Abstract
The conventional wisdom is that cell-surface receptors interact with ligands expressed on other cells to mediate cell-to-cell communication (trans interactions). Unexpectedly, it has recently been found that two classes of receptors specific for MHC class I molecules not only interact with MHC class I molecules expressed on opposing cells, but also with those on the same cell. These cis interactions are a feature of immunoreceptors that inhibit, rather than activate, cellular functions. Here, we review situations in which cis interactions have been observed, the characteristics of receptors that bind in trans and cis, and the biological roles of cis recognition.
As their name indicates, MHC molecules were discovered and characterized based on their role in inducing potent rejection of tissue grafts. However, the physiological role of MHC molecules remained enigmatic until ground-breaking studies demonstrated the fundamental importance of the MHC complex in T-cell-mediated immunity to infection1. A vast amount of subsequent work elucidated how T-cell receptors (TCRs) recognize self MHC molecules in complex with non-self peptides derived from microbial or other foreign proteins.
Transplantation biology provided early evidence for the existence of an alternative immune-recognition strategy. The laws of transplantation predicted that tissue grafts were accepted as long they displayed a subset of the recipient’s MHC alleles2. Indeed, MHC heterozygous offspring from two mouse strains that differed at the MHC locus (F1 hybrids) accepted solid tissue grafts from either parent. However, parental bone-marrow grafts were rejected by irradiated F1 hybrid mice, a phenomenon termed hybrid resistance3. This observation suggested a novel type of immune recognition, which was controlled by MHC genes. The basis for this phenomenon remained obscure until several lines of research converged on the proposal that the lack of (appropriate) MHC class I molecules on host cells resulted in natural killer (NK)-cell-mediated rejection (the ‘missing-self’ hypothesis)4. Subsequent work showed that NK cells expressed MHC-class-I-specific inhibitory receptors, which protected normal host cells from NK-cell-mediated attack. Consequently, the loss of MHC class I molecules, which arises owing to infection or transformation, and is likely selected for by cytolytic T cells, renders host cells susceptible to NK-cell-mediated lysis, as inhibitory receptors are no longer engaged. These studies led to the identification of families of MHC class I receptors expressed by NK cells. Hybrid resistance was eventually explained by the selectivity of inhibitory receptors for certain MHC class I alleles together with the differential expression of these receptors by subsets of NK cells. As a consequence, a subset of NK cells in MHC heterozygous hosts may fail to engage parental MHC class I molecules5, which results in NK-cell-mediated rejection of bone-marrow grafts.
‘Missing-self’ hypothesis
The concept that absence of MHC class I expression renders host cells sensitive to lysis by natural killer cells.
Inhibitory NK-cell receptors specific for classical MHC class I molecules belong to the mouse Ly49 (also known as KLRA) receptor family6,7 and the human killer immunoglobulin-like receptor (KIR) family8-10. The molecular cloning of NK-cell receptors led to the identification of additional MHC class I receptors, which had not been predicted on the basis of functional experiments: human leukocyte immunoglobulin-like receptors (LILRs; also known as LIRs, ILTs or MIRs), and their orthologues, mouse paired immunoglobulin-like receptors (PIRs), as well as the heterodimeric CD94-NKG2 (NK group 2) receptors11-14 (TABLE 1).
Table 1.
Inhibitory receptors specific for MHC class I molecules and viral proteins
| Receptor* | Species | Structure | Stalk length (amino acids) | ligand | Binding site‡ | Cis binding | Expression | Refs |
|---|---|---|---|---|---|---|---|---|
| KIR family (CD158)§ | ||||||||
| KIR2DL1, KIR2DL2, KIR2DL3 | Human | 2 immunoglobulin-like domains | 40 | HLA-C allotypes | Top | No∥; ND | Subsets of NK and effector T cells | |
| KIR2DL4 | Human | 2 immunoglobulin-like domains | 40 | HLA-G | ND | ND | Mostly NK cells | |
| KIR3DL1 | Human | 3 immunoglobulin-like domains | 40 | HLA-BBw4 | Top? | ND | Subsets of NK and effector T cells | |
| KIR3DL2 | Human | 3 immunoglobulin-like domains | 40 | HLA-A3, HLA-A11 | ND | ND | Subsets of NK and effector T cells | |
| Ly49 family (KLRA) | ||||||||
| Ly49A, Ly49C, Ly49F, Ly49G, Ly49I | Mouse | CTLD | 72-76 | Classical H2 molecules | Lateral | Yes | Subsets of NK and effector T cells | 15,21,25 |
| Ly49I129 | Mouse | CTLD | 76 | MCMV m157 | ND | ND | Subsets of NK and effector T cells | 75 |
| Ly49B | Mouse | CTLD | 78 | Classical H2 molecules | ND | No | Myeloid cells | 25,76 |
| Ly49Q | Mouse | CTLD | 77 | H2-K | ND | yes | DC and myeloid cells | 25,77 |
| LILR and PIR family (ILT, LIR, MIR, CD85)¶ | ||||||||
| LILRB1 (LIR1, ILT2) | Human | 4 immunoglobulin-like domains | 53 | Various HLA molecules | Lateral | ND | NK, T, B and myeloid cells | 13 |
| HCMV UL18 | Lateral | ND | 37 | |||||
| LILRB2 (LIR2, ILT4) | Human | 4 immunoglobulin-like domains | 42 | Various HLA molecules | ND | Yes | B, myeloid and mast cells | 16,64 |
| PIRB | Mouse | 6 immunoglobulin-like domains | 42 | Various H2 molecules | ND | Yes | B, myeloid and mast cells | 14,16,41 |
| CD94-NKG2§ | ||||||||
| CD94-NKG2A | Human | CTLD | 29 (CD94); 19 (NKG2A) | HLA-E | Top | ND | Subset of NK and effector T cells | 11 |
| CD94-NKG2A (KLRD1-KLRC1) | Mouse | CTLD | 29 (KLRD1); 26 (KLRC1) | Qa-1b | Top | No∥ | Subset of NK and effector T cells | 12 |
Receptor families include inhibitory and stimulatory receptors. Alternative names for receptors are provided in parentheses.
Top: α1 and α2 domains of MHC class I heavy chain and bound peptide; lateral: α3 domain and β2-microglobulin (plus α2 domain for Ly49).
For a review, see REF. 44.
Unpublished data.
For a review, see REF. 39. CTLD, C-type lectin-like domain; DC, dendritic cell; HCMV, human cytomegalovirus; ILT, immunoglobulin-like transcript; KIR, killer-cell immunoglobulin-like receptor; LILR, leukocyte immunoglobulin-like receptor; MCMV, mouse cytomegalovirus; MIR, monocyte/macrophage immunoglobulin-like receptor; ND, not determined; NK, natural killer; NKG2, NK group 2; PIR, paired immunoglobulin-like receptor.
All of these MHC class I receptors were identified based on their capacity to regulate immune-cell function on binding in trans to MHC ligands expressed by other cells (antigen-presenting cells (APCs) in the case of TCRs, and target cells in the case of NK-cell receptors). Recently, however, the structurally unrelated NK-cell receptors Ly49 and LILRs or PIRs were found to interact with MHC class I ligands expressed in the plane of the same membrane (in cis)15,16. Here, we examine the possible structural basis for cis versus trans binding of MHC molecules by these receptors and discuss the physiological relevance of cis and trans interactions for immune-cell function. we further review evidence that some receptors that recognize non-MHC ligands also act both in cis and trans, indicating that cis interactions may need to be considered alongside conventional trans interactions in the functional analysis of certain immunoreceptors.
MHC recognition in trans and cis
T-cell receptors
X-ray crystallographic studies of TCRs bound to peptide-MHC-class-I or peptide-MHC-class-II complexes have shown that the TCR is typically positioned diagonally across the composite surface created by the peptide and the MHC α-helices that flank the peptide-binding groove, with the variable-α (vα) domain situated over the N-terminal half of the peptide and the vβ domain over the C-terminal half17 (FIG. 1a). The affinity of this interaction determines the fate of T cells during thymocyte development and the type of immune response that T cells will elicit when mature. These effects are mediated by trans recognition of peptide-MHC complexes expressed by stromal cells or APCs. Although T cells express MHC class I molecules, this expression is not required for thymocyte positive selection18 or for the activation of peripheral T cells19. Thus, there is no evidence that TCRs functionally interact with MHC molecules in cis. Indeed, structural considerations would exclude this possibility, as the segments connecting both TCR and MHC molecules to their respective transmembrane domains appear too short (10-20 residues) to permit a TCR to engage an MHC molecule on the same cell. Moreover, segmental flexibility between the vαvβ and CαCβ modules of the TCR, as well as between the α1, α2 and α3 domains of an MHC class I molecule, is too restricted to compensate for the shortness of the connecting peptides.
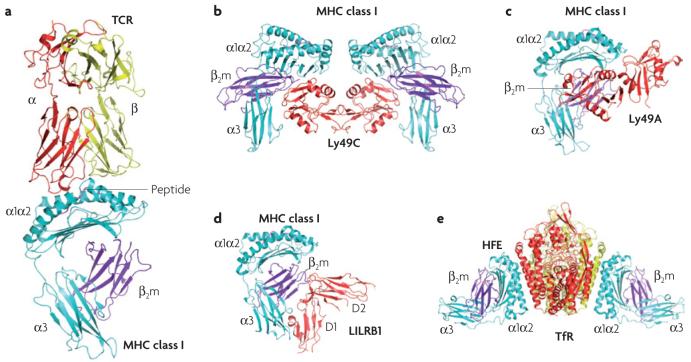
Figure 1. Structures of MHC class I receptors.
Ribbon diagrams showing the crystal structures of several MHC class I receptors bound to MHC class I ligands. The α1, α2 and α3 domains of the MHC class I heavy chain are shown in cyan, β2-microglobulin (β2m) is shown in purple and the MHC-bound peptide is shown in pink. a | A typical T-cell receptor (TCR)-peptide-MHC-class-I complex is shown (protein data bank (PDB) ID: 1MI5). The TCR α- and β-chains are shown in red and yellow, respectively. b | The Ly49C-H2-Kb complex is shown (PDB ID: 1P4L). The two C-type lectin domains (CTLDs) of Ly49C (shown in red) bind two H2-Kb molecules in a symmetrical arrangement. The Ly49C dimer is in the ‘open’ conformation. c | Representation of the Ly49A-H2-Dd complex (PDB ID: 1QO3). The two CTLDs of Ly49A (shown in red) bind a single H2-Dd molecule in an asymmetrical manner. The Ly49A dimer is in the ‘closed’ conformation. d | Representation of the LILRB1 (leukocyte immunoglobulin-like receptor B1)-HLA-A2 complex (PDB ID: 1P7Q). The D1 and D2 domains of LILRB1 are shown in red. e | The transferrin receptor (TfR)-haemochromatosis protein (HFE) complex is shown (PDB ID: 1A6Z). The two subunits of the TfR dimer are shown in red and yellow. HFE, a non-classical MHC class I molecule, must assume a supine orientation on the cell membrane to bind TfR in cis. By contrast, the MHC class I molecules in (a-d) are in upright orientations.
Positive selection
One step in the process of T-cell differentiation in the thymus. Thymocytes expressing T-cell receptors with moderate affinity for self-peptide-MHC complexes receive a survival signal and continue to develop towards becoming single positive (CD4+CD8- or CD4-CD8+) T cells. Positive selection is mediated by resident stromal cells in the thymic cortex.
Ly49 receptors
Ly49 receptors are homodimeric type II membrane proteins, with each chain composed of a C-type lectin-like domain (CTLD) connected by a stalk region of approximately 70 amino-acid residues to transmembrane and cytoplasmic domains. The Ly49 receptor family comprises at least 23 members in mice (Ly49A-Ly49w), 13 of which have an inhibitory function and 10 of which have an activating function20. This type of receptor is expressed by rodent but not human NK cells, which instead express KIRs to monitor MHC class I expression. Different Ly49 receptors exhibit distinct MHC-binding properties, ranging from the broad recognition of MHC class I molecules by Ly49C to the allelic specificity of Ly49A21. Bound peptide is necessary for MHC class I recognition by Ly49 receptors, although binding to Ly49A is independent of the peptide sequence22. Conversely, the Ly49C and Ly49I receptors are peptide selective, despite the absence of direct contacts with peptide23-26.
Type II membrane proteins
An integral membrane protein, such as Ly49, in which the carboxy terminus is extracellular.
C-type lectin-like domain
(CTLD). A protein module originally identified as a carbohydrate-recognition domain in a family of calcium-dependent lectins. The natural-killer-cell receptor group of C-type lectin-like receptors includes disulphide-linked homodimers or heterodimers that do not bind calcium and recognize proteins instead of carbohydrates.
In contrast to TCRs, which dock onto the top of MHC molecules, structural and mutagenesis studies of complexes of Ly49A with H2-Dd, and of Ly49C with H2-Kb, revealed that Ly49 receptors contact MHC class I molecules at a wide cavity underneath the peptide-binding platform26-28. This binding site, which partially overlaps that of CD8 (REF. 29), is formed by the α2 and α3 domains of MHC class I and β2-microglobulin (β2m). In the Ly49C-H2-Kb complex26, the Ly49C homodimer engages H2-Kb bivalently, such that each CTLD makes identical interactions with MHC class I to form a symmetrical, butterfly-shaped assembly (FIG. 1b). By contrast, in the Ly49A-H2-Dd complex28, the Ly49A dimer contacts H2-Dd asymmetrically, with only one of its subunits binding an MHC molecule (FIG. 1c). A second potential MHC-binding site, originally observed in the Ly49A-H2-Dd crystal structure27, was ruled out as not functionally relevant by subsequent mutagenesis studies30,31. The structural basis for the very different modes of MHC engagement in the Ly49C-H2-Kb and Ly49A-H2-Dd complexes is dependent on the different geometries of the Ly49C and Ly49A dimers. The Ly49C dimer adopts an ‘open’ conformation, in which the CTLD subunits are less closely juxtaposed than in the Ly49A dimer, which adopts a ‘closed’ conformation (FIG. 1b,c). Important steric clashes between MHC molecules would preclude the closed Ly49A dimer from simultaneously binding two MHC in the manner of the open Ly49C dimer. However, a nuclear magnetic resonance (NMR) study of unbound Ly49A revealed that the receptor exists predominantly in the open state in solution, similar to Ly49C, and that the open form of Ly49A can bind two MHC molecules32. Hence, Ly49A (and presumably other Ly49 receptors) can assume both open and closed conformations, resulting in differential engagement of MHC ligands that might correlate with trans and cis binding, respectively (see later).
The capacity of Ly49A to bind H2-Dd on other cells differs markedly between NK cells from mice that express H2-Dd and mice that lack H2-Dd expression33. This effect is due to the expression of H2-Dd on the NK cells themselves, which masks Ly49A and reduces its capacity to bind H2-Dd ligand on other cells15. Indeed H2-Dd occupies at least 75% of the Ly49A molecules expressed by individual NK cells, reducing the accessibility of Ly49A by approximately fourfold34,35. Similar to Ly49A, most other Ly49 receptors of known MHC specificity have the capacity to interact with MHC class I molecules in cis25,36. The notable exception is the more distantly related receptor Ly49B, which only binds MHC class I molecules in trans25.
The inhibitory interaction of Ly49A with H2-Dd on target cells (trans interaction) uses a binding site beneath the peptide-binding platform of H2-Dd (REFs 27,30,31). This same binding site mediates the interaction with H2-Dd in cis15. Consequently, Ly49 receptors must drastically reorient their CTLDs relative to the NK-cell surface to bind MHC class I in trans versus cis (FIG. 2a,b). It is most likely that the exceptionally long stalk regions linking the CTLDs of Ly49 molecules to the membrane provide the requisite flexibility. Although these proline-rich stalks have not been visualized in any Ly49 structure, they are predicted to consist of coiled-coil regions inter-spersed with several flexible segments that confer mobility to the ectodomains27. Based on the structure of the Ly49C-H2-Kb complex26, the stalks must adopt a back-folded, or bent, conformation for the Ly49 dimer to bind in trans, as the N-termini of the CTLDs point away from the NK-cell membrane (FIG. 2a). Conversely, cis binding would require the stalks to assume an extended conformation that orients the CTLDs with their N-termini directed towards the NK cell (FIG. 2b). Although it is unknown whether one Ly49 dimer can bind two MHC class I molecules in cis, simple modelling suggests that engagement of both CTLDs by MHC molecules on the NK cell is unlikely, owing to the orientation that binding of one MHC molecule would impose on the Ly49 dimer (FIG. 2b). If so, the (bivalent) Ly49C-H2-Kb and (monovalent) Ly49A-H2-Dd complexes would exemplify trans and cis recognition, respectively.
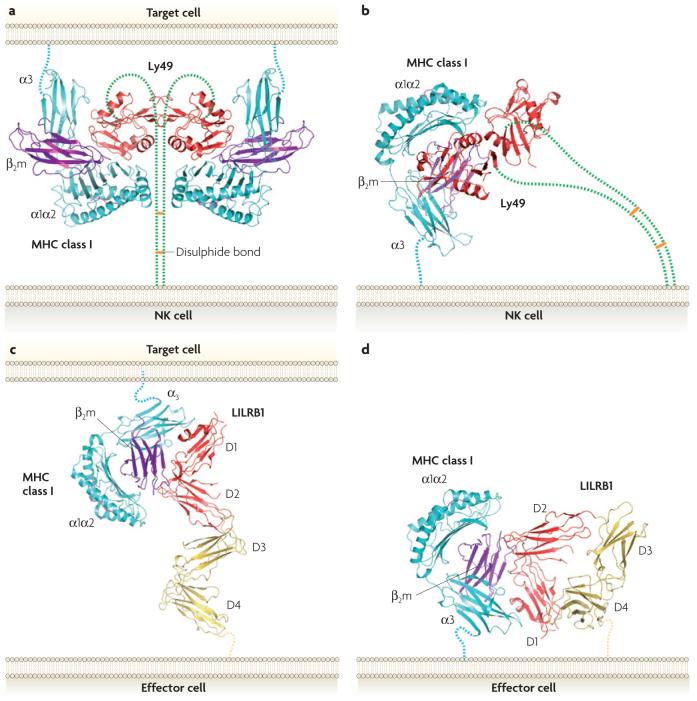
Figure 2. Hypothetical models for trans and cis interactions of ly49 receptors and LILRs with MHC class I ligands.
a| Trans interaction of a Ly49 receptor with two MHC class I molecules, based on the crystal structure of the Ly49C-H2-Kb complex26. The α1, α2 and α3 domains of the MHC class I heavy chain are shown in cyan, β2-microglobulin (β2m) is shown in purple and Ly49C is shown in red. The Ly49C homodimer on the natural killer (NK) cell binds two H2-Kb molecules on the target cell. The Ly49C-H2-Kb structure does not include the 70-amino-acid stalk regions that connect the Ly49C homodimer to the NK-cell membrane. The stalks are drawn arbitrarily here (in green), with horizontal orange bars to indicate disulphide bonds. To bind in trans, the stalks must adopt a back-folded conformation, as the N-termini of the Ly49C monomers point away from the NK-cell membrane (Ly49 receptors are type II transmembrane proteins). b | Cis interaction of Ly49 with MHC class I, based on the structure of the Ly49A-H2-Dd complex27. The Ly49A homodimer binds one H2-Dd molecule on the NK cell itself. In this case, the stalks may assume an extended conformation, as the N-termini of the Ly49A monomers point towards the NK cell. c | Trans interaction of a leukocyte immunoglobulin-like receptor (LILR) with MHC class I, based on the structure of the LILRB1-HLA-A2 complex40. The LILRB1-HLA-A2 structure includes the two N-terminal immunoglobulin-like domains of LILRB1 (D1 and D2), but not its two C-terminal immunoglobulin-like domains. Domains 3 and 4 (D3 and D4), which link the ligand-binding domains (shown in red) to the surface of the effector cell, are represented by homology models (shown in yellow). d | Cis interaction of LILRB1 with MHC class I. To bind to HLA-A2 on the same cell, LILRB1 must bend back on itself, presumably at the connecting region between D2 and D3.
LILRs and PIRs
After the C-type lectin-like Ly49 receptors, the immunoglobulin-like LILRs and PIRs were the second class of immunoreceptors that were found to interact with MHC class I in cis16. As LILRs and PIRs also recognize MHC class I in trans13, structurally unrelated receptors have evolved corresponding capacities to bind MHC ligands expressed on the same membrane, as well as on opposing membranes, indicating convergent evolution. This suggests that cis recognition of MHC class I may be of general importance as a mechanism for regulating immune-cell function (see later).
The human LILRs comprise at least five inhibitory receptors (LILRB1-LILRB5) and three activating receptors (LILRA1-LILRA3), which are characterized by two or four extracellular immunoglobulin-like domains13,37. The homologous mouse PIRs, which contain six immunoglobulin-like domains, include a single inhibitory receptor (PIRB). At least six genes encode activating receptors (PIRA)14,38. LILRB1 is broadly distributed, whereas LILRB2 and PIRB are more selectively expressed by B cells and myeloid cell types, but not by T cells and NK cells. These receptors bind a wide range of classical and non-classical MHC class I molecules with low affinity and without any apparent allelic specificity (reviewed in REF. 39).
Structural knowledge of LILRs and PIRs is based mainly on studies of LILRB1 (REF. 40). MHC class I binding is mediated by the two most membrane-distal immunoglobulin-like domains (D1 and D2) of this four immunoglobulin-like domain receptor. The structure of LILRB1 (D1 and D2 only) in complex with HLA-A2 showed that LILRB1 binds at a site formed by the α3 domain of HLA-A2 and β2m, such that the tip of D1 contacts the α3 domain and the D1-D2 interdomain hinge region contacts β2m (FIG. 1d). A significant contribution by β2m, which was also observed for PIRB41, helps to explain the broad MHC reactivity of LILRs and PIRs. Although the structure of the complete LILRB1 ectodomain (D1-D4) has not been determined, modelling indicates that the four immunoglobulin-like domains must adopt an extended conformation, similar to that reported for CD4 (REF. 42), to bind MHC class I on an opposing cell surface (FIG. 2c).
A similarly broad MHC recognition profile in cis and trans16 provides circumstantial evidence that the same binding site of LILRB2 mediates both interactions. If so, cis recognition would require D1 and D2 to reverse direction with respect to the effector-cell surface, resulting in a horseshoe-shaped configuration of D1-D4 in which LILRB1 bends back on itself (FIG. 2d). Although such a large reversal implies considerable flexibility in the peptide segment connecting D2 and D3, the structure of the N-terminal four immunoglobulin-like domains of the Drosophila melanogaster protein DSCAM (Down syndrome cell-adhesion molecule) showed a horseshoe arrangement, which is made possible by a five-residue hinge between the D2 and D3 immunoglobulin-like domains of DSCAM, that is very similar to that proposed here for cis-binding LILRB2 (REF. 43). In the case of PIRB, which has two more immunoglobulin-like domains proximal to the membrane compared with LILRB2, additional flexibility might derive from the D4-D5 or D5-D6 connecting segments. Thus, Ly49 receptors and LILRs or PIRs appear to have evolved two distinct strategies to achieve cis and trans MHC recognition: unusually long stalk regions in the case of Ly49 receptors, and very flexible (or multiple) interdomain hinges in the case of LILRs and PIRs.
KIRs and CD94-NKG2
Human NK cells use KIRs to monitor MHC class I expression on target cells (reviewed in REF. 44). These receptors, the role of which corresponds to that of Ly49 receptors in rodents, comprise two or three extracellular immunoglobulin-like domains, and hence are designated KIR2D and KIR3D, respectively. In contrast to both Ly49 receptors and LILRs, KIRs dock onto the top of an MHC class I molecule in a manner that resembles that of TCR binding45,46. There is currently no evidence that the function of KIR2DL is influenced by HLA-C expression in cis (N. Gardiol and W.H., unpublished observations). KIR3D contains an additional N-terminal immunoglobulin-like domain (D0), which enhances MHC class I binding by the D1 and D2 domains47. A possible effect of a cis ligand on KIR3DL function has not been tested.
The heterodimeric C-type lectin-like CD94-NKG2 receptors, which are expressed on NK cells and some effector T cells, bind the non-classical human HLA-E or mouse Qa-1b molecules11,12. Structural models based on the complex between NKG2D (NK group 2, member D) and one of its ligands, MICA (MHC-class-I-polypeptide-related sequence A)48, together with the structure of the CD94-NKG2 heterodimer49 and the role of specific residues in HLA-e and its bound peptide on recognition49,50, suggest that CD94-NKG2 binds to the α1-α2 platform domain of MHC class I, as do KIRs. Ligand binding by CD94-NKG2A is not influenced by Qa-1b expression by NK cells (L. Scarpellino and W.H., unpublished observations), suggesting that the inhibitory CD94-NKG2A receptor is not masked by cis ligands.
HLA-E
A non-classical MHC class I molecule with limited sequence variability. Its expression on the cell surface depends on the availability of peptides derived from the signal sequence of classical MHC class I molecules. HLA-E is recognized by CD94-NKG2 receptors.
Qa-1b
A functional mouse homologue of human HLA-E. similar to HLA-E, Qa-1b cell-surface expression depends on the binding of peptides derived from the signal sequence of classical MHC class I molecules and it is recognized by CD94-NKG2 receptors.
Although there is at present no evidence for cis recognition by inhibitory receptors that bind to the top of MHC class I ligands (KIRs, CD94-NKG2), an unusual feature initially described for the non-classical MHC molecule haemochromatosis protein (HFE) raises the formal possibility of cis interactions for at least some of these receptors. HFE contributes to the maintenance of iron homeostasis by binding to the transferrin receptor (TfR)51. The structure of the complex between HFE and TfR indicates that these molecules associate in cis on the same cell membrane, rather than in trans between opposing cell membranes52. To do so, HFE ‘lies down’ parallel to the cell membrane, to allow the HFE α-helices that form the peptide-binding groove in classical MHC class I molecules to contact TfR (FIG. 1e). Interestingly, HFE is recognized by αβ TCRs53, implying that it can also adopt an ‘upright’ configuration. If classical MHC molecules can assume a lying down orientation under certain circumstances, as has been suggested54, this may allow cis binding by immunoreceptors such as KIRs that engage MHC class I ligands via their α1-α2 platform domain. Irrespective of this possibility, however, the structure of the HFE-TfR complex52 suggests that cis association may predate the immunological functions of MHC molecules.
Haemochromatosis protein
(HFE). A non-classical MHC class I molecule that regulates iron metabolism by binding to the transferrin receptor. The HFE gene is mutated in hereditary haemochromatosis — an iron overload disease.
Role of MHC class I recognition in cis
To date, cis interactions have been documented exclusively for MHC class I receptors with inhibitory function. Such receptors are characterized by the presence of immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in their cytoplasmic domain. when phosphorylated by SRC family tyrosine kinases, such as LCK or LYN, ITIMs recruit phosphatases, such as SHP1 (SRC-homology 2 (SH2)-domain-containing protein tyrosine phosphatase 1), that subsequently suppress phosphorylation-based activation signalling (reviewed in REF. 55). Importantly, efficient ITIM phosphorylation, and consequently inhibitory signalling, requires co-ligation of inhibitory receptor(s) with activating receptor(s)56. Accordingly, the role of MHC class I recognition by inhibitory receptors in cis needs to be considered in the context of the activation receptor(s) or pathways that are co-engaged.
Immunoreceptor tyrosine-based inhibitory motif
(ITIM). A short amino-acid sequence (the consensus sequence of which is Val/Ile-X-Tyr-X-X-Val/Leu, where X denotes any amino acid) that is found in the cytoplasmic tail of inhibitory receptors. ITIMs are thought to mediate inhibitory signalling by recruiting phosphatases such as SHP1 (SRC-homology-2-domain-containing protein tyrosine phosphatase 1).
Ly49: regulation of NK-cell effector function
NK cells have essential direct and indirect roles in innate defence against infection and transformed cells. In response to diseased (as well as to normal) host cells, multiple distinct activation receptors induce the exocytosis of cytolytic granules and the production of immunoregulatory cytokines by NK cells. Inhibitory receptors, many of which are specific for MHC class I, counteract NK-cell activation. Target-cell lysis occurs when NK-cell activation exceeds inhibition signalling57. Consequently, inhibitory signals set the activation threshold required for NK-cell-mediated target-cell lysis.
Co-expression of Ly49A and H2-Dd on the NK-cell membrane does not lead to constitutive ITIM phosphorylation (L. Scarpellino and W.H., unpublished observations) and there is no evidence that the functionality of these NK cells is impaired15, indicating that Ly49A cis interactions do not generate tonic inhibition signals. Rather, the H2-Dd on the NK cells sequesters a significant fraction of Ly49A receptors, and this restricts the pool of Ly49A receptors that are available for functional interaction with MHC class I ligands on target cells. This is apparent at the site of NK-cell-target-cell contact, the so-called immunological synapse. when NK cells do not express H2-Dd themselves, Ly49A is efficiently recruited to the NK-cell synapse by the H2-Dd expressed on target cells34,58. By contrast, Ly49A accumulation is strongly reduced when NK cells express H2-Dd, suggesting that only the initially accessible Ly49A receptors can be recruited to the NK-cell synapse and that trans H2-Dd cannot compete with cis H2-Dd for Ly49A binding. This restricts the inhibitory capacity of Ly49A in the context of H2-Dd expression15,33.
Immunological synapse
A term derived from the similiarities to the synapses that occur in the nervous system, it defines a region that can form at the cell surface between two cells of the immune system that are in close contact, such as the interaction between a T cell and a natural killer cell with an antigen-presenting cell and a target cell, respectively. This interface involves adhesion molecules, as well as antigen receptors and cytokine receptors.
The moderation of the inhibitory signalling capacity renders NK cells more useful, as the activation threshold required for target-cell lysis is lowered. As an example, stress-associated expression of NKG2D ligands by host cells enhances NK-cell activation signalling. This is sufficient to induce target-cell lysis when NK-cell inhibition is moderate (FIG. 3a) but it may be insufficient when NK-cell inhibition is too potent (FIG. 3b). Likewise, when the number of available inhibitory receptors is limited, even a minor reduction of MHC class I ligand expression by host cells is sufficient to abrogate NK-cell inhibition and consequently lead to target-cell lysis59. Thus, the restriction of inhibitory signalling by cis interaction considerably improves the sensitivity of NK cells to react to diseased host cells. Despite the predominance of sequestered Ly49A, the number of residual Ly49A receptors that can functionally interact in trans is sufficient to prevent auto-aggression of non-stressed host cells expressing MHC class I molecules at physiological levels33,60.
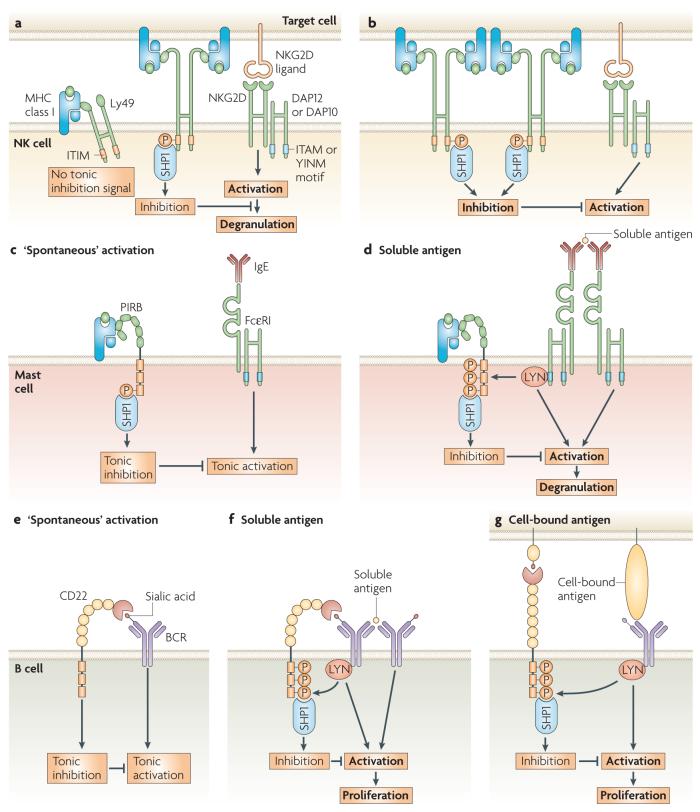
Figure 3. Physiological role of cis and trans interactions of Ly49A, PIRB and CD22.
a | Stable association of Ly49 with H2-Dd in the plane of the natural killer (NK)-cell membrane (in cis) reduces the pool of Ly49A receptors available to engage H2-Dd on target cells (in trans). When NK-cell inhibition is restricted, further NK-cell activation, upon the recognition of the NKG2D (NK group 2, member D) ligand on stressed host cells, results in target-cell lysis. b | In the absence of Ly49A-H2-Dd interactions in cis, all Ly49A receptors can interact with H2-Dd in trans, resulting in strong NK-cell inhibition. Limited additional NK-cell activation may not suffice to induce lysis of stressed host cells. Ly49A association with H2-Dd in cis decreases the NK-cell activation threshold. c | In the absence of deliberate mast-cell activation, paired immunoglobulin-like receptor B (PIRB) is constitutively phosphorylated and associated with MHC class I in the mast-cell membrane, suggesting that cis interactions mediate tonic suppression of spontaneous mast-cell degranulation. d | Mast-cell activation by crosslinking IgE bound to FcεRI (high-affinity Fc receptor for IgE), increases PIRB phosphorylation and the recruitment of SHP1 (SRC-homology 2 (SH2)-domain-containing protein tyrosine phosphatase 1). Unlike Ly49A, the inhibitory function of PIRB depends on cis interaction. PIRB cis interaction increases the threshold for mast-cell activation. e | CD22 is directly and indirectly associated with the B-cell receptor (BCR) by binding sialic-acid-modified glycoproteins. CD22 cis association dampens basal BCR signalling and prevents ‘spontaneous’ B-cell activation. f | Following B-cell activation with soluble antigen, the association of CD22 with the BCR is needed for the inhibitory function of CD22. CD22 cis association increases the threshold for B-cell activation. g | CD22 can switch from a cis- to a trans-bound state on the encounter of sialic-acid ligand on opposing membranes. The engagement of CD22 in trans is inhibitory and increases the threshold for B-cell activation. ITAM, immunoreceptor tyrosine-based activation motif; ITIM, immunoreceptor tyrosine-based inhibitory motif.
LILRB2 and PIRB: regulators of mast-cell effector function
Mast cells have essential protective roles in IgE-mediated immune responses, such as host protection against parasites. Mast cells are activated by the high-affinity Fc receptor for IgE (FcεRI), when bound IgE is crosslinked by multivalent antigens. Mast-cell activation triggers mast-cell degranulation, releasing vasoactive amines, such as histamine, and induces the synthesis of pro-inflammatory molecules and cytokines. excessive or aberrant mast-cell activation is associated with allergic reactions (reviewed in REF. 61).
Similar to NK cells, mast-cell activation is counter-regulated by several distinct inhibitory receptors, including PIRB16,62. Indeed, PIRB engagement by MHC class I dampens mast-cell activation16 (FIG. 3c). Cell mixing experiments have shown that mast-cell activation is not significantly attenuated when the PIRB-MHC class I interaction occurs exclusively in trans16. The inhibitory effect of PIRB on mast-cell activation therefore depends on MHC class I recognition in cis (FIG. 3d). even in the absence of deliberate mast-cell activation, PIRB is constitutively tyrosine phosphorylated and associated with SHP1 (REFs 16,63). Thus, in contrast to Ly49A, the PIRB-MHC class I cis interaction may generate cell-autonomous, tonic inhibitory signals (FIG. 3c). Tonic suppression seems to prevent ‘spontaneous’ mast-cell degranulation, which may occur by ‘accidental’ aggregation of FcεRI-bound IgE. MHC class I expression in cis might lead to PIRB aggregation and/or, according to the concept of co-ligation, mediate an association of PIRB with the activating receptor FcεRI.
The new findings regarding PIRB function in mast cells contrast with the role of LILRB1 and LILRB2 in NK cells and myeloid cells, respectively, in which receptor engagement by HLA ligands on other cells dampens cellular activation13,64. This provides evidence that the LILR and PIR family receptors can also functionally interact with MHC class I molecules in trans. The precise contribution of cis versus trans interactions involving LILRs and PIRs to inhibitory signalling therefore warrants further investigation. Similar to CD22 (see later), recognition of MHC class I by LILRs or PIRs in cis and/or in trans may depend on whether effector cells are activated by cell-bound ligands as opposed to soluble ligands.
In addition to MHC class I, LILRB1 recognizes the MHC-class-I-like molecule UL18 encoded by human cytomegalovirus (HCMV). Cis binding by LILRB2 raises the issue of whether UL18 interacts with LILRB1 on the surface of HCMV-infected cells. This could have a role for latent HCMV infection, which is established in myeloid lineage cells65, which often express LILRB1.
Siglecs: CD22 regulation of B-cell activation
Similar to the two-way recognition of MHC class I ligands, sialic-acid modifications of glycoproteins or glycolipids are recognized in cis and trans by several Siglecs (sialic-acid-binding immunoglobulin-like lectins). Siglecs are type I membrane proteins with an N-terminal V-type immunoglobulin domain that binds sialic acids and variable numbers (1-16) of C-type immunoglobulin domains (reviewed in REF. 66).
The most extensively characterized Siglec is CD22 (also known as Siglec-2), a negative regulator of B-cell receptor (BCR) signalling. CD22 is a seven-immunoglobulin domain receptor, which binds sialic acids linked to galactosidase (Sia α2-6Gal) typically found on N-linked glycans of glycoproteins. In B cells, CD22 is largely inaccessible to soluble, multivalent sialoside probes, owing to the interaction of CD22 in cis with sialic acids67,68. The crucial sialic-acid-modified cis ligand is unknown, even though CD22 can bind to membrane IgM, CD45 and CD22 itself. Access to the CD22 receptor is restored when sialic acids are removed by sialidase treatment67, or in mice lacking the sialyltransferase ST6GAL1 (REF. 68).
In the absence of deliberate antigen exposure, basal BCR signalling is increased in B cells lacking CD22, demonstrating that the primary function of CD22 is to dampen BCR signalling69. The direct and/or indirect association of CD22 with the BCR complex in cis therefore mediates tonic suppressive signals that prevent ‘spontaneous’ B-cell activation (FIG. 3e). On B-cell activation by soluble antigen, the CD22 ITIMs are phosphorylated by the BCR-associated SRC-family kinase LYN. The ensuing recruitment of SHP1 to CD22 dampens BCR signalling (FIG. 3f). CD22 variants that selectively lack sialic-acid-binding capacity mediate reduced SHP1 recruitment to CD22, and consequently increased BCR signalling69,70. Thus, direct or indirect association with the BCR complex in cis is essential for the inhibitory function of CD22. This is in contrast to Ly49 receptors, where cis association reduces the inhibitory function.
In addition to soluble antigens, B-cell activation by cell-bound antigen is also reduced when the stimulating cell displays sialic-acid modifications71. So, despite the fact that CD22 is normally masked, CD22 trans ligands negatively regulate BCR signalling. Indeed, unlike Ly49A34, sialic-acid-containing trans ligands efficiently recruit CD22 to sites of cell-cell contact72, indicating that masking is not stable, and that CD22 can switch from a cis- to a trans-bound state (FIG. 3g). This may be of importance to prevent B-cell activation by cell-bound self antigens.
Beyond the immune system
Receptors that can bind the equivalent ligand expressed in cis and trans via the same ligand-binding domain have also been found in biological situations other than the immune system. For example, ephrin receptors (Ephs) mediate cellular repulsion on interaction with ephrin ligands expressed by other cells. This permits the sorting of distinct cell types or directs the growth of axons. Recent data suggest that Eph activity is influenced by ephrins expressed in the plane of the same membrane73. Indeed, Ephs can bind ephrin ligands expressed in cis via the ligand-binding domain of the receptor74. Interestingly, cis interactions did not induce Eph tyrosine kinase activity; rather, signalling by Eph to mediate cellular repulsion was reduced74. Eph therefore provides the first example of an activating receptor that is subject to regulation by a ligand interaction in cis.
Prospects and predictions
So far, immunoreceptors that can bind the same ligand in cis and trans are receptors with inhibitory function. The emerging theme is that cis interactions modify inhibitory signalling. Whereas trans interactions are always inhibitory, significant differences exist with regard to the precise role of cis interactions. Cis interactions are required for the inhibitory function of CD22 and probably also PIRB, whereas they reduce the overall inhibitory capacity of Ly49 receptors. Consequently, cis interactions can serve to increase or decrease the threshold at which cellular activation signalling produces a biological response.
Following the discovery of inhibitory MHC class I receptors on NK cells, a plethora of inhibitory immunoreceptors has been identified55. Many of these receptors show wide tissue distribution whereby ligands may be co-expressed with their receptors on the same cell. It therefore seems likely that the activity of additional inhibitory receptors and perhaps also activating receptors is controlled via cis interactions, a possibility that should be considered along with trans interactions in the study of immunoreceptors.
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
CD22 | CD94 | Ly49A | NKG2D | PIRB
Protein Data Bank: http://www.rcsb.org/pdb/home/home.do
1A6Z | 1MI5 | 1P4L | 1P7Q | 1QO3
FURTHER INFORMATION
Werner Held’s homepage: http://www.lau.licr.org/pages/RG-NKTCDG.htm
Roy Mariuzza’s homepage: https://carb.umbi.umd.edu/user/mariuzza
Acknowledgements
Work in the authors’ laboratories is supported in part by grants from the Swiss National Science Foundation and Oncosuissse (to W.H.) and the National Institutes of Health, USA (AI047990 to R.A.M.). We thank S. Cho for assistance with preparation of the original figures.
References
- 1.Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- 2.Snell GD. The genetics of transplantation. J. Natl Cancer Inst. 1953;14:691–700. [PubMed] [Google Scholar]
- 3.Cudkowicz G, Bennett M. Peculiar immunobiology of bone marrow allografts. I. Graft rejection by irradiated responder mice. J. Exp. Med. 1971;134:83–102. doi: 10.1084/jem.134.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 5.Yu YY, et al. The role of Ly49A and 5E6(Ly49C) molecules in hybrid resistance mediated by murine natural killer cells against normal T cell blasts. Immunity. 1996;4:67–76. doi: 10.1016/s1074-7613(00)80299-x. [DOI] [PubMed] [Google Scholar]
- 6.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 7.Smith HRC, Karlhofer FM, Yokoyama WM. Ly-49 multigene family expressed by IL-2-activated NK cells. J. Immunol. 1994;153:1068–1079. [PubMed] [Google Scholar]
- 8.Moretta A, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J. Exp. Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 10.Wagtmann N, et al. Molecular clones of the p58 natural killer cell receptor reveal Ig-related molecules with diversity in both the extra- and intra-cellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 11.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 12.Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1. J. Exp. Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colonna M, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubagawa H, Burrows PD, Cooper MD. A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells. Proc. Natl Acad. Sci. USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doucey MA, et al. Cis-association of Ly49A with MHC class I restricts natural killer cell inhibition. Nature Immunol. 2004;5:328–336. doi: 10.1038/ni1043.This report provides the first direct evidence that an MHC class I receptor (Ly49A) not only binds to a ligand on an opposing cell membrane but also interacts with a ligand expressed on the same membrane. Cis interaction is shown to inhibit inhibition, thereby facilitating NK-cell activation.
- 16.Masuda A, Nakamura A, Maeda T, Sakamoto Y, Takai T. Cis binding between inhibitory receptors and MHC class I can regulate mast cell activation. J. Exp. Med. 2007;204:907–920. doi: 10.1084/jem.20060631.This paper demonstrates cis binding of MHC class I ligands by immunoglobulin-like LILRB1 and PIRB. In these cases, the cis interaction is thought to dampen mast-cell activation.
- 17.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 18.Bix M, Raulet D. Inefficient positive selection of T cells directed by hematopoietic cells. Nature. 1992;359:330–333. doi: 10.1038/359330a0. [DOI] [PubMed] [Google Scholar]
- 19.Schott E, Bertho N, Ge Q, Maurice MM, Ploegh HL. Class I negative CD8 T cells reveal the confounding role of peptide-transfer onto CD8 T cells stimulated with soluble H2-Kb molecules. Proc. Natl Acad. Sci. USA. 2002;99:13735–13740. doi: 10.1073/pnas.212515399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson SK, Ortaldo JR, McVicar DW. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol. Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- 21.Hanke T, et al. Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity. 1999;11:67–77. doi: 10.1016/s1074-7613(00)80082-5. [DOI] [PubMed] [Google Scholar]
- 22.Correa I, Raulet DH. Binding of diverse peptides to MHC class I molecules inhibits target cell lysis by activated natural killer cells. Immunity. 1995;2:61–71. doi: 10.1016/1074-7613(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 23.Franksson L, et al. Peptide dependency and selectivity of the NK cell inhibitory receptor Ly-49C. Eur. J. Immunol. 1999;29:2748–2758. doi: 10.1002/(SICI)1521-4141(199909)29:09<2748::AID-IMMU2748>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 24.Hanke T, Raulet DH. Cumulative inhibition of NK cells and T cells resulting from engagement of multiple inhibitory Ly49 receptors. J. Immunol. 2001;166:3002–3007. doi: 10.4049/jimmunol.166.5.3002. [DOI] [PubMed] [Google Scholar]
- 25.Scarpellino L, et al. Interactions of Ly49 family receptors with MHC class I ligands in trans and cis. J. Immunol. 2007;178:1277–1284. doi: 10.4049/jimmunol.178.3.1277. [DOI] [PubMed] [Google Scholar]
- 26.Dam J, et al. Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2Kb. Nature Immunol. 2003;4:1213–1222. doi: 10.1038/ni1006.The structures described in references 26, 27 and 32 reveal significant variability in Ly49-receptor dimerization and engagement of MHC class I ligands. The bivalent Ly49C-H2-K and monovalent Ly49A-H2-D complexes might represent trans and cis recognition, respectively.
- 27.Tormo J, Natarajan K, Margulies DH, Mariuzza RA. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature. 1999;402:623–631. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- 28.Deng L, Mariuzza RA. Structural basis for recognition of MHC and MHC-like ligands by natural killer cell receptors. Semin. Immunol. 2006;18:159–166. doi: 10.1016/j.smim.2006.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao GF, et al. Crystal structure of the complex between human CD8αα and HLA-A2. Nature. 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto N, Mitsuki M, Tajima K, Yokoyama WM, Yamamoto K. The functional binding site for the C-type lectin-like natural killer cell receptor Ly49A spans three domains of its major histocompatibility complex class I ligand. J. Exp. Med. 2001;193:147–157. doi: 10.1084/jem.193.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, et al. Binding of the natural killer cell inhibitory receptor Ly49A to its major histocompatibility complex class I ligand. J. Biol. Chem. 2002;277:1433–1442. doi: 10.1074/jbc.M110316200. [DOI] [PubMed] [Google Scholar]
- 32.Dam J, et al. Variable dimerization of the Ly49A natural killer cell receptor results in differential engagement of its MHC class I ligand. J. Mol. Biol. 2006;362:102–113. doi: 10.1016/j.jmb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Zimmer J, Ioannidis V, Held W. H-2D ligand expression by Ly49A+ natural killer (NK) cells precludes ligand uptake from environmental cells: implications for NK cell function. J. Exp. Med. 2001;194:1531–1539. doi: 10.1084/jem.194.10.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Back J, Chalifour A, Scarpellino L, Held W. Stable masking by H-2Ddcis ligand limits Ly49A relocalization to the site of NK cell/target cell contact. Proc. Natl Acad. Sci. USA. 2007;104:3978–3983. doi: 10.1073/pnas.0607418104.This paper shows that Ly49A remains stably associated with cis H2-D during NK-cell-target-cell interactions, thereby reducing trans H2-Dd-driven Ly49A recruitment to the NK-cell synapse.
- 35.Andersson KE, Williams GS, Davis DM, Hoglund P. Quantifying the reduction in accessibility of the inhibitory NK cell receptor Ly49A caused by binding MHC class I proteins in cis. Eur. J. Immunol. 2007;37:516–527. doi: 10.1002/eji.200636693. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 37.Cosman D, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 38.Hayami K, et al. Molecular cloning of a novel murine cell-surface glycoprotein homologous to killer cell inhibitory receptors. J. Biol. Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 39.Takai T. A novel recognition system for MHC class I molecules constituted by PIR. Adv. Immunol. 2005;88:161–192. doi: 10.1016/S0065-2776(05)88005-8. [DOI] [PubMed] [Google Scholar]
- 40.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nature Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura A, Kobayashi E, Takai T. Exacerbated graft-versus-host disease in Pirb-/- mice. Nature Immunol. 2004;5:623–629. doi: 10.1038/ni1074. [DOI] [PubMed] [Google Scholar]
- 42.Wu H, Kwong PD, Hendrickson WA. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 43.Meijers R, et al. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- 44.Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 45.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 46.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nature Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 47.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 48.Li P, et al. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nature Immunol. 2001;2:443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan LC, et al. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 2007;27:900–911. doi: 10.1016/j.immuni.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 50.Wada H, Matsumoto N, Maenaka K, Suzuki K, Yamamoto K. The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA-E through mostly shared but partially distinct sets of HLA-E residues. Eur. J. Immunol. 2004;34:81–90. doi: 10.1002/eji.200324432. [DOI] [PubMed] [Google Scholar]
- 51.Lebron JA, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 52.Bennett MJ, Lebron JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417.This paper reports a co-crystal structure that reveals an association of the non-classical MHC class I molecule HFE with the TfR on the same membrane.
- 53.Rohrlich PS, et al. Direct recognition by αβ cytolytic T cells of Hfe, a MHC class Ib molecule without antigen-presenting function. Proc. Natl Acad. Sci. USA. 2005;102:12855–12860. doi: 10.1073/pnas.0502309102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitra AK, et al. Supine orientation of a murine MHC class I molecule on the membrane bilayer. Curr. Biol. 2004;14:718–724. doi: 10.1016/j.cub.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Veillette A, Latour S, Davidson D. Negative regulation of immunoreceptor signaling. Annu. Rev. Immunol. 2002;20:669–707. doi: 10.1146/annurev.immunol.20.081501.130710. [DOI] [PubMed] [Google Scholar]
- 56.Blery M, et al. Reconstituted killer cell inhibitory receptors for major histocompatibility complex class I molecules control mast cell activation induced via immunoreceptor tyrosine-based activation motifs. J. Biol. Chem. 1997;272:8989–8996. doi: 10.1074/jbc.272.14.8989. [DOI] [PubMed] [Google Scholar]
- 57.Lanier LL, Corliss B, Phillips JH. Arousal and inhibition of human NK cells. Immunol. Rev. 1997;155:145–154. doi: 10.1111/j.1600-065x.1997.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 58.Eriksson M, Ryan JC, Nakamura MC, Sentman CL. Ly49A inhibitory receptors redistribute on natural killer cells during target cell interaction. Immunology. 1999;97:341–347. doi: 10.1046/j.1365-2567.1999.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsson-Alheim MY, Salcedo M, Ljunggren H-G, Kärre K, Sentman CL. NK cell receptor calibration. Effects of MHC class I induction on killing by Ly49Ahigh and Ly49Alow NK cells. J. Immunol. 1997;159:3189–3194. [PubMed] [Google Scholar]
- 60.Olsson MY, Karre K, Sentman CL. Altered phenotype and function of natural killer cells expressing the major histocompatibility complex receptor Ly-49 in mice transgenic for its ligand. Proc. Natl Acad. Sci. USA. 1995;92:1649–1653. doi: 10.1073/pnas.92.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawakami T, Galli SJ. Regulation of mast-cell and basophil function and survival by IgE. Nature Rev. Immunol. 2002;2:773–786. doi: 10.1038/nri914. [DOI] [PubMed] [Google Scholar]
- 62.Katz HR. Inhibitory receptors and allergy. Curr. Opin. Immunol. 2002;14:698–704. doi: 10.1016/s0952-7915(02)00400-4. [DOI] [PubMed] [Google Scholar]
- 63.Ho LH, Uehara T, Chen CC, Kubagawa H, Cooper MD. Constitutive tyrosine phosphorylation of the inhibitory paired Ig-like receptor PIR-B. Proc. Natl Acad. Sci. USA. 1999;96:15086–15090. doi: 10.1073/pnas.96.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colonna M, et al. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J. Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- 65.Soderberg-Naucler C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 66.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nature Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 67.Razi N, Varki A. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl Acad. Sci. USA. 1998;95:7469–7474. doi: 10.1073/pnas.95.13.7469.This is one of the first demonstrations that immunoreceptors (CD22) can be masked by ligand (sialic acid) expression on the same cell.
- 68.Collins BE, et al. Constitutively unmasked CD22 on B cells of ST6Gal I knockout mice: novel sialoside probe for murine CD22. Glycobiology. 2002;12:563–571. doi: 10.1093/glycob/cwf067.In this paper it is shown that CD22 can switch from a cis- to a trans-bound state on cell-cell interactions. This is not observed with cis-bound Ly49A (see reference 34).
- 69.O’Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science. 1996;274:798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 70.Poe JC, et al. CD22 regulates B lymphocyte function in vivo through both ligand-dependent and ligand-independent mechanisms. Nature Immunol. 2004;5:1078–1087. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 71.Lanoue A, Batista FD, Stewart M, Neuberger MS. Interaction of CD22 with α2,6-linked sialoglycoconjugates: innate recognition of self to dampen B cell autoreactivity? Eur. J. Immunol. 2002;32:348–355. doi: 10.1002/1521-4141(200202)32:2<348::AID-IMMU348>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 72.Collins BE, et al. Masking of CD22 by cis ligands does not prevent redistribution of CD22 to sites of cell contact. Proc. Natl Acad. Sci. USA. 2004;101:6104–6109. doi: 10.1073/pnas.0400851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007;17:230–238. doi: 10.1016/j.tcb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 74.Yin Y, et al. EphA receptor tyrosine kinases interact with co-expressed ephrin-A ligands in cis. Neurosci. Res. 2004;48:285–296. doi: 10.1016/j.neures.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 76.Gays F, et al. Ly49B is expressed on multiple subpopulations of myeloid cells. J. Immunol. 2006;177:5840–5851. doi: 10.4049/jimmunol.177.9.5840. [DOI] [PubMed] [Google Scholar]
- 77.Toyama-Sorimachi N, et al. Ly49Q, a member of the Ly49 family that is selectively expressed on myeloid lineage cells and involved in regulation of cytoskeletal architecture. Proc. Natl Acad. Sci. USA. 2004;101:1016–1021. doi: 10.1073/pnas.0305400101. [DOI] [PMC free article] [PubMed] [Google Scholar]