Abstract
Infection by HIV-1 involves the fusion of viral and cellular membranes with subsequent transfer of viral genetic material into the cell. The HIV-1 envelope glycoprotein that mediates fusion consists of the surface subunit gp120 and the transmembrane subunit gp41. gp120 directs virion attachment to the cell–surface receptors, and gp41 then promotes viral–cell membrane fusion. A soluble, α-helical, trimeric complex within gp41 composed of N-terminal and C-terminal extraviral segments has been proposed to represent the core of the fusion-active conformation of the HIV-1 envelope. A thermostable subdomain denoted N34(L6)C28 can be formed by the N-34 and C-28 peptides connected by a flexible linker in place of the disulfide-bonded loop region. Three-dimensional structure of N34(L6)C28 reveals that three molecules fold into a six-stranded helical bundle. Three N-terminal helices within the bundle form a central, parallel, trimeric coiled coil, whereas three C-terminal helices pack in the reverse direction into three hydrophobic grooves on the surface of the N-terminal trimer. This thermostable subdomain displays the salient features of the core structure of the isolated gp41 subunit and thus provides a possible target for therapeutics designed selectively to block HIV-1 entry.
Infection of target cells by HIV-1 is initiated by the fusion of viral and cellular membranes, leading to release of viral genetic material into the cell. The HIV-1 envelope glycoprotein (Env) mediates the specific attachment of virions to cell-surface receptors and promotes membrane fusion (for a recent review, see ref. 1). The Env protein is also a major target for the human immune response for HIV-1 infection (2). The HIV-1 Env is synthesized as a polypeptide precursor gp160, which subsequently is cleaved to yield two noncovalently associated subunits, gp120 and gp41 (1, 3). The surface subunit gp120 determines viral tropism through interaction with the primary cellular receptor CD4 and particular chemokine receptors (for a review, see ref. 4). The transmembrane subunit gp41 mediates direct fusion of the viral envelope with the cellular membrane.
Many viral membrane fusion proteins are known to undergo conformational changes to become active in mediating viral entry into target cells. The best characterized example is influenza virus hemagglutinin (HA) (5). In this case, the low pH environment in endosomes triggers a major structural change within native HA, resulting in the transformation of a loop region into a coiled coil and thereby delivering the N-terminal fusion peptide about 100 Å toward the target membrane (6, 7). Several lines of evidence suggest that gp120 binding to both CD4 and a coreceptor results in the conformational change in gp120/gp41 required for initiating membrane fusion (8–10). First, binding of soluble CD4 to laboratory-adapted isolates of HIV-1 induces dissociation of gp120 (shedding) from the viral surface (11, 12). Second, new antigenic epitopes of gp41 often are exposed upon binding of soluble CD4 to primary isolates of virus (10). Moreover, soluble CD4 increases the infectivity of primary isolates at low concentrations, but inhibits infectivity at higher concentrations, presumably as gp120 sheds from the virus (13, 14). Third, although single point mutations introduced at the N-terminal coiled coil region of gp41 abrogate Env-mediated membrane fusion, these changes do not interfere with the formation of a complex of gp120 and gp41 (15–18). Additionally, the fusion activity of these mutant proteins correlates with the hydrophobicity of the side chain at the substitution site (15, 18). Therefore, these mutations may inhibit a conformational change involving the refolding of a coiled coil essential for activating the fusion potential of gp120/gp41 (6, 18, 19).
The ectodomain (that is, the extraviral portion) of gp41 is the most conserved region in HIV-1 Env, which otherwise exhibits considerable genetic diversity, even among closely related isolates. gp41 contains two 4-3 hydrophobic repeat sequences within the ectodomain that are predicted to form a coiled coil (20–22). The N-terminal 4-3 hydrophobic repeat is located adjacent to the fusion peptide that is essential for membrane fusion, whereas the C-terminal repeat precedes the transmembrane segment. Interestingly, synthetic peptides corresponding to the sequences of the heptad repeat regions of gp41 are potent inhibitors of viral infectivity and syncytium formation (22–24). A synthetic peptide corresponding to the N-terminal heptad repeat sequence of gp41 is highly helical in solution, and mutations that disrupt the α-helical structure of this peptide abolish its antiviral activity (22). In addition, single proline substitutions within the N-terminal 4-3 hydrophobic sequence of gp41 result in Env proteins that are completely defective in mediating fusion (15–18). Taken together, these results suggest that the two heptad repeat regions in gp41 and, hence, formation of the coiled coil structure are critical for HIV-1 Env-mediated membrane fusion (21, 25). In the case of influenza HA, the extension of α-helical coiled coils underlies the “spring-loaded mechanism” for the HA conformational change (6, 7).
Protein dissection experiments have been used to generate a soluble, protease-resistant complex within gp41 composed of two peptides denoted N-51 and C-43 that are derived from the N- and C-terminal regions of the ectodomain, respectively (26). The N-51 and C-43 peptides associate to form a stable, α-helical trimeric complex of heterodimers, with the N-51 and C-43 helices oriented in an antiparallel fashion (26). A similar soluble complex has been identified by protein dissection of the gp41 ectodomain from the simian immunodeficiency virus (SIV) (27). Proteolysis of a single polypeptide model for the N-51/C-43 complex has led to the identification of N-36 and C-34 peptides, which correspond to the central regions of N-51 and C-43, respectively (M.L. and P. S. Kim, unpublished data). The biophysical properties of N-36/C-34 and N-51/C-43 are very similar (M.L. and P. S. Kim, unpublished data).
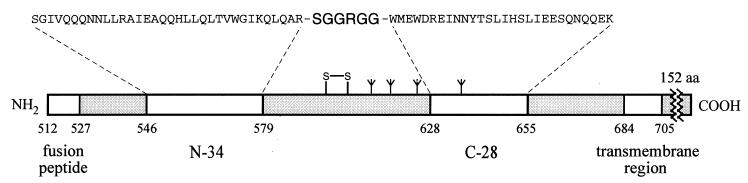
A minimal, but thermostable, subdomain within gp41 can be constructed from two further truncated peptides, N-34 and C-28, connected by a six-residue hydrophilic linker Ser-Gly-Gly-Arg-Gly-Gly in place of the disulfide-bonded “loop” region (Fig. 1) (M.L. and S.S, unpublished data). The single-chain polypeptide designated N34(L6)C28 forms a fully helical, trimeric structure that retains high thermostability with a Tm of 70°C under physiological conditions (M.L. and S.S., unpublished data). Here we report the crystal structure of the linked complex N34(L6)C28 determined at 2.4-Å resolution. Very recently, independent results on two crystal structures of α-helical domains from gp41 have been reported (30, 31). Comparison of these crystal structures shows that the minimal N34(L6)C28 subdomain imparts the key determinants of the core structure of gp41. Thus, this single-chain, thermostable polypeptide model may aid in the design and discovery of small-molecule drugs of HIV-1 infection.
Figure 1.
A thermostable, α-helical subdomain designated N34(L6)C28 within the extraviral portion of HIV-1 gp41. The single-chain recombinant N34(L6)C28 polypeptide consists of residues 546–579 (N-34) and 628–655 (C-28) of gp41 plus a linker of six hydrophilic residues (Ser-Gly-Gly-Arg-Gly-Gly). The important functional features of gp41 are shown. Expansion above the N-34 and C-28 peptides shows the amino acid sequence in single letter code. The disulfide bond and four potential N glycosylation sites are depicted. The residues are numbered according to their position in gp160.
MATERIALS AND METHODS
Peptide Production and Purification.
Plasmid pN34/C28-L6 for the expression of the N34(L6)C28 polypeptide was derived from the construct pN41/C34-L6 (M.L. and P. S. Kim, unpublished data). A leucine-to-methionine mutation at the residue 568 (L568M) of gp41 was introduced into pN34/C28-L6 by oligonucleotide-directed mutagenesis (32). Standard recombinant DNA techniques were used (33). The recombinant proteins were expressed in Escherichia coli BL21(DE3) pLysS with the T7 system (34) and purified from the soluble fraction (M.L. and P. S. Kim, unpublished data). All proteins were purified to homogeneity by reverse-phase HPLC with a Vydac (Hesperia, CA) C-18 preparative column and a linear gradient of acetonitrile containing 0.1% trifluoracetic acid. The identity of each HPLC-purified peptide was confirmed by laser desorption mass spectrometry.
Crystallization and Data Collection.
Crystallization was carried out at 20°C with the conventional hanging-droplet vapor diffusion method. A 15 mg/ml protein stock in 0.1 M NaAc at pH 5.0 was mixed with equal volumes of a reservoir solution of 20% polyethylene glycol 4000/0.2 M (NH4)2SO4/10% glycerol/0.1 M NaAc, pH 5.0, and allowed to equilibrate against the reservoir. Including 10% glycerol was the key to obtain single crystals of high diffraction quality. X-ray diffraction data were collected from a single crystal at room temperature on a MAR-Research (Hamburg, Germany) area detector equipped with Rigaku (Tokyo) RU200 rotating-anode x-ray generator. Reflections were integrated and scaled by using programs denzo and scalepack (HKL Research, Charlottesville, VA). Subsequent data analysis was performed by using the CCP4 suite (35).
Structure Determination.
The structure was determined by single isomorphous replacement with anomalous scattering (SIRAS). The platinum derivative was acquired by soaking crystals overnight in the crystallization buffer containing 1 mM K2PtCl4. Heavy-atom positions were determined from difference Patterson and difference Fourier maps. The heavy atom parameter refinement and the experimental phase calculation were performed with program mlphare within the CCP4 suite. At the early stage, the maps were always subjected to density modification procedure implemented in the program dm within the CCP4 suite for improvement. Only solvent flattening and histogram matching were applied. The model was built on Silicon Graphics (Mountain View, CA) solid impact color graphics workstation by using program o (36) and refined by using x-plor (37).
RESULTS
Structure Solution and Refinement.
The crystals belong to the space group R3, a= b = 53.07 Å, c = 60.58 Å, containing one N34(L6)C28 molecule per asymmetric unit and 43% solvent. For the platinum derivative, both isomorphous and anomalous difference Pattersons calculated at 12–3.5 Å showed a single site. The dm-modified siras map based on this derivative at the same resolution clearly showed two α-helical regions in the asymmetric unit. Nevertheless, the map quality was not good enough for determination of the helices’ polarity and the assignment of side chains unambiguously. The data sets from an L568M mutant crystal and its derivative soaked with the same K2PtCl4 compound then were collected. The difference Fourier of this mutant pair phased by the wild-type siras data gave an additional Pt site, apparently because of the binding to the methionine at the mutated position 568. This helped resolve all of the ambiguity in tracing. Because the mutant crystal is not well isomorphous to the wild-type one, only wild-type data were used for phasing. Table 1 gives the statistics on the crystallographic data.
Table 1.
Crystallographic and refinement statistics
| Data set | Resolution, Å | Unique reflections | Redundancy | Completeness, % | Rmerge, % | Riso, % | PP | PPano | Rcullis, % | FOM, % |
|---|---|---|---|---|---|---|---|---|---|---|
| Native | 2.4 | 2481 | 8.9 | 99.6 | 5.7 | |||||
| K2PtCl4 | 3.1 | 1116 | 6.9 | 100.0 | 7.0 | 21.6 | 1.69 | 0.38 | 0.70 | 40.9 |
| to 3.5 Å | ||||||||||
| Mutant | 2.5 | 2143 | 6.9 | 99.6 | 6.1 | 13.9 | ||||
| K2PtCl4 | 3.1 | 1106 | 9.4 | 99.8 | 9.3 | 25.9 | 1.46 | 0.76 | 32.0 |
Rmerge = 100 × ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) and 〈I(h)〉 are the ith and mean measurement of the intensity of reflection h.
Riso = Σ|Fph − Fp|/ΣFp, where Fph and Fp are derivative and native structure factor amplitudes, respectively.
PP (phasing power) = 〈Fh〉/E, where 〈Fh〉 is the rms heavy atom structure factor amplitude and E is the residual lack of closure error.
Rcullis (Cullis R factor) = Σ∥Fph ± Fp| − Fh(calc)|/Σ|Fph ± Fp|, where Fh(calc) is the calculated heavy atom structure factor amplitude.
FOM, the mean figure of merit.
An initial model with two α-helical polyalanine fragments, 25 alanines in N helix and 16 in C helix, was built on the dm-modified siras map. After three rounds of refinement, phase combination, and density modification, 85% of the residues were assigned with their side chains except for those in the linker region and the termini of the two helices. Subsequent cycles of refinement and model building eventually allowed the completion of the structure of 68 amino acid residues and the placement of 42 water molecules. Omit maps were calculated to confirm the assignment of the side chains. The final free and working R factors are 26.9% and 20.8%, respectively (F > 3σ), in the resolution range 15–2.4 Å. The rms deviation of bond lengths and bond angles from the ideal values are 0.010 Å and 0.999°, respectively. The quality of the refined model was checked by procheck (38). The Ramachandran plot revealed that 97% of the nonglycine residues lie in the most favored region and none in the disallowed region. The refined coordinates have been deposited in the Protein Data Bank (Brookhaven National Laboratory, Upton, NY).
Structure of the N34(L6)C28 Molecule.
The polypeptide chain of N34(L6)C28 folds into an intramolecular, hairpin-like structure in which the N- and C-terminal helices are connected by a six-residue hydrophilic linker (Ser-Gly-Gly-Arg-Gly-Gly) (Fig. 2). The C-terminal helix (residues 628–655) packs in an antiparallel orientation against the N-terminal helix (residues 546–579). Interhelical interactions are largely hydrophobic. Residues involved in interhelical contacts include Ile-559, Vla-570, and Ile-573 from the N-terminal helix, and Trp-631, Ile-635, and Ile-646 from the C-terminal helix. Moreover, six hydrogen bonds and one salt bridge are uniformly distributed along the helices and interdigitated with the hydrophobic contacts. Buried polar residues have been shown to determine structural specificity in the folding of coiled coils (39–41). Thus, polar interactions are likely to be crucial in maintaining the helix packing and hairpin-like structure of N34(L6)C28.
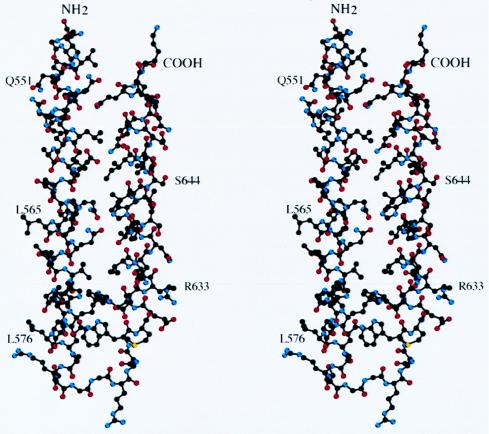
Figure 2.
Structure of the N34(L6)C28 monomer. A stereo drawing of the N34(L6)C28 monomer. The carbon atoms are black, oxygen atoms are red, and nitrogen atoms are blue. The N and C termini of the molecule are labeled. The residues are shown in single letter code and numbered according to their position in gp160. Diagrams were prepared by using the program molscript (48).
The “flexible” hydrophilic linker (42) Ser-Gly-Gly-Arg-Gly-Gly within N34(L6)C28 is evident, except for a main-chain break at the third residue Gly, in the electron density map. Interestingly, the first linker residue Ser utilizes its hydroxyl group to form a hydrogen bond with the carbonyl oxygen of Leu-576 in the N-terminal helix. Additionally, the fourth linker residue Arg stretches into a hydrophilic environment formed by the symmetry related molecules that pack into a crystal. In the crystal environment, these polar interactions appear to make the six-residue linker well ordered.
The N34(L6)C28 Trimer.
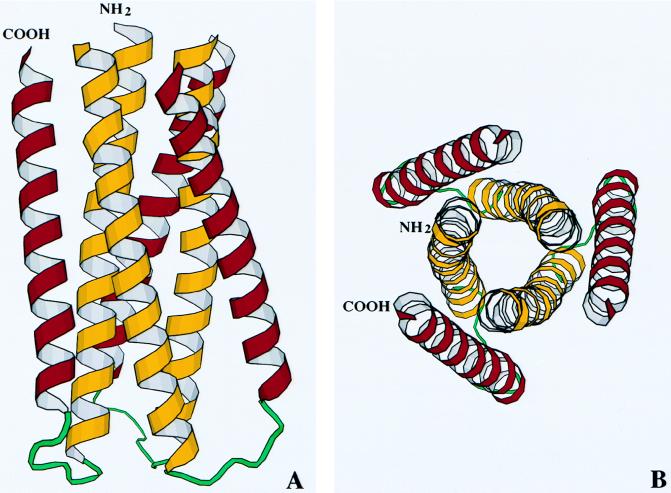
The major feature of the crystal structure of N34(L6)C28 is that three hairpin-like molecules pack together around the crystallographic three-fold axis to form a trimer, resulting in a six-stranded helical bundle (Fig. 3). At the center of the bundle, the side chains at the interface position of three N-terminal helices display typical “knobs-into-holes” packing (43). Three C-terminal helices wrap in the reverse direction into three hydrophobic grooves on the surface of the N-terminal trimer. This structural architecture and core packing confirm many aspects of the model for the structure of the N-51/C-43 complex proposed on the basis of biophysical studies and sequence analysis (26). We have noticed that the distance between the Cα of N helix’s last residue Gly 585 and Cα of C helix’s first residue Trp-628 of the same molecule (14.6 Å) is even larger than that between the Cα of Gly-585 and the Cα of Trp-585 of the symmetry-related neighboring molecule (14.1 Å). This is a good indication that the folded gp41 molecules pack to form trimer very stably, assuming that the linker does not dominate the structure.
Figure 3.
Structure of the N34(L)C28 trimer. (A) Ribbon drawing of the trimer with the N terminus and C terminus of one molecule labeled. The N-terminal helices are colored yellow, whereas the C-terminal helices are purple. (B) An end-on view of N34(L6)C28 looking down the 3-fold axis of the trimer. The figure was generated with the program molscript (48).
Classical coiled coil proteins have a characteristic heptad repeat sequence labeled a-b-c-d-e-f-g, with hydrophobic residues at the positions a and d and predominantly charged residues at the positions e and g (44, 45). In the N-terminal heptad repeat region of gp41, the a, d, e, and g positions are of similar hydrophobicity, with few charged amino acids at the positions e and g (26). Consistent with the extended hydrophobic surface of the molecule, isolated peptides corresponding to the N-terminal heptad sequence tend to aggregate in solution (refs. 22 and 26; M.L. and P. S. Kim, unpublished data). In the trimeric coiled coil with the N terminus of N34(L6)C28, the a and d residues pack in characteristic “acute” packing geometry, with the Cα-Cβ bond in the side chain making an acute angle with respect to the recipient hole (refs. 39 and 46; for a review, see ref. 47). The core of this N-terminal trimer contains nine layers of homotrimeric packing at the a and d positions (Fig. 4A). In the layer around Ile-559 (Fig. 4B), for example, residues from both N- and C-terminal helices, including Ala-558, Leu-645, and Ile-646 pack together, forming a hydrophobic layer. Although these homotrimeric contacts are largely hydrophobic, three polar residues are buried in the core packing. In the layer around Thr-569 at the d position (Fig. 4A), the residue has its hydroxyl group hydrogen-bonded to the carbonyl oxygen of Leu 565 and still uses its hydrophobic methyl group pointing toward the center of the trimer. By contrast, two glutamines at the positions 552 and 562 located at the other two layers interact to their 3-fold related counterparts with their polar atoms, respectively. These polar residues are conserved in HIV-1 and SIV and may prove to be critical in maintaining the trimeric coiled coil structure.
Figure 4.
Helix packing in the structure of the N34(L6)C28 trimer. (A) Packing in the interior, trimeric coiled coil. Successive a and d positions of the three N-terminal helices form nine layers of homotrimeric contacts. The side chains of the residues at the a and d positions are shown. The carbon atoms are black, oxygen atoms are red, and nitrogen atoms are blue. (B) Packing in the hydrophobic layer between Ala-558 and Ile-559 in the interior trimer and Leu-645 and Ile-646 in the outside C-terminal helices. Atoms are colored the same as in A. (C) Packing in the hydrophilic layer including Gln-550, Gln-551, Gln-552, and Asn-553 from the interior trimer, and Gln-652 and Gln-653 from the outside C-terminal helices. Residues are shown in single letter code. Only Q550, N553, Q652, and Q654 are labeled. Atoms are colored the same as in A. The figure was created with the program molscript (48).
Three C-terminal helices pack in the reverse direction into three hydrophobic grooves outside the interior coiled coil trimer and are tilted relative to the N-terminal superhelix by approximately 13 degrees (Fig. 3). Residues in the a and d positions of the C-terminal helix pack against residues at the e and g positions of the interior N-terminal trimer. For example, Leu-645 and Ile-646 of the C-terminal helix make contact with Ile-559 at the a position of the N-terminal helix and Ala-558 at the g position of the neighboring N-terminal helix (Fig. 4B). In addition, the increase in interhelix spacing at the base of the bundle is due to a hydrophobic cluster formed by the bulky residues Trp-571, Trp-628, and Trp-631. Although these interhelical contacts are mainly hydrophobic, interhelical hydrogen bonds and salt bridges are also apparent in the structure of the N34(L6)C28 trimer. An intricate network of hydrogen bonds, for example, is formed by two prominent patches of conserved polar residues, including Gln-551, Gln-552, Asn-553 of the N-terminal helix and Gln-652 and Gln-653 of the C-terminal helix (Fig. 4C). Although residues at the exterior of the bundle are divergent between HIV-1 and SIV, residues in the interhelical core packing are strongly conserved. It is therefore likely that the interactions between the N- and C-terminal helices are crucial for HIV-1-mediated membrane fusion.
DISCUSSION
Comparison of the Crystal Structures of an α-Helical Domain of gp41.
Very recently, Kim and coworkers (30) have determined the crystal structure of a trimeric subdomain within gp41, composed of the N-36 (residues 546–581) and C-34 (residues 628–661) peptides (M.L. and P. S. Kim, unpublished data), whereas Wiley and coworkers (31) have reported the structure of a larger complex designated GCN4/gp41 , composed of an N-terminal of gp41 segment (residues 541–590) fused to a GCN4-derived trimeric zipper, plus a C-terminal segment of gp41 (residues 624–665). We report here the structure of a minimal, yet thermostable, subdomain of gp41, in which the N-34 (residues 546–579) and C-28 peptides (residues 628–655) are connected by a six-residue flexible linker (Fig. 1). All three molecules fold into a six-stranded helical bundle. To compare the structures of the gp41 ectodomain seen in our minimal model (62 residues) and that of the larger complex (94 residues) (31), models of the two structures were overlaid by aligning their corresponding α-carbon atoms. The helical regions of the two structures can be superimposed, with a rms difference of 0.7 Å. There is essentially no difference in the N-terminal helices of the central coiled coil trimer between the two structures. However, the outer layer C-terminal helices in GCN4/gp41 are tilted with respect to the core helices by about 2 degrees more than those in N34(L6)C28. It is striking that the information required to specify the fold of the core structure from HIV-1 gp41 is contained within the small N-34/C-28 subdomain. Additionally, the short peptide linker within N34(L6)C28 stabilizes the α-helical subdomain, presumably for entropy reasons, without perturbing its structure. At neutral pH and 10 μM peptide, the apparent Tm values of N-36/C-34 and N34(L6)C28 are 62 and 70°C, respectively (M.L. and P. S. Kim, unpublished data and M.L. and S.S., unpublished data). The minimal N34(L6)C28 subdomain (62 residues) is considerably more stable than the slightly larger N-36/C-34 complex (70 residues) because N-36 and C-34 are synthetic peptides with blocked termini, which have a major effect on helix stability (49, 50). In essence, the N34(L6)C28 polypeptide provides a highly simplified and tractable model for investigating the determinants of the structure and function of HIV-1 gp41.
We have measured the distance between the center of an individual α-helix and the center of a trimeric coiled coil, defined as the radius by Crick (43), in N34(L6)C28 and other molecules. The radius is 6.4 Å for N34(L6)C28 (residues 546–579), 6.6 Å for influenza HA (residues 41–60) (7), 6.6 Å for the transmembrane subunit of the Moloney murine leukemia virus (residues 46–78) (51), and 6.7 Å for a GCN4 leucine zipper (residues 2–29) (52). Interestingly, among these known coiled coil structures, the radius for N34(L6)C28 is the smallest, indicating a compact fold of the N-terminal coiled coil within gp41. This compactness may be related to the high thermostablity of N34(L6)C28 (M.L. and S.S., unpublished data), which shows extensive interactions among three N-terminal helices as well as between the N- and C-terminal helices.
HIV-1 Inhibition by Peptides.
Isolated peptides that correspond to, or overlap, the N- and C-terminal heptad repeat regions of HIV-1 gp41 have been found to be potent inhibitors of HIV-1 infection and syncytium formation (22–24, 26). Although much is known about the relationships between sequence and activity for these peptide inhibitors (22–24, 53, 54), the mechanism of inhibition has not been fully established. Nevertheless, several lines of evidence support the proposal that these peptide inhibitors block membrane fusion by binding to viral gp41, in a dominant-negative manner (55), to disrupt HIV-1 Env-mediated viral entry (26). First, inhibition of HIV-1 Env-induced cell fusion by the C-43 peptide is markedly reduced when stoichiometric amounts of the N-51 peptide are also present (26). Second, when a gp41 ectodomain maltose-binding chimeric protein contains a proline mutation in the N-terminal heptad repeat region of gp41, the resulting mutant is a potent inhibitor of HIV-1 fusion, although the wild-type protein and a truncated chimeric protein lacking the C-terminal heptad sequence do not have inhibitory activity (19). Third, the antiviral activity of a synthetic peptide corresponding to the C-terminal heptad region of gp41 correlates well with its ability to interact with the N-terminal peptide (53). X-ray crystallographic studies of the soluble, trimeric domain within gp41 composed of two interacting peptides, presented here and by other groups (30, 31), strongly support the proposed dominant-negative mechanism for peptide inhibition. These crystal structures also provide detailed three-dimensional structural information defining the interaction between the N- and C-terminal peptides of gp41, and therefore may lead to new therapeutic and prophylactic strategies targeting HIV-1 entry.
Implications for Viral Membrane Fusion.
Influenza HA is known to undergo a dramatic conformational change to initiate membrane fusion (5). Crystallographic studies showed dramatic differences between the native and fusogenic structures of HA (7, 56). The receptor-binding subunit HA1 serves as a clamp that inhibits structural rearrangements in the transmembrane subunit HA2 from the native to the fusogenic conformation (6, 7). Interestingly, the crystal structure of an α-helical domain of HIV-1 gp41 shares several features in common with those of low pH-converted influenza virus HA2 and the transmembrane subunit of Moloney murine leukemia virus (7, 30, 31, 51). All three viral membrane fusion proteins have α-helical, three-stranded coiled coils adjacent to the N-terminal fusion peptide that is known, at least in the case of HA, to insert into the host bilayer during the fusion process (28, 29). Moreover, the polypeptide chain reverses direction at the end of the N-terminal coiled coil and forms an α-helix that proceeds toward the N terminus of the molecule. These structural similarities suggest that there may be a common structural basis for viral membrane fusion mechanisms (30, 31).
Numerous studies suggest that target–cell receptor-induced release of gp120 from the viral surface is accompanied by a conformational change in gp41 from a native to a fusogenic state (8–10). By analogy with the influenza HA protein, it is likely that the structure of an α-helical, trimeric domain of HIV-1 gp41 comprises the core of the HIV-1 membrane fusion machinery, similar to the low pH induced conformation of influenza virus HA2 (26, 30, 31). Nevertheless, studies to date have not ruled out the possibility that this structure also corresponds to the native state of gp41. Further experiments are required to understand the nature of the conformational change in gp41, its role in membrane fusion, and the mechanism of HIV-1 entry into cells.
Acknowledgments
We thank Ellis Reinherz and Neville Kallenbach for critical reading of the manuscript. We also thank Don Wiley for making their models available for comparison. This work was supported by the start-up fund from the Cornell University Medical College to M.L. and in part by National Institutes of Health Grant AI27336 to Ellis Reinherz.
ABBREVIATIONS
- Env
envelope glycoprotein
- SIV
simian immunodeficiency virus
- HA
hemagglutinin
- SIRAS
single isomorphous replacement with anomalous scattering
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference ISTZ).
References
- 1.Luciw P A. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melinick J L, Monath T P, Roizman B, Straus S E, editors. Philadelphia: Lippincott; 1996. pp. 1881–1952. [Google Scholar]
- 2.Nara P L, Garrity R R, Goudsmit J. FASEB J. 1991;5:2437–2455. doi: 10.1096/fasebj.5.10.1712328. [DOI] [PubMed] [Google Scholar]
- 3.Freed E Q, Martin M A. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson D. Curr Biol. 1996;6:1051–1053. doi: 10.1016/s0960-9822(02)70661-1. [DOI] [PubMed] [Google Scholar]
- 5.Wiley D C, Skehel J J. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 6.Carr C M, Kim P S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 7.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 8.D’Souza M P, Harden V A. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 9.Moore J P, McKeating J A, Weiss R A, Clapham P R, Sattentau Q J. Science. 1991;252:1322–1323. [Google Scholar]
- 10.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 12.Hart T K, Kirsch R, Ellens H, Sweet R W, Lambert D M, Petteway S R, Jr, Bugelski P J. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan J S. Science. 1991;252:1322–1323. doi: 10.1126/science.1925547. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubay J W, Roberts S J, Brody B, Hunter E. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S S, Lee C N, Lee W R, McIntosh K, Lee T H. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wild C T, Dubay J W, Greenwell T, Baird T, Oas T G, McDanal C, Hunter E, Matthews T J. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delwart E J, Mosialos G, Gilmore T. AIDS Res Hum Retroviruses. 1990;6:703–706. doi: 10.1089/aid.1990.6.703. [DOI] [PubMed] [Google Scholar]
- 21.Chambers P, Pringle C R, Easton A J. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 22.Wild C T, Oas T, McDanal C B, Bolognesi D, Matthews T J. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S, Lin K, Strick N, Neurath A R. Nature (London) 1993;365:113. doi: 10.1038/365113a0. (letter). [DOI] [PubMed] [Google Scholar]
- 25.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 26.Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 27.Blacklow S C, Lu M, Kim P S. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 28.Stegmann T, Delfino J M, Richards F M, Helenius A. J Biol Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 29.Tsurudome M, Gluck R, Graf R, Falchetto R, Schaller U, Brunner J. J Biol Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- 30.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 31.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 35.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;39:813–817. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Brünger A T. A System for X-Ray Crystallography and NMR: x-plor. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 38.Laskowski R A, MacArthur M V, Moss D D, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 39.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 40.Lumb K J, Kim P S. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 41.Gonzaelez L, Woolfson D N, Alber T. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 42.Kaiser E T, Kezdy F J. Proc Natl Acad Sci USA. 1983;80:1137–1143. doi: 10.1073/pnas.80.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crick F H C. Acta Crystallogr. 1953;6:685–689. [Google Scholar]
- 44.Hodges R S, Sodek J, Smillie L B, Jurasek L. Cold Spring Harbor Symp Quant Biol. 1972;37:299–310. [Google Scholar]
- 45.McLachlan A D, Stewart M. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 46.Harbury P B, Kim P S, Alber T. Nature (London) 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 47.Betz S F, Bryson J W, DeGrado W F. Curr Opin Struct Biol. 1995;5:457–463. doi: 10.1016/0959-440x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 48.Kraulis P. J Appl Crystallogr. 1991;24:924–950. [Google Scholar]
- 49.Chakrabartty A, Doig A J, Baldwin R L. Proc Natl Acad Sci USA. 1993;90:10907–10908. doi: 10.1073/pnas.90.23.11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong Y, Zhou H X, Kallenbach N R. Protein Sci. 1995;4:1446–1456. doi: 10.1002/pro.5560040802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 52.O’Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 53.Wild C T, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T. AIDS Res Hum Retroviruses. 1995;11:323–325. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 54.Neurath A R, Lin K, Strick N, Jiang S. AIDS Res Hum Retroviruses. 1995;11:189–190. doi: 10.1089/aid.1995.11.189. [DOI] [PubMed] [Google Scholar]
- 55.Herskowitz I. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 56.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]