Abstract
We previously proposed that mutations in the connection subdomain (cn) of HIV-1 reverse transcriptase increase AZT resistance by altering the balance between nucleotide excision and template RNA degradation. To test the predictions of this model, we analyzed the effects of previously identified cn mutations in combination with thymidine analog mutations (D67N, K70R, T215Y, and K219Q) on in vitro RNase H activity and AZT monophosphate (AZTMP) excision. We found that cn mutations G335C/D, N348I, A360I/V, V365I, and A376S decreased primary and secondary RNase H cleavages. The patient-derived cns increased ATP- and PPi-mediated AZTMP excision on an RNA template compared with a DNA template. One of 5 cns caused an increase in ATP-mediated AZTMP excision on a DNA template, whereas three cns showed a higher ratio of ATP- to PPi-mediated excision, indicating that some cn mutations also affect excision on a DNA substrate. Overall, the results strongly support the model that cn mutations increase AZT resistance by reducing template RNA degradation, thereby providing additional time for RT to excise AZTMP.
Keywords: drug resistance, RNase H, TAMs, NRTI, NNRTI
To date, >33 million people worldwide are infected with HIV-1, with an estimated 6,800 new HIV-1 infections occurring daily (www.who.int). This worldwide pandemic has provided great impetus to find drug treatments that are effective against HIV-1. Most standard drug therapies include inhibitors of reverse transcriptase (RT), an enzyme necessary for HIV-1 to complete its life cycle. Reverse transcriptase is a multifunctional enzyme containing a DNA polymerase, which can copy either RNA or DNA templates, and an RNase H, which degrades RNA only when it is in a heteroduplex with DNA (1). These activities allow HIV-1 to copy the HIV-1 genomic RNA into double-stranded DNA.
High rates of HIV-1 production (2–4) and high levels of genetic variation in viral populations (5–7) lead to the emergence of drug-resistant variants (8). Mutations in RT that confer resistance to nucleoside reverse transcriptase inhibitors (NRTIs) either decrease the incorporation of the NRTI (discrimination) or increase ATP-mediated excision of the incorporated NRTI (9–11). Resistance to AZT is associated with mutations in RT (M41L, D67N, K70R, L210W, T215F/Y, and K219Q/E/N); these mutations also confer much lower levels of resistance to some other NRTIs (12, 13). Previous characterization of drug resistance mutations in RT has focused primarily on the N-terminal 300 aa of RT; the connection subdomain (cn), which consists of amino acids 312–425 (also called the connection domain), as well as the RNase H domain (amino acids 426–560) are routinely excluded from the analysis of drug resistance mutations.
We recently showed that mutations in the cn (E312Q, G335C/D, N348I, A360I/V, V365I, and A376S) are selected in response to antiviral therapy [see supplemental Table 3 in Nikolenko et al. (14)] and that these mutations can significantly enhance AZT resistance (AZT-R) in the presence of thymidine analog mutations (TAMs: D67N, K70R, T215Y, and K219Q) (14). We also showed that mutations in the RNase H primer grip residues in the cn and RNase H domain increased the level of AZT-R (15). Other studies have also confirmed the role of N348I and G333D in NRTI and non-NRTI resistance (16–19). Furthermore, A371V + Q509L mutations were selected upon passage of HIV-1 in vitro in the presence of AZT and shown to increase AZT-R (20).
The mechanism by which cn mutations enhance AZT-R has not been determined. We previously proposed a model in which cn mutations alter the balance between polymerization and RNase H activity, and this imbalance leads to an overall increase in drug resistance (14, 21). We proposed that reducing RNase H activity stabilizes the RNA/DNA duplex, giving RT more time to excise the AZT from the terminated primer. The results of these studies strongly support the predictions of this model and show that mutations in the cn decrease RNase H cleavages and increase ATP- and PPi-mediated AZT monophosphate (AZTMP) excision on RNA templates compared with DNA templates.
Results
Characterization of cn Mutations.
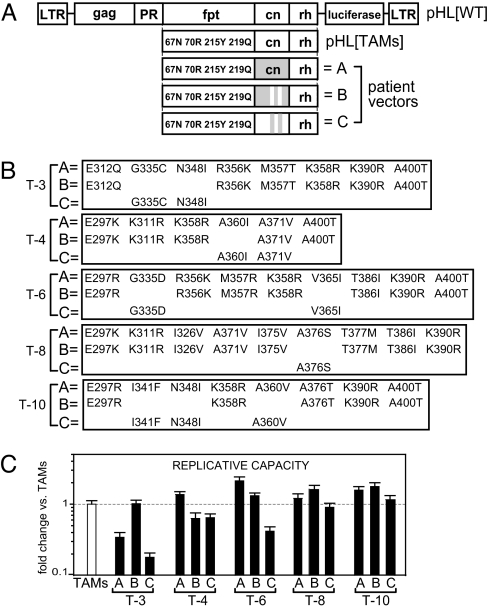
We sought to characterize the biochemical properties of RTs containing the cn mutations G335C/D, N348I, A360I/V, V365I, and A376S to determine their role in AZT-R. Previously, using an HIV-1 vector (pHL[TAMs]) with RT-containing TAMs (D67N, K70R, T215Y, and K219Q), we constructed viral vectors by using polymerase chain reaction products obtained from patients who had failed NRTI-containing therapy (patients T-3, T-4, T-5, T-6, T-8, and T-10). These products contained either the entire cn isolated from patients (Fig. 1 A and B; “A” vectors) or the cn isolated from patients in whom the cn-derived AZT-R mutations were reverted back to WT (Fig. 1 A and B; “B” vectors) (14). We also generated viral vectors in which the cn-derived AZT-R mutations present in the patient cns were introduced into a WT cn in the context of pHL[TAMs] (Fig. 1 A and B; “C” vectors).
Fig. 1.
Schematic representation of HIV-1-based constructs and their replicative capacity. (A) Structure of pHL[WT] and pHL[TAMs]. Three vectors were constructed with viruses from each patient: the A vectors contain the entire cn (gray box); the B vectors contain the previously identified AZT resistance mutations in cn reverted back to WT amino acids (gray box with white stripes); the C vectors contain the previously identified AZT resistance mutations from the cn added back into a WT cn (white box with gray stripes). (B) cn sequences for vectors A– C from viruses from each treatment-experienced patient (T-3, T-4, T-6, T-8, and T-10). Vector A lists all patient-derived cn mutations that were different from pNL43. Vector B lists the mutations remaining after reversion of the AZT resistance mutations back to WT amino acids. Vector C lists the cn mutations obtained from patient-derived viruses that were added back into a WT cn. Note: Only the A360I substitution in T-4 was reverted in the B vector; when both A360I and A371V substitutions were reverted in the T-4 A vector, RNase H activity was reduced in a similar manner (data not shown). (C) Replicative capacities relative to the TAMs control (dotted line) are shown for each virus. Error bars represent SEM from at least two independent experiments. LTR, long terminal repeat; fpt, fingers, palm, and thumb region (amino acids 1–287); PR, protease; rh, RNase H domain. Note: Replicative capacity for the A vectors was previously described (14).
Replicative capacities (Fig. 1C) for each vector virus were determined and normalized to the control TAMs virus. T-3A and T-3C showed significantly reduced levels of replicative capacity (3- and 5.5-fold, respectively; P < 0.007) compared with T-3B and the TAMs virus, suggesting that the cn mutations present in both T-3A and T-3C (G335C and N348I) negatively affected viral fitness. For the remaining vectors, replicative capacities were within 2-fold of a vector that contained only the TAMs.
cn Mutations in the Context of TAMs Increase AZT-R.
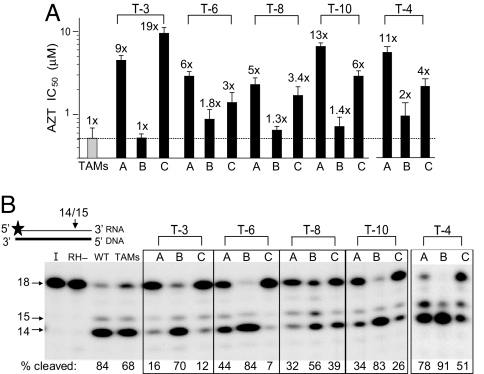
We previously reported the fold changes in AZT-R (14) associated with vectors A and B (see Fig. 1). Single-cycle drug susceptibility assays were also performed to determine AZT IC50s for the C vectors (Fig. 2A). The AZT IC50 was 0.52 ± 0.17 μM for the TAMs virus, a 10-fold increase in resistance relative to WT (0.05 ± 0.01 μM). All of the B vectors had AZT IC50s that were within 2-fold of the IC50 of the TAMs virus, indicating that the specific cn mutations were largely responsible for the increase in resistance seen in the A vectors. Furthermore, like the A vectors, the C vectors exhibited high levels of AZT-R, confirming that, in the presence of TAMs, the cn mutations G335C/D, N348I, A360I/V, V365I, and A376S were primarily responsible for the increased AZT-R.
Fig. 2.
Association of AZT-R with RNase H primary cleavage for cn mutations. (A) AZT IC50s were measured for each virus and compared with the TAMs control (dotted line). Fold changes in AZT-R compared with the TAMs control are shown above each bar. Error bars represent the SEM from two to five independent experiments. Note: AZT IC50s for vectors A and B were previously reported (14). (B) Primary RNase H cleavages were assayed on a substrate containing a 32P-labeled (star) 18-nt RNA (thin black line) annealed to a complementary 18-nt DNA (thick black line). The percentages of substrate cleaved (14- and 15-nt bands) are listed below each lane in the autoradiogram and represent the average of two independent experiments. I, input control; RH−, RNase H mutant D498N.
Prevalence of cn Mutations in Viruses from Treatment-Experienced Patients.
The Stanford University HIV Drug Resistance Database contains 1667 RT sequences that extend to amino acid 312 and 972 sequences that extend to amino acid 376 [supporting information (SI) Table S1]. These sequences were used to analyze the prevalence of the cn mutations E312Q, G335C/D, N348I, A360I/V, V365I, and A376S in viruses present in treatment-experienced patients and their association with TAMs. The overall frequency of the sequences containing one or more of the eight cn mutations was 21% (217/972), which increased to 27% in viral sequences that contained one or more TAMs (147/537; P < 0.02). Among the sequences containing one of these cn mutations, 68% contained one or more TAMs (147/217; P < 1 × 10−7). The association of N348I, A360V, or V365I with TAMs was highly significant (P < 0.01, P < 1 × 10−6, and P < 0.035, respectively). These data strongly suggest that there is a positive association between TAMs and cn mutations.
cn Mutations Associated with AZT-R Reduce RNase H Cleavage.
One central prediction of our model is that cn mutations increase AZT-R by decreasing RNase H activity. To test this prediction, we first asked whether the cn mutations affect primary RNase H cleavages by using a previously described 18-nt RNA/DNA substrate (22). Primary RNase H cleavages on this template are expected to occur at the −3 and −4 position from the 3′ end of the RNA. The WT and TAMs RTs cleaved 84% and 68% of the RNA substrate to a 15- and 14-nt fragment, respectively (Fig. 2B). For each of the virion-derived RTs (from vectors A–C), there was an inverse association between the efficiency of the RNase H cleavage and AZT-R: All A and C vectors, which exhibited higher AZT-R levels than the B vectors, had lower levels of RNase H cleavage compared with the corresponding B vectors.
To verify that the reductions in primary RNase H cleavage were not influenced by the specific combination of TAMs in the vector, we replaced the region of RT containing TAMs with that present in the patient-derived RT. Each patient-derived RT contained a unique set of mutations, including a different combination of TAMs [sequences listed in Nikolenko et al. (14)]. RNase H cleavage analysis on the 18-nt RNA/DNA substrate showed that virion-derived RTs containing the fingers, palm, and thumb region and the cn from the same patient showed 2–3 times less primary RNase H cleavage than the corresponding virion-derived RTs that did not have the patient-derived cn (data not shown). Therefore, the reductions in the primary RNase H cleavages were associated with mutations in the cn and not with other mutations in RT.
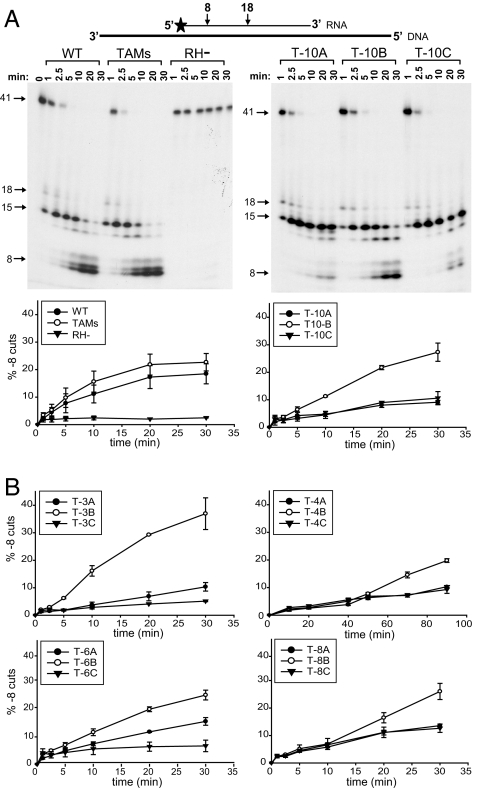
To further characterize the effects of the cn mutations on RNase H cleavage, the patient-derived virion RTs were assayed with an RNA/DNA substrate that was previously used to measure RNase H secondary (−8) cleavages (23). In these assays, the primary RNase H (−18) cleavage was too rapid to allow accurate quantitation. In a 30-min time course virion-derived WT and TAMs RTs cleaved 23% and 18%, respectively, of the RNA/DNA substrate to the −8 product (Fig. 3A). As expected, RNase H defective mutant D498N showed little or no secondary cleavage at 30 min (≈2%). All of the mutant RTs from the A and C vectors reduced secondary cleavages compared with the RTs from the B vectors (Fig. 3B), a result that is consistent with the results of the assay of the primary cleavage (see Fig. 2B). Thus, the vectors with higher levels of AZT-R exhibited lower levels of primary and secondary RNase H cleavages.
Fig. 3.
RNase H secondary cleavages. (A) Secondary (−8 cut) RNase H cleavages were assayed on a substrate containing a 32P-labeled (star) 41-nt RNA primer (thin black line) annealed to a 77-nt DNA template (thick black line). The percentages of secondary cleavages over time are graphically shown below each autoradiogram; error bars represent the SEM from at least two independent experiments. (B) The percentage of secondary cleavages for T-3, T-4, T-6, and T-8. RH−, RNase H mutant D498N.
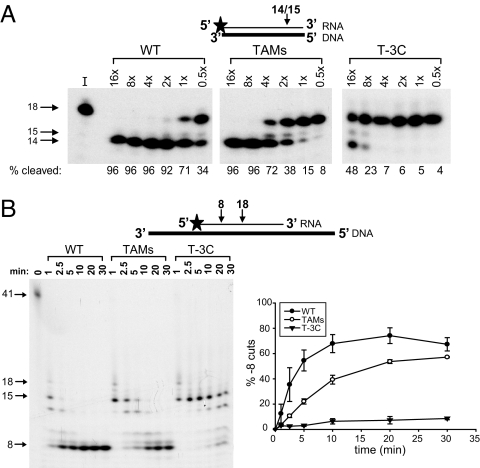
We tested whether the reductions in RNase H cleavages seen with virion-derived RT would also be seen with purified recombinant T-3C RT. Purified WT and TAMs RT cleaved 96% and 72% of the 18-nt RNA/DNA substrate, respectively, if the RNase H assays included equivalent amounts of DNA polymerase activity (Fig. 4A; compare lanes labeled 4×). Purified TAMs RT showed a reduction in primary RNase H cleavages that was similar to the results with virion-derived TAMs RT (see Fig. 2B). Much higher amounts of purified T-3C RT (16×) were required to achieve at least 48% primary RNase H cleavage. Furthermore, the T-3C RT was inefficient in making secondary RNase H cleavages, producing only 9% of the −8 product after 30 min, compared with WT (67%) and TAMs (57%) RT (Fig. 4B).
Fig. 4.
Purified RT RNase H primary and secondary cleavages. (A) Primary RNase H cleavages from purified WT, TAMs, and T-3C RTs were assayed on a substrate containing a 32P-labeled (star) 18-nt RNA (thin black line) annealed to a complementary 18-nt DNA (thick black line). The percentage of substrate cleaved (14- and 15-nt bands) with serial dilutions of the purified RTs (16× to 0.5×) is listed below each lane in the autoradiogram and represents the average of two independent experiments. (B) Secondary (−8 cut) RNase H cleavages for purified WT, TAMs, and T-3C RTs were assayed over time on a substrate containing a 32P-labeled (star) 41-nt RNA primer (thin black line) annealed to a 77-nt DNA template (thick black line). The percentages of secondary cleavages are graphically shown below each autoradiogram; error bars represent the SEM from two independent experiments. I, input control.
cn Mutations Enhance ATP- and PPi-Mediated Excision and Extension of AZTMP Terminated Primers.
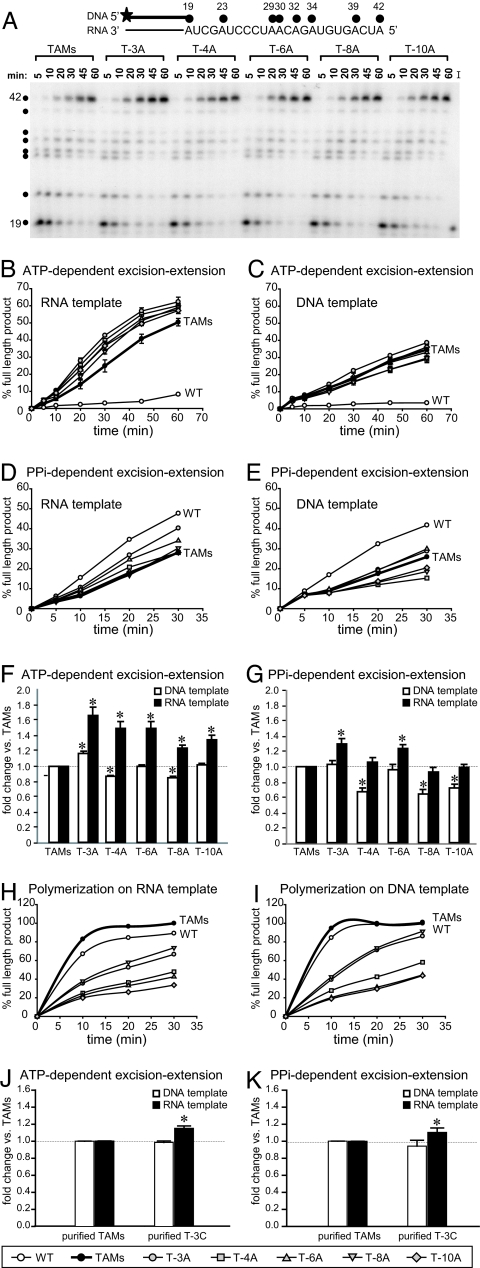
A key prediction of our model is that decreased levels of RNase H activity will result in increased levels of AZTMP excision from a terminated primer on an RNA template. We tested this prediction by carrying out ATP-mediated AZTMP excision–extension assays by using virion-derived RTs from the A vectors on an RNA template (Fig. 5 A and B) and on a DNA template (Fig. 5C). To determine whether the effects of cn mutations on AZTMP excision are specific to ATP-mediated AZTMP excision, we also carried out PPi-mediated excision–extension assays on the same RNA (Fig. 5D) and DNA (Fig. 5E) template. The results of the ATP- and PPi-dependent excision–extension assays obtained for all patient-derived RTs were quantified and normalized to the TAMs RT (Fig. 5 F and G, respectively).
Fig. 5.
AZTMP excision–extension assays. (A) Representative autoradiogram of the kinetics of DNA synthesis catalyzed by patient-derived cn virion-derived RTs (A vectors) evaluated by using a blocked primer (19mer) on an RNA template. The 32P-labeled (star) blocked DNA primer (thick black line) bound to the 42-nt RNA template is shown above the autoradiogram, with black dots representing sites of AZTTP incorporation. I, input control. (B and C) Efficiency of ATP-mediated excision–extension on an RNA template (B) or a DNA template (C) in the presence of AZTTP and 3.3 mM ATP. (D and E) Efficiency of PPi-mediated excision–extension on an RNA template (D) or a DNA template (E) in the presence of AZTTP and 100 μM PPi. (F and G) Comparison of ATP-mediated (F) and PPi-mediated AZTMP excision–extension (G) on RNA and DNA templates. The percentages of full-length products for the TAMs RT were set to 1.0 for 20-, 30-, 45-, and 60-min time points, and the fold change in the percentage full-length products obtained for each A vector are represented as an average value relative to the TAMs control. (H and I) Polymerization rates on the RNA (H) and DNA (I) templates were normalized to TAMs control (TAMs full-length product at 30 min was 100%). (J and K) Comparison of ATP-mediated AZTMP excision–extension (J) and PPi-mediated AZTMP excision–extension (K) on RNA and DNA templates for purified TAMs RT and purified T-3C RT. Analyses were performed as described for F and G. Error bars represent the SEM from at least two independent experiments. Asterisks represent statistically significant differences from the TAMs control (P < 0.006).
The results showed that the virion-derived RTs from all five A vectors were more efficient at ATP-mediated AZTMP excision and extended the primer to generate more full-length DNA than the TAMs RT on an RNA template (Fig. 5F; P < 0.002). In contrast, the RTs from the A vectors had minor effects on AZTMP excision and extension on a DNA template compared with the TAMs RT. All 5 virion-derived RTs exhibited more efficient ATP-mediated AZTMP excision–extension on the RNA template compared with the DNA template (Fig. 5F). By contrast, only the T-3A RT exhibited a higher level of AZTMP excision on the DNA template compared with TAMs (Fig. 5F; P < 0.0005), indicating that cn mutations can directly increase ATP-mediated AZTMP excision through a mechanism not mediated by RNA degradation.
The results of PPi-mediated excision–extension assays showed that, as expected, WT RT outperformed TAMs RT (10, 11, 24). On the RNA template, excision activity was enhanced for T-3A and T-6A RTs compared with the TAMs RT (Fig. 5G; paired t test, P < 0.006). On the DNA template, the level of PPi-mediated excision was lower than the TAMs RT for T-4A, T-8A, and T-10A RTs, indicating that some cn mutations affect the ratio of ATP-mediated to PPi-mediated AZTMP excision. All five of the virion-derived RTs exhibited higher levels of PPi-mediated excision–extension activity on the RNA template compared with the DNA template (Fig. 5G).
To verify that the differences observed in AZTMP excision–extension by the patient-derived RTs were not due to a higher efficiency of DNA polymerization, we determined the rates of DNA polymerization on the same RNA and DNA templates used in the excision and extension assay (Fig. 5 H and I, respectively). On both templates, the patient-derived RTs exhibited lower rates of polymerization compared with WT and TAMs RTs, indicating that the observed increases in AZTMP excision and extension (Fig. 5 A–G) were not due to increased polymerization activity. However, in the polymerization assay, all RTs (except T-6A) were more active on the DNA template, consistent with the idea that the increase seen in the activity of the patient-derived RTs on an RNA template in the excision–extension assay is due to an effect on excision.
We also compared the ATP- and PPi-mediated excision activities of purified WT, TAMs, and T-3C RTs (Fig. 5 J and K). Purified WT RT was inefficient at ATP-mediated excision on both RNA and DNA templates (data not shown). The T-3C RT was more efficient at ATP- and PPi-mediated excision compared with the TAMs RT on an RNA template (P < 0.007) but not a DNA template. Together, these observations support the hypothesis that cn mutations reduce RNase H activity and thereby increase ATP- and PPi-mediated AZTMP excision on an RNA template. In addition to their effects on RNase H cleavage, some cn mutations also affect the ratio of ATP-mediated excision to PPi-mediated excision.
Discussion
The studies described here strongly support the model that mutations in the cn increase AZT-R by reducing template RNA degradation, thereby providing more time for AZTMP excision to take place on an RNA template (14). The first key prediction of this model is that the cn mutations associated with AZT-R will decrease RNase H activity and template RNA degradation. Our results clearly show that the cn mutations associated with AZT-R reduce the primary and secondary RNase H cleavage of template RNA. These in vitro studies are consistent with our previous in vivo observations that the cn mutations and RNase H primer grip mutations reduce RT template-switching frequencies, a result of decreased template RNA degradation (14, 15). Other recent studies support a relationship between RNase H activity and AZT-R (16, 24). Although it is likely that the shorter RNA–DNA hybrid is less stable and reduces the time available for AZTMP excision, it is also possible that the shorter RNA–DNA hybrid is a less efficient substrate for NRTI excision (25). It is important to point out that the stability of the shorter RNA–DNA hybrid is likely to depend on the nucleotide sequence of the hybrid and the conditions of the assay.
The second key prediction of this model is that cn mutations associated with AZT-R will preferentially increase AZTMP excision on an RNA template compared with a DNA template. These studies showed that all five of the cns obtained from treatment-experienced patients significantly increased ATP- and PPi-mediated AZTMP excision on an RNA template compared with a DNA template. The increase in both ATP- and PPi-mediated AZTMP excision on the RNA indicated that the effect of the cn mutations on AZT-R was likely to be related to the effect of these mutations on reducing RNA template degradation. Although the increase in excision efficiency was <2-fold, AZT triphosphate (AZTTP) incorporation and chain termination occurred at only 7 positions on this template, compared with the ≈2,500 possible sites of chain termination on the viral genome. Thus, the effect of this increase in excision efficiency on AZT-R is likely to be considerable when extrapolated to the viral genome.
In addition to the increase in AZTMP excision on an RNA template, some of the cns from treatment-experienced patients also influenced AZTMP excision on a DNA template. The cn from one patient-derived RT increased ATP-mediated AZTMP excision, whereas three others showed a higher level of ATP-mediated excision relative to PPi-mediated excision on a DNA template. PPi-mediated AZTMP excision is the reverse of polymerization: AZTTP + DNA⇆DNA·AZTMP + PPi. Because of the reversibility of this reaction, the efficiency of PPi-mediated AZTMP excision reflects the efficiency with which RT catalyzes the incorporation of AZTMP into the growing DNA strand. A comparison of the ATP-mediated excision with the PPi-mediated excision reactions on a DNA template showed that, relative to PPi-mediated excision, ATP-mediated AZTMP excision is more efficient for all patient-derived RTs except for T-6A RT (Fig. 5 F and G; compare excision activities on DNA templates). Mutations in the cn could affect the fingers, palm, and thumb region and excision activity by influencing the positioning of the template-primer at the polymerase active site (26). Consistent with this hypothesis, it was recently observed that the G333D substitution in combination with M184V increases the ability of RT to discriminate between deoxycytidine triphosphate and 3TC triphosphate (19).
Examination of the Stanford University HIV Drug Resistance Database indicates that cn mutations play a significant role in drug resistance during therapy. Our analysis indicated that >50% of the sequences that contained either E312Q, G335C, N348I, A360I/V, V365I, or A376S also contained at least one TAM. Only the cn mutations N348I, A360V, and V365I showed statistically significant positive correlations with TAMs. These results agree with other treatment-experienced patient database analyses done with N348I (17, 18) and A360V (27).
Overall, this study has shown that the cn mutations E312Q, G335C/D, N348I, A360I/V, V365I, and A376S (i) are present in viruses from treatment-experienced patients at high frequencies, (ii) enhance AZT-R in association with TAMs, (iii) increase RT's ability to excise chain-terminating nucleotide analogs on an RNA template, and (iv) reduce the level of RNase H primary and secondary cleavage. In addition, some of the patient-derived cns increased ATP-mediated AZTMP excision relative to their effect on PPi-mediated AZTMP excision. Overall, these data support our model that cn mutations affect the balance between polymerase and RNase H activity, which results in increased AZT-R. These studies strongly suggest that inclusion of the C-terminal portions of RT in clinical genotypic and phenotypic assays could lead to more accurate determination of AZT drug resistance.
Materials and Methods
Plasmids, Cloning and Mutagenesis.
Construction of luciferase-expressing HIV-1 vectors pHL[WT] and pHL[TAMs] (contains mutations D67N, K70R, T215Y, and K219Q) was previously described (14). pHCMV-G expresses the G glycoprotein of vesicular stomatitis virus (28). Site-directed mutagenesis was performed by using the QuikChange XL site-directed mutagenesis kit (Stratagene).
Patient Sequences and Database Analysis.
The cn sequences from viruses from treatment-experienced patients T-3, T-4, T-6, T-8, and T-10 were previously described (14). Clinical relevance of the cn mutations was analyzed by using the Stanford University HIV Drug Resistance Database (http://hivdb.Stanford.edu).
Purified RTs.
An Escherichia coli expression plasmid containing WT HIV-1 RT (11) was used to subclone the region of RT containing TAMs (D67N, K70R, T215Y, and K219Q) and subclone mutations from patient T-3 (G335C and N348I) in the context of TAMs. Reverse transcriptases were purified as previously described (11).
Cells, Transfection, and Virus Production.
293T cells were transfected with the vector of interest in the presence of pHCMV-G using GenJet (SignalDT Biosystems). Viral supernatants were clarified by centrifugation 48 h later (2 K, 15 min, 4°C) and then filtered through a Millex GS 0.45-μm-pore-size filter (Nalge). Virus was collected by centrifugation through a 30% sucrose cushion (25 K) for 90 min at 4°C and concentrated 100-fold in PBS. The final 100× viral stocks were verified to be free of nucleases and used for RT activity assays as previously described (15) and in vitro reactions. Replicative capacity and AZT (Sigma-Aldrich) IC50 determinations were as previously described (21).
Substrate Preparation.
RNA or DNA oligonucleotides were 5′ end-labeled with [γ-32P]ATP (Perkin-Elmer) and purified by using CENTRI-SEP columns (Princeton Separations). RNA/DNA hybrids were prepared by annealing labeled and unlabeled oligonucleotides in a 1:2 ratio. AZT terminated primers were created by incubating 500 nM hybrid (final concentration) with MLV-RT (RH−) (2 units; Stratagene) for 30 min at 37°C in the presence of 20 μM AZTTP/50 mM Tris·HCl (pH 7.5)/50 mM KCl/10 mM MgCl2.
RNase H Assays.
Primary RNase H cleavages were assayed on an RNA/DNA hybrid consisting of an 18-nt DNA 5′-AGCTCCCAGGCTCAGATC-3′ annealed to a 32P-labeled 18-nt RNA 5′-GAUCUGAGCCUGGGAGCU-3′ (22). Final reaction mixtures contained 50 mM Tris·HCl (pH 8), 60 mM KCl, 10 mM DTT, 2.5 mM EGTA, 0.26% Nonidet P-40, 10 units of RNaseOUT (Invitrogen), 5 mM MgCl2, and 100 nM hybrid. Cell-free viruses (100 ng of p24) or purified RTs (varying concentrations) were resuspended in an equal volume of reaction buffer lacking MgCl2 and hybrid at room temperature for 5 min. Reactions were initiated by the addition of MgCl2 and hybrid and incubated for 30 min at 37°C. Reactions were stopped with an equal volume of formamide dye (Gel Buffer II; Ambion). Samples were heated (95°C, 5 min) and loaded onto a 15% denaturing PAGE. Band intensities were scanned by using a PhosphorImager (BioRad Laboratories), and density analysis was performed by using Quantity One Software (Bio-Rad Laboratories). Secondary RNase H cleavages were assayed by using the previously described hybrid: a 32P-labeled 41-nt RNA primer 5′-GGGCGAAUUCGAGCUCGGUACCCGGGGAUCCUCUAGAGUCG-3′ annealed to a 77-nt DNA 5′-TGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCGCCCTATAGTGAGTCGTATTACAAT-3′ template (23).
ATP- or PPi-Mediated Excision and/or Extension Assays.
A previously described (29) 32P-labeled 18-nt DNA primer (5′- GTCACTGTTCGAGCACCA-3′) was annealed to either a 42-nt RNA (5′-AUCAGUGUAGACAAUCCCUAGCUAUGGUGCUCGAACAGUGAC-3′) or a 42-nt DNA (5′-ATCAGTGTAGACAATCCCTAGCTATGGTGCTCGAACAGTGAC-3′) template and blocked with AZTTP. Final reaction mixtures, for ATP-mediated excision and extension, contained 50 mM Tris·HCl (pH 8), 60 mM KCl, 10 mM DTT, 2.5 mM EGTA, 0.26% Nonidet P-40, 5 units RNase OUT, 10 mM MgCl2, 5 μM dNTPs, 2.5 μM AZTTP, 3.3 mM ATP, and 5 nM hybrid. Viral or purified RTs (normalized by using RT activity) were incubated with hybrid (24°C, 5 min) in an equal volume of reaction buffer lacking MgCl2, dNTPs, AZTTP, and ATP. Reactions were initiated by the addition of MgCl2, dNTPs, AZTTP, and ATP and incubated over time at 37°C. Reactions were analyzed as described above. For reactions in which extension alone was measured, the reactions were carried out in the absence of AZTTP and ATP with an unblocked 32P-labeled 18-nt DNA primer annealed to either the 42-nt RNA or DNA template. For PPi-mediated excision and extension reactions, ATP in the above reactions was replaced with 100 μM PPi (final concentration).
Statistical Analysis.
Student's t test or paired two-sample t test was used to determine statistically significant differences in some experiments (SigmaPlot 8.0; Systat Software). Bonferroni's adjustment was used where appropriate to control for type I errors. Two proportions analysis was performedby using NCSS 2000 Statistical Software (NCSS).
Supplementary Material
Acknowledgments.
We thank Wei-Shau Hu for intellectual input throughout the project, Frank Maldarelli for intellectual input and critical discussion of the manuscript, and Wei Shao for help with database analysis. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804660105/DCSupplemental.
References
- 1.Telesnisky A, Goff SP. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Plainview, NY: Cold Spring Harbor Lab Press; 1997. pp. 121–160. [Google Scholar]
- 2.Ho DD, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 3.Wei X, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 4.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 5.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 7.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 8.Johnson VA, et al. Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med. 2007;15:119–125. [PubMed] [Google Scholar]
- 9.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): Increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 10.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: An increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 11.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucher CA, et al. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 13.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 14.Nikolenko GN, et al. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc Natl Acad Sci USA. 2007;104:317–322. doi: 10.1073/pnas.0609642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delviks-Frankenberry KA, Nikolenko GN, Barr R, Pathak VK. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3′-azido-3′-deoxythymidine resistance. J Virol. 2007;81:6837–6845. doi: 10.1128/JVI.02820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp SD, et al. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and L-2′,3′-dideoxy-3′-thiacytidine. J Virol. 1998;72:5093–5098. doi: 10.1128/jvi.72.6.5093-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap SH, et al. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 2007;4:e335. doi: 10.1371/journal.pmed.0040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hachiya A, et al. Amino acid mutation N348I in the connection subdomain of HIV-1 reverse transcriptase confers multi-class resistance to nucleoside and non-nucleoside reverse transcriptase inhibitors. J Virol. 2008;82:3261–3270. doi: 10.1128/JVI.01154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelina S, Sheen CW, Radzio J, Mellors JW, Sluis-Cremer N. Mechanisms by which the G333D mutation in human immunodeficiency virus type 1 reverse transcriptase facilitates dual resistance to zidovudine and lamivudine. Antimicrob Agents Chemother. 2008;52:157–163. doi: 10.1128/AAC.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brehm JH, et al. Selection of mutations in the connection and RNase H domains of human immunodeficiency virus type 1 reverse transcriptase that increase resistance to 3′-azido-3′-dideoxythymidine. J Virol. 2007;81:7852–7859. doi: 10.1128/JVI.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolenko GN, et al. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: Balance between RNase H activity and nucleotide excision. Proc Natl Acad Sci USA. 2005;102:2093–2098. doi: 10.1073/pnas.0409823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Wisniewski M, et al. Substrate requirements for secondary cleavage by HIV-1 reverse transcriptase RNase H. J Biol Chem. 2002;277:28400–28410. doi: 10.1074/jbc.M201645200. [DOI] [PubMed] [Google Scholar]
- 24.Meyer PR, Matsuura SE, Schinazi RF, So AG, Scott WA. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob Agents Chemother. 2000;44:3465–3472. doi: 10.1128/aac.44.12.3465-3472.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radzio J, Sluis-Cremer N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol Pharmacol. 2008;73:601–606. doi: 10.1124/mol.107.038596. [DOI] [PubMed] [Google Scholar]
- 26.Zhang WH, Svarovskaia ES, Barr R, Pathak VK. Y586F mutation in murine leukemia virus reverse transcriptase decreases fidelity of DNA synthesis in regions associated with adenine-thymine tracts. Proc Natl Acad Sci USA. 2002;99:10090–10095. doi: 10.1073/pnas.152186199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos AF, et al. Conservation patterns of HIV-1 RT connection and RNase H domains: Identification of new mutations in NRTI-treated patients. PLoS ONE. 2008;3:e1781. doi: 10.1371/journal.pone.0001781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yee JK, et al. A general method for the generation of high-titer, pantropic retroviral vectors: Highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyer PL, Imamich T, Sarafianos SG, Arnold E, Hughes SH. Effects of the Delta67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision. J Virol. 2004;78:9987–9997. doi: 10.1128/JVI.78.18.9987-9997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.