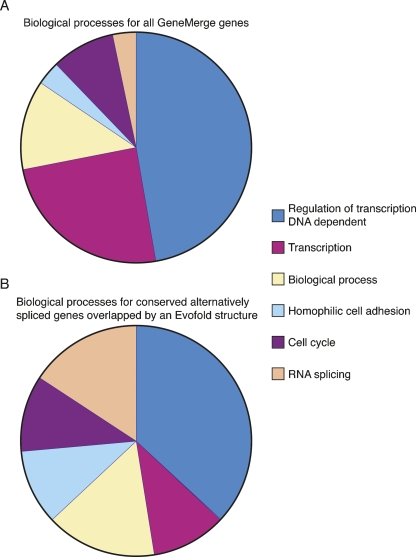
To investigate whether the conserved alternative splicing events that associate with an Evofold structure are enriched for certain biological processes, a GO ontology analysis was performed using the gene entries listed in Supplemental Tables 1–3 (Beissbarth 2006). Compared to the unbiased control group, a statistically significant enrichment of genes that are associated with RNA splicing (false discovery rate corrected P value = 0.0024) and homophilic cell adhesion (false discovery rate corrected P value = 0.038) are observed within the group of conserved alternative splicing events that are overlapped by an Evofold structure (Fig. 4). Ironically, among this group are components of the spliceosome itself. These observations suggest that some evolutionarily conserved alternative splicing events associated with Evofold structures may modulate the efficiency of the spliceosome, thus potentially influencing pre-mRNA splicing on a more global level.
FIGURE 4.
GO ontology analysis of conserved alternative splicing events associated with Evofold structures. Using Supplemental Tables 1–3, a GO ontology analysis was performed for (A) all gene ontology entries and (B) all conserved alternative splicing events associated with an Evofold structure. Each data set was evaluated for the fractional representation of biological processes identified within the group of conserved alternative splicing events associated with an Evofold structure (B). Biological processes are color coded and are defined to the right of the graphs.
DISCUSSION
The genome-wide analysis described here demonstrates that a significant fraction of alternative splicing associates with RNA secondary structure formation. Using an unbiased approach we showed that alternative 5′- and 3′-splice-site events are enriched for their ability to mask splice sites in stable secondary structures. This result is highly significant when evaluated statistically (P values < 10−6) and when compared to the control group. Furthermore, an identical analysis of alternative exon inclusion did not result in a striking visual separation between constitutive and alternative events as was observed for alternative 5′- and 3′-splice-site events, suggesting that not all alternative splicing events are subject to similar mechanisms of induction. To provide support for the argument that the putative secondary structures have functional significance, we took advantage of Evofold, an RNA folding algorithm that combines phylogeny between eight species and RNA structure formation (Pedersen et al. 2006). Evofold identifies putative RNA structures conserved across several species. Importantly, the algorithm greatly enhances the folding potential of a putative RNA structure if the genome alignment identifies compensatory base changes that preserve it. Given these criteria, Evofold preferentially identifies RNA structures for which primary sequence differences exist between species and that have been tested by base changes preserving the secondary structure. When combining these stringent selection criteria with conserved alternative splicing we were able to provide the estimate that up to 4% of alternative splicing events are likely to be modulated through RNA secondary structure (Fig. 3).
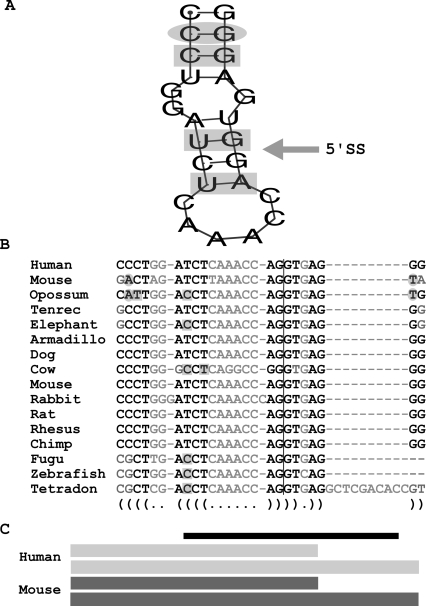
The principle of this approach is illustrated in Figure 5. The protein retinoblastoma 1 (RB1)-inducible coiled-coil 1 (RB1CC1) undergoes frame-preserving alternative 5′ splice-site selection. RB1CC1 has recently been identified as a key nuclear regulator of the tumor-suppressor gene RB1, and truncation mutations in RB1CC1 have frequently been associated with breast cancer (Chano et al. 2002). Alternative 5′-splice-site selection of this classical tumor-suppressor gene is conserved between humans and six other species. Evofold predicts the formation of a conserved hairpin loop that masks one of the alternative 5′ splice sites (Fig. 5A). Importantly, the comparison of the RNA secondary structures among the species evaluated indicates that compensatory base changes have maintained the integrity of the hairpin stem while accepting changes within the primary sequence (Fig. 5). The 5′ splice site contained within the conserved RNA secondary structure (CAG/gugagg) competes with the alternative downstream splice site GTG/gtaagt. Based on its ability to pair with U1 snRNA and other computational predictions, the upstream 5′ splice site is the stronger splice site (Yeo and Burge 2004). However, EST analysis demonstrates that the downstream 5′ splice site is used slightly more often that the upstream 5′ splice site. These observations are in agreement with the proposal that the conserved RNA secondary structure promotes alternative 5′-splice-site selection by interfering with the recognition of the stronger upstream 5′ splice site.
FIGURE 5.
Representation of an Evofold secondary structure spanning the 5′ splice site of a conserved alternatively splicing event within RB1CC1. The RNA energy minimization program MFOLD was used to generate a representation of the folded conserved secondary structure (A). Base pairs shaded by a gray oval indicate a double substitution that maintains complementarity while base pairs shaded by a gray rectangle indicate single substitutions that maintain base pairing. (B) The nucleotide alignment of the putative structure is shown for 16 vertebrates. Bases in black indicate that nucleotides participate in base pairing, bases in gray do not, bases within gray circles represent nucleotides involved in a double substitution that maintains base pairing, and bases within gray squares represent single base substitutions that maintain base pairing. The 5′ splice site is indicated by the vertical line. (C) Cartoon showing the resulting alternatively spliced exons in humans (light gray) and the mouse (blue gray) with one of the splice sites overlapped by a secondary structure (black bar). ESTs representing these two isoforms are conserved in six other species. The competing 5′ splice site lies outside of the conserved secondary structure.
A similar analysis of the calsyntenin gene serves as an example for RNA secondary structure associated exon skipping (Supplemental Fig. 5). Calsyntenins (CLSTN1) are type-1 neuronal transmembrane proteins of the cadherin superfamily found in the postsynaptic membrane of the adult brain (Vogt et al. 2001). The alternatively spliced exon is unique in that it is only 30-nt long. Significantly, an Evofold structure spans the entire exon masking both splice junctions. As predicted from the secondary structure analysis, the EST representation of calsyntenin demonstrates that the alternative exon is approximately seven times more likely to be skipped than included into the final mRNA.
Taken together, our analysis strongly supports the notion that RNA secondary structures frequently modulate pre-mRNA splicing in human and suggests that they play a more significant role in modulating splice-site recognition than currently appreciated. The influence of RNA secondary structure on alternative splicing is likely an underestimate because our analysis concentrated on identifying local RNA structures that overlap splice sites, neglecting the possibility that local RNA secondary structures could interfere with the recognition of splicing repressors and silencers. It is unclear whether the presence of a stable RNA secondary structure induces stochastic or regulated alternative splicing. However, given the tight association of RNA synthesis and pre-mRNA processing, it is likely that minor changes in the kinetics of RNA transcription influence the formation of local RNA secondary structures, thus inducing alternative splicing (Eperon et al. 1988). Regardless of the mechanisms of induction, the strong structure/function correlation demonstrated here suggests that RNA secondary structure formation is an important mechanism for gene regulation.
MATERIALS AND METHODS
Constructing a data set of alternative and constitutively spliced genes
The UCSC Genome Browser was used as the source for sequence and alternative splicing information. For the analysis of the human genome, genomic coordinates for all alternative 5′, 3′, and cassette splicing events were downloaded from the UCSC Genome Browser alternative splicing track assembly hg18 (Sugnet) (Karolchik et al. 2008). For each of the 2642 alternative 3′ splicing events (5284 splice sites) >7 nt apart, 2051 alternative 5′ splicing events (4102 spice sites) >4 nt apart, 8385 skipping events (8385 5′ + 8385 3′ splice sites), and 2470 skipping events (2470 5′ + 2470 3′ splice sites) with exons >200 nt, 30 nt were added upstream of and downstream from each splice-site coordinate and the corresponding sequence was extracted from the UCSC Table Browser. A list of internal 13,523 constitutively spliced exons was compiled by taking the overlapping subset of all 66,518 exons of unique isoforms from the UCSC Genome Browser and a set of 43,775 constitutively spliced exons provided by Jim Kent at the UCSC genome browser (personal communication). For each of the 13,523 constitutively spiced exons, 30 nucleotides were added upstream and downstream of both the 5′- and 3′-splice-site coordinates and the corresponding sequence was extracted from the UCSC Table Browser.
Conserved alternative splicing events
The ASAP2 database http://www.bioinformatics.ucla.edu/ASAP2/ was used as a source for conserved 5′, 3′, and cassette alternative splicing events (Kim et al. 2007). MYSQL tables were downloaded from the ASAP2 and SQL queries were used to extract alternative 5′, 3′, and cassette splice sites conserved in humans and at least one of 14 other animal species. These queries retrieved 230 conserved 5′ alternate splicing events, 306 conserved alternate 3′ splicing events, and 992 conserved cassette events.
Determining free energy of sequence flanking splice sites
The RNA free energy minimization program RNAfold was downloaded locally from http://www.tbi.univie.ac.at/RNA/, and used to find the minimum free energy of 60-mers (Zuker 2003). A Perl script was used as a wrapper around the RNAfold program to iteratively parse and send the sequences to the MFOLD algorithm.
Conserved secondary structure
Evofold is a comparative method for identifying functional RNA structures in multiple-sequence alignments. An eight-way human-referenced genomic vertebrate-alignment (which includes human, chimpanzee, mouse, rat, dog, chicken, pufferfish, and zebrafish) was used to predict 47,511 Evofold sequences (Pedersen et al. 2006). The hg18 bed format coordinates were downloaded from the UCSC Table Browser. A Perl script was used to find Evofold sequences that overlapped splice sites.
Gene ontology
To determine if certain biological processes are overrepresented in our data set we compiled two gene lists. List 1 contains Uniprot IDs corresponding to 58 genes with a conserved alternative exon cassette event, a conserved alternative 5′-splice-site selection event, or a conserved alternative 3′-splice-site selection event overlapped by an Evofold structure that was downloaded from the UCSC Table Browser. A second list of all GO Biological Process terms for all 25,098 genes was obtained from the Homo sapiens Biological Process data set (list 2) provided by GeneMerge (http://genemerge.bioteam.net/). List 1 was pruned to contain only Uniprot Ids that have biological process ontologies contained in the GeneMerge list. This resulted in a list of 31 Uniprot Ids (list 1a) all for unique gene loci from list 1. The relative representation of GO biological Processes of Uniprot ids from list 1a was compared to list 2 using the stand-alone PERL script GeneMerge http://genemerge.bioteam.net/. Because Gene Ontology is hierarchical, and because genes may have duplicate biological processes, many genes can appear more than once in an ontology. To more clearly represent the data and to avoid skewing the analysis, each Uniprot id is allowed to appear only once in our analysis; furthermore, only ontologies with two or more biological processes are represented. For each ontology in list 1a, we compared the number of Uniprot IDs to the normalized number of Uniprot ontologies in list 2.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by NIH Grants GM62287 (K.J.H.) and GM079413 (K.J.H.). P.J.S. was supported by a Biomedical Informatics Training Grant. We thank members of the Hertel and Baldi Laboratories for discussions and critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1069408.
REFERENCES
- Beissbarth T. Interpreting experimental results using gene ontologies. Methods Enzymol. 2006;411:340–352. doi: 10.1016/S0076-6879(06)11018-6. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Chano T., Kontani K., Teramoto K., Okabe H., Ikegawa S. Truncating mutations of RB1CC1 in human breast cancer. Nat. Genet. 2002;31:285–288. doi: 10.1038/ng911. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Eperon L.P., Graham I.R., Griffiths A.D., Eperon I.C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh K.L., Dou Y., Lam B.J., Hung S.P., Baldi P.F., Hertel K.J. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel K.J. Combinatorial control of exon recognition. J. Biol. Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- Hiller M., Zhang Z., Backofen R., Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Alekseyenko A.V., Roy M., Lee C. The ASAP II database: Analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–D98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.N., Singh R.N., Androphy E.J. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt L., Schrimpf S.P., Meskenaite V., Frischknecht R., Kinter J., Leone D.P., Ziegler U., Sonderegger P. Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol. Cell. Neurosci. 2001;17:151–166. doi: 10.1006/mcne.2000.0937. [DOI] [PubMed] [Google Scholar]
- Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]