Abstract
Background
Genetic variation and rapid evolution are hallmarks of RNA viruses, the result of high mutation rates in RNA replication and selection of mutants that enhance viral adaptation, including the escape from host immune responses. Variability is uneven across the genome because mutations resulting in a deleterious effect on viral fitness are restricted. RNA viruses are thus marked by protein sites permissive to multiple mutations and sites critical to viral structure-function that are evolutionarily robust and highly conserved. Identification and characterization of the historical dynamics of the conserved sites have relevance to multiple applications, including potential targets for diagnosis, and prophylactic and therapeutic purposes.
Methodology/Principal Findings
We describe a large-scale identification and analysis of evolutionarily highly conserved amino acid sequences of the entire dengue virus (DENV) proteome, with a focus on sequences of 9 amino acids or more, and thus immune-relevant as potential T-cell determinants. DENV protein sequence data were collected from the NCBI Entrez protein database in 2005 (9,512 sequences) and again in 2007 (12,404 sequences). Forty-four (44) sequences (pan-DENV sequences), mainly those of nonstructural proteins and representing ∼15% of the DENV polyprotein length, were identical in 80% or more of all recorded DENV sequences. Of these 44 sequences, 34 (∼77%) were present in ≥95% of sequences of each DENV type, and 27 (∼61%) were conserved in other Flaviviruses. The frequencies of variants of the pan-DENV sequences were low (0 to ∼5%), as compared to variant frequencies of ∼60 to ∼85% in the non pan-DENV sequence regions. We further showed that the majority of the conserved sequences were immunologically relevant: 34 contained numerous predicted human leukocyte antigen (HLA) supertype-restricted peptide sequences, and 26 contained T-cell determinants identified by studies with HLA-transgenic mice and/or reported to be immunogenic in humans.
Conclusions/Significance
Forty-four (44) pan-DENV sequences of at least 9 amino acids were highly conserved and identical in 80% or more of all recorded DENV sequences, and the majority were found to be immune-relevant by their correspondence to known or putative HLA-restricted T-cell determinants. The conservation of these sequences through the entire recorded DENV genetic history supports their possible value for diagnosis, prophylactic and/or therapeutic applications. The combination of bioinformatics and experimental approaches applied herein provides a framework for large-scale and systematic analysis of conserved and variable sequences of other pathogens, in particular, for rapidly mutating viruses, such as influenza A virus and HIV.
Author Summary
Dengue viruses (DENVs) circulate in nature as a population of 4 distinct types, each with multiple genotypes and variants, and represent an increasing global public health issue with no prophylactic and therapeutic formulations currently available. Viral genomes contain sites that are evolutionarily stable and therefore highly conserved, presumably because changes in these sites have deleterious effects on viral fitness and survival. The identification and characterization of the historical dynamics of these sites in DENV have relevance to several applications such as diagnosis and drug and vaccine development. In this study, we have identified sequence fragments that were conserved across the majority of available DENV sequences, analyzed their historical dynamics, and evaluated their relevance as candidate vaccine targets, using various bioinformatics-based methods and immune assay in human leukocyte antigen (HLA) transgenic mice. This approach provides a framework for large-scale and systematic analysis of other human pathogens.
Introduction
Dengue viruses (DENVs) are mosquito-borne pathogens of the family Flaviviridae, genus Flavivirus, which are phylogenetically related to other important human pathogens, such as Yellow fever (YFV), Japanese encephalitis (JEV), and West Nile (WNV) viruses, among others. DENVs are enveloped, single-stranded RNA (+) viruses coding for a polyprotein precursor of approximately 3,400 amino acids, which is cleaved into three structural (capsid, C; precursor membrane and membrane, prM/M; envelope, E) and seven nonstructural proteins (NS1, 2a, 2b, 3, 4a, 4b and 5). Viral replication occurs in the cytoplasm in association with virus-induced membrane structures and involves the NS proteins. There are 4 genetically distinct DENV types, referred to as DENV-1 to -4, with multiple genotypic variants [1],[2]. DENVs are transmitted to humans primarily by Aedes aegypti mosquitoes and cause a wide range of symptoms from an unapparent or mild dengue fever (DF) to severe dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS) that may be fatal. It is estimated that more than 100 million people are infected each year, with up to several hundred thousand DHF/DSS cases [3]. To date, there is no licensed prophylactic vaccine and no specific therapeutic formulation available.
Adaptive immune responses include cellular responses to short peptides derived from self and foreign proteins by proteolysis. The peptides are presented to T-cell receptors (TCRs) by major histocompatibility complex (MHC) molecules, referred to as human leukocyte antigen (HLA) molecules in humans. HLA class I and class II molecules bind and present peptides to CD8 and CD4 T-cells, respectively, that play a critical role in antigen (Ag)-specific cytotoxic responses and the induction and maintenance of Ag-specific memory responses [4]–[6]. Peptides that are recognized by the T cells and trigger an immune response are referred to as T-cell determinants. One problem in developing a tetravalent DENV vaccine is the viral diversity [7], with rather low intra-type, but high inter-type variability, resulting in type-specific and type cross-reactive T-cell determinants [8]. This variability of related structures gives rise to a large number of variant peptide sequences with one or more amino acid differences that may function as alternative determinants, or altered peptide ligands [9], and affect anti-DENV host immunity [10],[11]. There is abundant evidence that interactions of memory T cells with peptide ligands bearing amino acid substitutions at TCR contact residues may alter T-cell activation and effector function [9], [12]–[15]. Even a single amino acid substitution can impair the function of T cells in a variety of ways, producing profoundly different phenotypes that range from modified stimulatory function to complete inhibition [14]. These findings suggest that infection or immunization with multiple DENV types, as is the case with some tetravalent vaccines, may lead to T-cell responses to variant peptides that might be deleterious. There is also the possibility that the altered-ligand phenomenon and cross-reactive T-cell responses, referred to as original antigenic sin, may play a role in DHF/DSS [7],[11],[16],[17]. Although the etiology of DHF and DSS is only partially understood, this consideration may have profound implications for the safety and efficiency of candidate vaccines.
The objective of this study was to search for sequence regions conserved across the majority of DENVs and representing potential immune targets [18]. Bioinformatics-based approaches were used to (a) extract all DENV sequences available in public databases, (b) identify and examine the structure-function relationship and distribution in nature of sequences that are highly conserved in the majority of DENVs (referred to as pan-DENV sequences), (c) analyze the variability of DENV sequences, and (d) examine the immune relevance of the conserved sequences as potential T-cell determinants that would be applicable to the majority of the human population worldwide [19]. We have also correlated the conserved DENV sequences to previously reported T-cell determinants and further identified novel candidate T-cell determinants by analyzing HLA-restricted immune responses in HLA transgenic mice.
Methods
Methodology overview
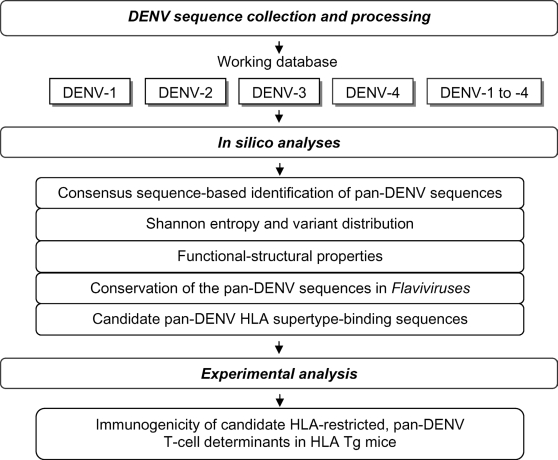
The bioinformatics approaches and rationale for the methodology adopted in this study have been previously described [20] and are summarized in Figure 1 .
Figure 1. Overview of the bioinformatics and experimental approaches employed for the identification and analysis of the pan-DENV sequences.
Data collection and sequence organization
DENV protein sequences were retrieved from the NCBI Entrez protein database in December 2005, and again in December 2007 for validation purposes, by use of a taxonomy ID search via the NCBI taxonomy browser [21]. The taxonomy IDs for DENV-1 to -4 were 11053, 11060, 11069 and 11070, respectively. The data for 2007 were processed separately from the 2005 dataset, but using identical procedures.
The sequences of the DENV proteins C, prM, E, NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5 were extracted from the database records ( Dataset S1 ) by multiple sequence alignments, and application of the known cleavage sites obtained from the annotation of the GenPept [21] reference polyprotein sequences of DENV-1 to -4 (AAF59976, P14340, AAM51537, AAG45437, respectively), and from the literature [22]. Grouping of the sequences of each DENV type was performed by BLAST [23] followed by CLUSTALX 1.83 [24] multiple sequence alignments. Both full-length and partial sequences of each DENV protein were used for analysis, and identical sequences were not removed from datasets, unless otherwise indicated. All multiple sequence alignments were manually inspected and corrected for misalignments.
Identification of pan-DENV sequences
The DENV protein sequences were examined by a consensus-sequence based approach [25] to identify sequence fragments that were common across the 4 types. The consensus sequences for the proteins of each type (intra-type consensus) were first derived by multiple sequence alignments to select the predominant residue at each amino acid position. The 4 intra-type consensus sequences for a given protein (one from each type) were then aligned to reveal sequence fragments identical across each of the types that were at least 9 amino acids long. This minimum length was chosen because it represents the binding core length of a majority of HLA-restricted T-cell determinants [26]. Only sequence fragments that were identical in at least 80% of the sequences of each of the 4 types were retained for further analyses. Peptides with residue X in the alignment were ignored from the percentage representation (i.e. frequency) computation. The 80% intra-type representation cut-off was chosen because 44 of the 46 sequence fragments that were common across the 4 DENV types exhibited intra-type representation of ≥81%, and those two that did not had significantly lower representation (∼56–67%) in one of the 4 types.
Information entropy analysis of pan-DENV sequences
Shannon information entropy [20],[27] was used to study the diversity of DENV protein sequences within each type (intra-type diversity) and across all DENVs (pan-DENV diversity) and to assess the predicted evolutionary stability of the identified pan-DENV sequences. All entropy analyses were carried out by using the in-house developed Antigenic Variability Analyser tool (AVANA) [28]. For immunological applications, the entropy measure for antigenic sequences was based on nonamer peptides [26], centered at any given position in the alignment. Applying Shannon's formula, the nonamer peptide entropy H(x) at any given position x in the alignment was computed by
where p(i, x) is the probability of a particular nonamer peptide i being centered at position x. The entropy value increases with n(x), the total number of peptides observed at position x; it is also sensitive to the relative frequency of the peptides; such that it decreases when one peptide is clearly dominant (i.e. the position is conserved). Only sequences that contain a valid amino acid at position x were used for the entropy computation, and the alignment gaps were ignored. Although gaps tend to occur in high-diversity regions, proteins that have a high fraction of gaps have reduced statistical support, yielding an artificially low entropy value; for this reason, positions where more than 50% of sequences contained a gap were discarded. Because of the statistical nature of the entropy measure, both complete protein and shorter fragment sequences were used in this computation. The first and last 4 positions in the alignment of each protein were not assigned any peptide entropy value as they cannot be the center of a nonamer.
In theory, nonamer entropy values can range from 0, for a completely conserved nonamer peptide in all sequences analyzed, to 39 (log2 209); in practice, however, the upper bound is very much lower for alignments of closely related sequences. For finite-size sets of sequences, entropy computations are affected by the sequence count in the alignment. For an alignment of N sequences, alignment size bias is proportional to 1/N [29]. This relationship allows a correction for size bias by applying to each alignment a statistical adjustment that estimates entropy values for an infinitely-sized alignment with analogous peptide distribution. To obtain such an estimate, the alignment was repeatedly randomly sampled to create smaller alignments of varying size, whose entropy was measured. At each alignment position, the entropy of these subset alignments of size N was plotted against 1/N, using a linear regression to extrapolate the entropy estimate for N→∞. The regression's coefficient of determination (r2) was used as a goodness-of-fit of the resulting estimate. In this study, size bias correction was applied to all entropy calculations, so that alignment sequence counts could be ignored in comparisons. All entropy values reported are therefore infinite-size set estimates, rather than the values directly computed from the alignments.
Nonamer variant analysis of pan-DENV sequences
Data from information entropy analysis were used to study the distribution of the representation of nonamer variant peptides in DENV sequences, within and across the types. For any given position x in the alignment, the combined representation of all nonamers, excluding the predominant peptide, was computed. The predominant nonamer was the peptide that was contained in the majority of the sequences at the position in the alignment. All the other peptides that differed by at least one amino acid from the predominant nonamer were defined as variants.
Functional and structural analyses of pan-DENV sequences
The known and putative structural and functional properties of pan-DENV sequences were searched in the literature and by use of the Prosite [30], via ScanProsite [31], and Pfam [32] databases. When possible, the sequences were mapped on the three-dimensional (3-D) structures of available DENV Ag in the PDB database [33] by use of ICM-Browser version 3.3 (www.molsoft.com). X-ray diffraction 3-D structures were visualized by use of the Corey, Pauling and Koltun (cpk) representation in the ICM-Browser.
Identification of pan-DENV sequences common to other viruses and organisms
Pan-DENV sequences that overlapped at least 9 consecutive amino acid sequences of other viruses and organisms were identified by performing BLAST search against viral protein sequences reported at NCBI (as of July 2007), excluding DENV sequences (parameters set: limit by Entrez query “txid10239[Organism:exp] NOT txid12637[Organism:exp]”; automatically adjust parameters for short sequences option enabled; low-complexity filter disabled; alignments: 20,000), and against protein sequences of all organisms excluding viruses (parameters set: limit by Entrez query “Root[ORGN] NOT Viruses[ORGN] NOT txid81077[ORGN]”; automatically adjust parameters for short sequences option enabled; low-complexity filter disabled; alignments: 20,000). The keyword “NOT txid81077 [ORGN]” was used to remove artificial sequence hits.
Identification of known and predicted pan-DENV HLA supertype binding sequences
Both literature search and query against the Immune Epitope Database [34] (www.immuneepitope.org) were performed to detect reported immunogenic, human T-cell determinants (both class I and II) of DENV that either fully or partially overlapped with the pan-DENV sequences. In addition, dedicated algorithms based on several prediction models were used to identify candidate putative HLA-binding sequences to multiple HLA class I and II supertype alleles within the pan-DENV sequences. Putative HLA supertypes class I-restricted peptides were identified by use of NetCTL [35], Multipred [36], ARB [37], and class II-restricted peptides by Multipred and TEPITOPE [38]. Further, the intra-type representation of the putative T-cell determinants was analyzed.
The NetCTL 1.2 algorithm (www.cbs.dtu.dk/services/NetCTL/) predicts peptides restricted by 12 HLA class I supertypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58 and B62). The algorithm integrates the predictions of HLA binding, proteasomal C-terminal cleavage and transport efficiency by the transporter associated with antigen processing (TAP) molecules. HLA binding and proteasomal cleavage predictions are performed by an artificial neural networks (ANN) method, while TAP transport efficiency is predicted using a weight matrix method. The parameters used for NetCTL prediction were: 0.15 weight on C terminal cleavage (default), 0.05 weight on TAP transport efficiency (default), and 0.5 threshold for HLA supertype binding, which was reported to be optimal (sensitivity (SN), 0.89 and specificity (SP), 0.94) in a large benchmark study containing more than 800 known class I T-cell determinants [35].
The TEPITOPE software (2000 beta version; courtesy of J. Hammer) utilizes quantitative matrix-based motifs, obtained from experimental scanning of the binding of P1-anchored designer peptides to soluble HLA-DR molecules in in-vitro competition assays, to predict peptides binding to 25 common HLA-DR alleles (DRB1*0101, *0102, *0301, *0401, *0402, *0404, *0405, *0410, *0421, *0701, *0801, *0802, *0804, *0806, *1101, *1104, *1106, *1107, *1305, *1307, *1311, *1321, *1501, *1502, and DRB5*0101) [38],[39]. The parameters for TEPITOPE predictions were: 5% quantitative threshold and putative determinants with a 10-fold inhibitory residue included. Nonamer peptides predicted to bind at least 10 out of the 25 HLA-DR alleles were selected as putative supertype-restricted determinants.
Multipred (research.i2r.a-star.edu.sg/multipred/) is a computational system for the prediction of peptides that bind to HLA class I supertypes A2 and A3 and class II HLA-DR supertype [36]. The HLA alleles selected to represent these supertypes by Multipred were as follows: A2 supertype, A*0201, *0202, *0203, *0204, *0205, *0206, *0207 and *0209; A3 supertype, A*0301, *0302, *1101, *1102, *3101, *3301 and *6801; DR supertype, DRB1*0101, *0301, *0401, *0701, *0801, *1101, *1301, and *1501. Hidden Markov model (HMM) and ANN methods are the predictive models of Multipred; both have been optimized and show similar performances [36]. The sum thresholds used for prediction of peptides restricted to the three HLA supertypes by ANN and HMM methods were: A2, 31.33 (ANN; SN = 0.80 and SP = 0.83) and 47.08 (HMM; SN = 0.80 and SP = 0.78); A3, 24.53 (ANN; SN = 0.90 and SP = 0.95) and 37.58 (HMM; SN = 0.80 and SP = 0.87); and DR, 23.42 (ANN; SN = 0.90 and SP = 0.92) and 51.08 (HMM; SN = 0.90 and SP = 1.00). Consensus predictions of the two methods were taken as final predictions for each HLA supertype.
The ARB matrix method (epitope.liai.org:8080/matrix/matrix_prediction.jsp) is based on a matrix of coefficients to predict IC50 values [37]. The HLA class I alleles predicted by ARB were grouped according to the current supertype classification [19],[40] and supertypes containing more than two alleles were selected, namely A2 (A*0201, *0202, *0203, *0206, and *6802), A3 (A*0301, *1101, *3101, *3301 and *6801), B7 (B*0702, A*3501, *5101, *5301, and *5401), and B44 supertypes (B*4001, *4002, *4402, *4403, and *4501). The prediction threshold value chosen for optimum sensitivity and specificity was IC50≤1000 nM and nonamer peptides predicted to bind 3 or more alleles of the supertype were considered as putative promiscuous HLA supertype-restricted determinants.
ELISpot analysis of HLA-DR restricted determinants in pan-DENV sequences
All experiments were approved by the Johns Hopkins University Institutional Animal Care and Use Committee. Murine H-2 class II-deficient, HLA-DR2 [41], HLA-DR3 [42],[43], HLA-DR4 (referred to as DR4/IE) [44] and HLA-DR4/human CD4 (huCD4) [45],[46] Tg mice were used, bred and maintained in the Johns Hopkins University School of Medicine Animal Facility. Specific pathogen-free (SFP) colonies were maintained in a helicobacter-negative mice facility. The HLA-DR expression of the experimental transgenic mice was evaluated by flow cytometry.
Mice were immunized subcutaneously at the base of the tail, twice at two weeks interval, with pools of overlapping peptides covering the DENV-3 protein (15–17 aa, overlapping by 10–11 aa) (Schafer-N Inc., Copenhagen, Denmark; BEI Resources, Manassas, VA). Peptide pools (73–155 peptides per pool) contained 1 µg of each peptide and were emulsified (1∶1) in TiterMax adjuvant (TiterMax USA, Inc.). An aqueous preparation of TiterMax (1∶1) was used as a negative control. Two weeks after the second immunization, the mice were sacrificed and HLA-DR-restricted CD4 T cell reponses were assessed by ex vivo IFN-γ ELISpot assay using CD8-depleted splenocytes. Each target peptide was tested in duplicate. Spot-forming cell (SFC) counts were normalized to 106 cells. The results were considered significant when the average SFC minus two standard deviations (SD) was greater than the average of the background plus two SD; and the average values were greater than 10 SFC per 106 splenocytes. The initial screening assays were performed with peptide matrices [47], followed by assays with the relevant individual peptides (Nascimento et al., manuscript in preparation).
Results
Dengue virus type protein datasets
A total of 9,512 and 12,404 complete and partial DENV protein sequences were collected from the NCBI Entrez protein database of December 2005 and 2007, respectively, representing an increase of approximately 30% (2892 sequences) in the 24-months interval (Table 1). The total number of sequences (2007) varied from 4,011 for DENV-2 to 1,415 for DENV-4 and from 3,845 for E to 523 for NS4a proteins. Most of the individual protein sequences originated from DENV strains that were unique variants with respect to the entire polyprotein, but were identical to other strains with respect to individual proteins [48].
Table 1. Number and distribution of reported DENV protein sequences.
| DENV proteinb | No. of sequencesa | ||||||||||
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | Total | |||||||
| 2005 | 2007 | 2005 | 2007 | 2005 | 2007 | 2005 | 2007 | 2005 | 2007 | Increase | |
| C | 194 | 298 | 266 | 311 | 414 | 547 | 117 | 122 | 991 | 1278 | 287 |
| prM | 206 | 311 | 353 | 404 | 458 | 590 | 207 | 225 | 1224 | 1530 | 306 |
| E | 852 | 1051 | 1277 | 1518 | 716 | 910 | 338 | 366 | 3183 | 3845 | 662 |
| NS1 | 410 | 565 | 640 | 752 | 201 | 308 | 142 | 159 | 1393 | 1784 | 391 |
| NS2a | 150 | 238 | 132 | 173 | 90 | 169 | 121 | 125 | 493 | 705 | 212 |
| NS2b | 136 | 224 | 130 | 163 | 104 | 183 | 40 | 44 | 410 | 614 | 204 |
| NS3 | 98 | 186 | 145 | 178 | 216 | 297 | 30 | 34 | 489 | 695 | 206 |
| NS4a | 91 | 178 | 128 | 162 | 70 | 151 | 28 | 32 | 317 | 523 | 206 |
| NS4b | 89 | 176 | 129 | 163 | 70 | 150 | 109 | 113 | 397 | 602 | 205 |
| NS5 | 92 | 179 | 151 | 187 | 181 | 267 | 191 | 195 | 615 | 828 | 213 |
| Total | 2318 | 3406 | 3351 | 4011 | 2520 | 3572 | 1323 | 1415 | 9512 | 12404 | 2892 |
Collected from the NCBI Entrez protein database
Manually processed after multiple sequence alignments and use of the known DENV cleavage sites
Conserved pan-DENV sequences
The consensus-sequence approach [20],[25] identified a total of 44 pan-DENV sequences of at least 9 amino acids that were present in ≥80% of all sequences of each DENV type for both 2005 and 2007 datasets ( Figure 2 ; Table S1). Strikingly, 34 of the 44 (∼77%) were conserved in ≥95% of all reported DENV sequences. The size of the pan-DENV sequences ranged from 9 to 22 amino acids, with a combined size of 514 residues, corresponding approximately to 15% of the complete DENV polyprotein (∼3390 amino acids) ( Table 2 ). The vast majority (42/44) of the pan-DENV sequences were localized in the NS proteins, with 17, 12, 7 and 5 sequences found in NS5, NS3, NS1 and NS4b, respectively, and 1 in the NS4a protein. Notably, the remaining two pan-DENV sequences were localized in the E protein. No region of at least 9 amino acids and conserved in ≥80% of the sequences of each DENV type was found in the C, prM, NS2a and NS2b proteins. The largest size of the combined pan-DENV sequences was in the NS5 protein, representing a total of 215 amino acid positions covering ∼24% of the protein, followed by NS3, NS1 and NS4b with 122, 74 and 69 amino acid positions covering ∼20, ∼21 and ∼28% of the corresponding proteins, respectively. The two pan-DENV sequences in the E protein had a combined size of only 25 amino acids, corresponding to ∼5% of the protein.
Figure 2. Pan-DENV sequences and their representations in the 4 DENV types.
The 44 pan-DENV sequences of at least 9 amino acids that were found present in ≥80% of the recorded sequences of each DENV type are shown. The representation values are shown for the 2005 dataset; see Table S1 for values of both 2005 and 2007 datasets. Amino acid positions were numbered according to the sequence alignments of the 4 DENV types. The corresponding proteins are indicated on the left.
Table 2. Distribution and size of the pan-DENV sequences.
| DENV protein | Size (aa) | Pan-DENV sequencesa | ||
| No. | Sizeb | % of proteinc | ||
| C | 113–115 | 0 | 0 | 0 |
| prM | 166 | 0 | 0 | 0 |
| E | 493–495 | 2 | 25 | 5 |
| NS1 | 352 | 7 | 74 | 21 |
| NS2a | 218 | 0 | 0 | 0 |
| NS2b | 130 | 0 | 0 | 0 |
| NS3 | 618–619 | 12 | 122 | 20 |
| NS4a | 150 | 1 | 9 | 6 |
| NS4b | 245–249 | 5 | 69 | 28 |
| NS5 | 900–904 | 17 | 215 | 24 |
| Total | 3387–3398 | 44 | 514 | 15 |
Sequences of at least 9 amino acids that were represented in ≥80% of all DENV sequences of each type
Combined amino acid size of all pan-DENV sequences in the protein
Percentage of the combined pan-DENV sequence size over that of the corresponding protein size
In large-scale genomic analyses such as this study, biases may result from the collection of completely or partially overlapping redundant sequences, corresponding to identical or highly similar circulating DENV isolates sequenced by various dengue surveillance programs in different countries. Although to some extent this redundancy may be accepted as a reflection of the incidence of the corresponding DENV isolates in nature, we assessed its potential bias effect by repeating the analysis of conservation after discarding duplicate sequences from the datasets. The analysis of unique sequences identified all the pan-DENV sequences that were identified when including duplicates ( Figure 2 ), except for NS112–20, NS125–35 and NS5597–616. Therefore, the presence of duplicates in the DENV datasets did not significantly affect the results. Although the removal of duplicates does not fully compensate for biases in the datasets, the removal of highly similar sequences, which may have been generated from relatively large sequencing efforts in single outbreaks, was deemed undesirable, since such arbitrary selection would introduce additional biases.
Evolutionary diversity of DENV protein nonamer peptide sequences
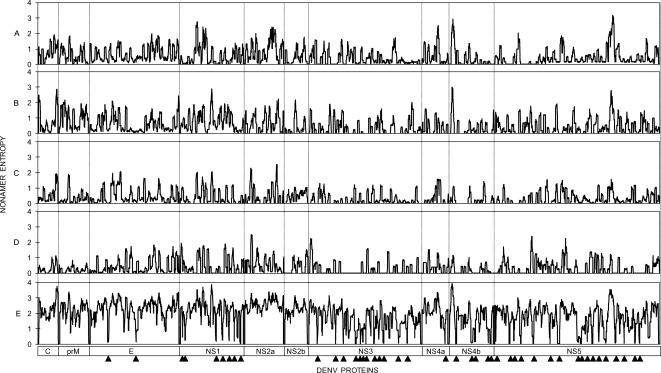
The evolutionary diversity of each DENV type, and the 4 types combined, was studied by use of Shannon information entropy [27], modified to examine the variability of nonamer peptide sequences, as described in the Methods . The entropy of the proteome of the recorded viruses of each type showed numerous long regions of low entropy (≤1), reflecting the relatively high degree of intra-type sequence conservation, in particular in the NS3, NS4b and NS5 proteins ( Figure 3A–D ). Overall, the average intra-type nonamer entropy values of the individual protein sequences of DENV-1, -2, -3 and -4 ranged from 0.2 for the DENV-4 NS4b to 1.0 for DENV-2 prM ( Figure S1 ). Of note, however, were the marked differences in the relative degree of entropy of each protein between the 4 DENV types. For example, NS4b had the least diversity of the proteins of 3 types, but was replaced in DENV-2 by NS2b, which was the second most variable in DENV-3. The consequence of the differences in the sequences of each protein between the 4 types was a marked increase in the peptide entropy across the DENV 1-4 proteomes ( Figure 3E ), with average peptide entropy ranging from 1.6 for NS3 to 2.6 for NS2a (Figure S1), except for 44 sharply defined regions of low nonamer entropy (≤0.5) where the sequences were highly conserved in all DENVs ( Figure 3E ), with no significant difference between the 2005 and 2007 datasets ( Table S2 ). Majority of the pan-DENV sequences had entropy values of ≤0.3, corresponding to the intra-type representation of ≥90%. Thus, the congruent consensus- and entropy-based analyses of the DENV nonamer peptides revealed highly conserved and evolutionarily stable pan-DENV sequences distributed in several viral proteins, despite the marked viral diversity defining multiple DENV types, genotypes and variants [49].
Figure 3. Shannon entropy of nonamer peptides within and across DENV types sequences.
The entropy values were computed from the alignments of DENV sequences using the Antigenic Variability Analyzer software, as described in the Methods . Values were plotted for DENV-1 (A), DENV-2 (B), DENV-3 (C), DENV-4 (D), and all 4 DENV types (E) sequences (2005 dataset). Entropy values around protein cleavage sites are non significant, since the corresponding positions cannot be the center of a nonamer (see Methods ). The triangles below indicate the locations of the pan-DENV sequences in the corresponding proteins.
Representation of DENV variant nonamer peptide sequences
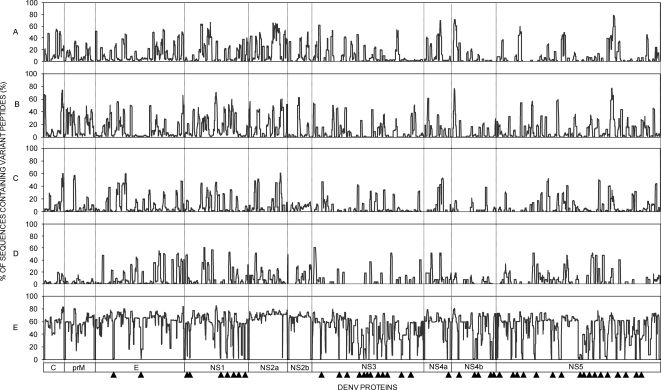
The combined representation of variant peptides that differed by at least one amino acid from the predominant peptide was also analyzed at each nonamer position. Examples of this analysis for DENV-3 proteins are shown in Table 3 . Nonamers that lack entropy (zero entropy) have one sequence in all of the recorded virus isolates, and therefore have no variants. Positions with high entropy can contain many different variant peptides, each at lesser (or equal) frequency than the predominant peptide. The combined representation of variant peptides at each nonamer position across the proteome of each individual DENV type was generally low, representing less than 10% of the corresponding sequences, except for some positions where it was more than 50% ( Figure 4A–D ). Notably, the nonamer position with the highest combined variant representation for each DENV type was found in the nonstructural proteins and not the structural ones, with representation values ranging from ∼61 to ∼78% (DENV-1 NS5, DENV-2 NS5, DENV-3 NS2a, and DENV-4 NS1 and NS3 proteins). When representations of variants across all DENVs were calculated, the majority of all nonamer sites contained variants that together represented ∼60–85% of the total DENV sequences at that site (the highest representation of ∼85% was in the NS1 protein) ( Figure 4E ). This was in striking contrast to the 0 to ∼5% combined representation of variants at each nonamer position in the pan-DENV sequences, with no significant difference between the 2005 and 2007 datasets ( Table S2 ). The majority of all nonamer sites in the pan-DENV sequences lacked variant or contained variants that together represented <1% of all recorded DENVs. These data further illustrate the extremely high genetic stability of the 44 pan-DENV sequences, among all recorded DENV sequences and demonstrate that irrespective of the high variability between the sequences of the 4 DENV types, the representation of variants in the pan-DENV sequences was almost negligible.
Table 3. Examples of the distribution of variant nonamer peptides in DENV-3.
| DENV-3 protein | Nonamer position | No. of sequences | Nonamer peptidesa | Representation of peptides | Combined % representation of variantsb | Nonamer entropyc |
| E | 14 | 479 | DFVEGLSGA | 479 (100%) | 0 | 0 |
| NS2a | 176 | 64 | LAGISLLPV | 25 (39%) | 61 | 2.4 |
| LAGVSLLPV | 11 (17%) | |||||
| LAGVSLLPL | 9 (14%) | |||||
| LAVISLLPV | 9 (14%) | |||||
| LAGISLLPL | 6 (9%) | |||||
| LAGISLFPV | 2 (3%) | |||||
| LAGISLMPV | 2 (3%) | |||||
| NS4a | 86 | 68 | SIGLICVVA | 39 (57%) | 43 | 1.5 |
| SIGLICVIA | 19 (28%) | |||||
| SIGLICVIV | 8 (13%) | |||||
| SIGLICVAA | 2 (3%) |
The predominant peptide is underlined
Variants include all the peptides at the position, except the predominant
Entropy value of all the peptides at the position (predominant peptide included)
Figure 4. Percentage representations of variant nonamer peptides within and across DENV types sequences.
The percentage of sequences that contained variant peptides at each nonamer position are shown for DENV-1 (A), DENV-2 (B), DENV-3 (C), DENV-4 (D), and all 4 DENV types (E) (2005 dataset). Values around protein cleavage sites are non significant (see Figure 3). The triangles below indicate the locations of the pan-DENV sequences in the corresponding proteins.
Functional and structural correlates of the pan-DENV sequences
Highly conserved protein sequences are likely to represent critical sites and domains [50]. A search of the literature and the Prosite and Pfam databases [30],[32] revealed that 27 of the 44 pan-DENV sequences were associated with biological activities ( Table S3 ); the functional significance of the remaining 17 pan-DENV sequences was not known. The two pan-DENV sequences in the E protein corresponded to the fusion peptide (positions 98 to 110) and dimerisation domain [51],[52]. In NS3, one pan-DENV sequence corresponded to the peptidase family S7 (Flavivirus serine protease) domain and comprised the His-51 catalytic residue [53], 3 sequences corresponded to known/putative Flavivirus Asp-Glu-Ala-Asp/His (DEAD/H) domain associated with ATP-dependent helicase activity [54], and two sequences were predicted to be required for cell attachment and targeting signal for microbodies. In NS5, one pan-DENV sequence corresponded to the conserved methyltransferase (MTase) S-adenosyl-L-methionine binding motif I (positons 77–86) involved in viral RNA capping [55], and two sequences corresponded to RNA dependent RNA polymerase (RdRp) domain [56]. Furthermore, 6 of the 27 pan-DENV sequences were predicted to exhibit post-translational modification(s), including N-glycosylation, protein kinase C and casein kinase II phosphorylation, N-myristoylation and/or amidation ( Table S3 ).
It is generally recognized that amino acids buried inside proteins are subject to greater interactions and packing constraints [57] than those exposed on the outer surface. Although none of the DENV protein structures in the protein data bank (PDB) [33] was full-length, 19 of the 44 pan-DENV sequences could be mapped on the available crystallographic models of the E ectodomain (Accession No. 1OAN; 394 out of 493–495 residues), NS3 (1BEF and 2BMF, 181 and 451 out of 618–619 residues, respectively) and NS5 fragments (1R6A, 295 out of 900–904 residues). Eleven of the 19 pan-DENV sequences were buried, 2 partially exposed and 6 exposed at the surface of the corresponding structures ( Figure S2 ). However, these results should be considered preliminary until full-length 3-D structures are available.
Distribution of pan-DENV sequences in nature
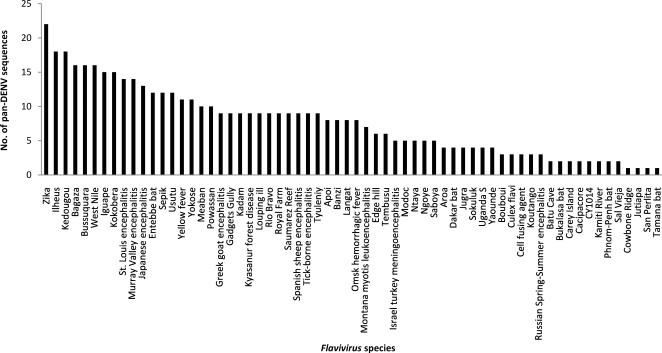
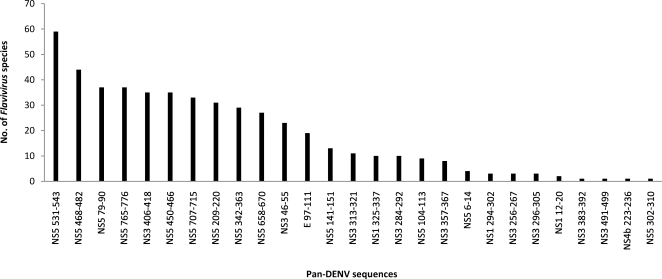
Twenty-seven (27) of the 44 pan-DENV sequences overlapped at least 9 amino acid sequences of as many as 64 other viruses of the family Flaviviridae, genus Flavivirus ( Figure 5 ). Zika virus shared 22 of the 27 sequences; Ilheus and Kedougou viruses, 18; and representatives of some of the significant human pathogens, West Nile, St. Louis encephalitis, Japanese encephalitis, Yellow fever and Tick-borne encephalitis viruses, shared from 16 to 9 pan-DENV sequences. Thirteen (13) of the 27 sequences represented NS5, of which 9 were present in at least 27 Flavivirus species; 9 represented NS3, of which two were found in 35 and 23 species; one E sequence was found in 19 species; and the remaining were in NS1 and NS4b ( Figure 6 ; Table S4). Five (5) of the 27 were associated with known biological activities (NS579–90 MTase, NS5658–670 RdRp, NS346–55 peptidase S7, NS3284–292 DEAD/H and E97–111 dimerisation/fusion domains). Interestingly, two sequences, NS3406–418 and NS5597–616, overlapped 9 amino acid sequences of the cell fusing agent virus polyprotein-like protein from the mosquito Aedes albopictus [58], and the phage-related tail fibre protein-like protein from the bacteria Chromohalobacter salexigens DSM 3043, respectively.
Figure 5. Number of pan-DENV sequences conserved in the different Flaviviruses.
Figure 6. Number of Flaviviruses shared by the pan-DENV sequences.
The representation of many of the pan-DENV sequences was high among known sequences of several of the highly studied Flaviviruses ( Table S4 ): St. Louis encephalitis, West Nile, Japanese encephalitis, Murray Valley encephalitis, Usutu, Kokobera, Ilheus, Tick-borne encephalitis, Langat, Omsk hemorrhagic fever, Louping ill, Powassan, Kyasanur forest disease and Yellow fever viruses. Protein sequence data for the rest of the Flaviviruses that shared pan-DENV sequences was limited (<10 sequences) in the public database. Seven of the 27 pan-DENV sequences, NS112–20, NS3256–267, NS3383–392, NS3491–499, NS4b223–236, NS56–14 and NS5302–310, were present in a few species with less than 10 reported total sequences ( Table S4 ).
Known and predicted HLA supertype-restricted, pan-DENV T-cell determinants
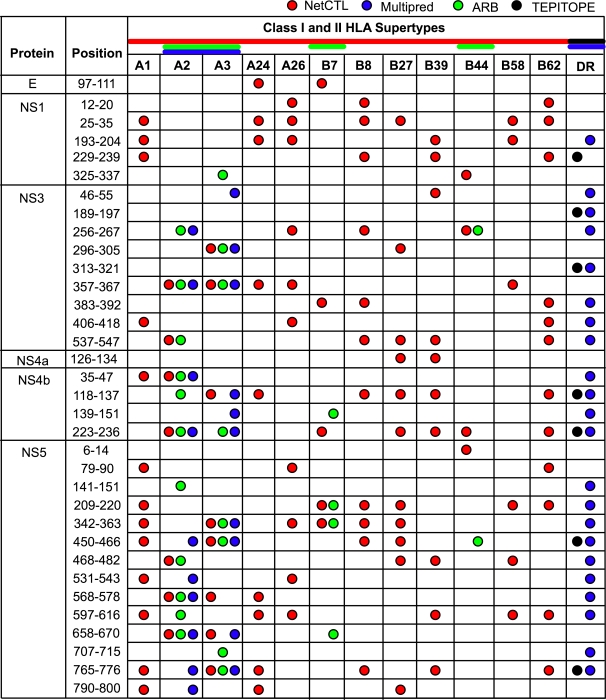
Literature survey and database search revealed that 10 of the pan-DENV sequences (9 in NS3, one in E) overlapped at least 9 amino acids of 15 previously reported DENV T-cell determinants immunogenic in human, with their HLA restriction, when known, showed both class II (DR*15, DPw2) and class I (A*11) specificities ( Table 4 ). Further evaluation of the immune-relevance of the pan-DENV sequences included a search for candidate putative promiscuous HLA supertype-restricted T-cell determinants within these regions by use of several computational algorithms: NetCTL [35], Multipred [36], ARB [37] and TEPITOPE [38]. Overall, 34 of 44 (∼77%) pan-DENV sequences ( Figure 7 ), identified in the NS5, NS3, NS1, E and NS4a proteins were predicted to contain 100 supertype-restricted binding nonamers ( Table S5 ). The majority (88/100) of the predicted promiscuous HLA-binding nonamers were present in ≥95% of the sequences of each DENV type ( Table S6 ). Thirty-one (∼91%) of the 34 putative supertype pan-DENV sequences contained HLA-binding nonamers for multiple HLA supertypes. Clusters (hotspots) of two or more overlapping HLA-binder nonamer core peptides were present in 27 (∼79%) of the 34 putative supertype pan-DENV sequences. About half (14/27) of these clusters contained three or more nonamer binders overlapping by 8 amino acids, covering most or the entire corresponding conserved region.
Table 4. Reported human T-cell determinants in the pan-DENV sequences.
| DENV protein | Pan-DENV sequencea | Immunogenic T-cell determinantsb | |||
| Sequencec | T subset | HLA Ag | Reference(s) | ||
| E | 252VLGSQEGAMH261 | KKQDVVVLGSQEGAM | - | - | [76] |
| NS3 | 46FHTMWHVTRG55 | TFHTMWHVTRGAVLM | CD4 | - | [76] |
| 148GLYGNGVVT156 | KVVGLYGNGVVTRSG | CD4 | DR*15 | [76] | |
| 189LTIMDLHPG197 | KRLTIMDLHPGAGKT | CD4 | - | [72] | |
| RKLTIMDLHPGSGKT | CD4 | - | [72] | ||
| RKLTIMDLHPGAGKT | CD4 | - | [72] | ||
| RNLTIMDLHPGSGKT | CD4 | - | [72] | ||
| 256EIVDLMCHATFT267 | EHTGREIVDLMCHAT | CD4 | - | [76] | |
| EIVDLMCHATFTMRL | CD4 | - | [76] | ||
| EIVDLMCHAT | CD4 | DPw2 | [77],[78] | ||
| 284MDEAHFTDP292 | LIIMDEAHFTDPASI | - | - | [76] | |
| 313IFMTATPPG321 | AGIFMTATPPGSRDP | - | - | [76] | |
| 357GKTVWFVPSIK367 | TVWFVPSIK | CD8 | A*11 | [16] | |
| 383VIQLSRKTFD392 | KKVIQLSRKTFDSEY | - | - | [76] | |
| 406VVTTDISEMGANF418 | NDWDFVVTTDISEMG | - | - | [76] | |
Amino acid positions numbered according to the sequence alignments of the 4 DENV types
Dashes, not determined
Sequences present in the pan-DENV sequences are underlined
Figure 7. Candidate putative HLA supertype-restricted, pan-DENV T-cell determinants predicted by computational algorithms.
Amino acid positions of the pan-DENV sequences are numbered according to the sequence alignments of the 4 DENV types; the corresponding DENV proteins are indicated on the left. Predicted HLA-restricted T-cell determinants were identified using NetCTL, Multipred, ARB and TEPITOPE algorithms (see Methods).
Immunogenicity of HLA-DR-restricted pan-DENV sequences in HLA Tg mice
The immunogenicity of the pan-DENV sequences was also analyzed by assay of peptide-specific HLA-restricted T-cell responses in murine H-2 class II-deficient, HLA-DR Tg mice expressing 3 prototypic HLA-DR alleles, corresponding to the divergent subgroups HLA-DR2 (DRB1*1501), HLA-DR3 (DRB1*0301), and HLA-DR4 (DRB1*0401). Mice were immunized with pools of overlapping peptides covering the sequences of the E, NS1, NS3, and NS5 proteins of DENV-3, and HLA-DR-restricted CD4 T-cell responses were assessed by IFN-γ ELISpot assays using CD8-depleted splenocytes. Thirty peptides eliciting positive T-cell responses in the HLA Tg mice contained 9 or more consecutive amino acids of 22 pan-DENV sequences, that were localized in the NS5 (11), NS3 (6), NS1 (4), and E proteins (one) ( Table 5 ). Overall, 9, 10 and 18 peptides elicited positive responses in HLA-DR2, -DR3, and/or -DR4 Tg mice, respectively; 20 corresponded to sequences of NS5, 10 of NS3, 6 of NS1, and one of E. Furthermore, at least 7 of the pan-DENV sequences, all localized in the NS5 and NS1 proteins, contained promiscuous T-cell determinants for multiple HLA-DR alleles ( Table 5 ). These data, together with those previously reported ( Table 4 ), showed that a minimum of 26 of the 44 pan-DENV sequences, distributed predominantly in the NS5 and NS3 proteins, and to a lesser extent in NS1 and E, contained numerous HLA-restricted class II and/or class I determinants demonstrated by assays of T-cell responses in vivo.
Table 5. Immunogenicity of the pan-DENV sequences in HLA-DR transgenic mice.
| DENV protein | Pan-DENV sequenceb | Ag-specific CD4 T-cell responsesa | |||
| Peptide sequences (DENV-3)c | IFN-γ-SFC/106splenocytes±SDd | ||||
| DR2 | DR3 | DR4 | |||
| E | 252VLGSQEGAMH261 | PEVVVLGSQEGAMHT | - | - | 88±34 |
| NS1 | 193AVHADMGYWIES204 | AVHADMGYWIESQKN | - | 17±1 | - |
| 229HTLWSNGVLES239 | WPKSHTLWSNGVLES | - | 129±3* | - | |
| HTLWSNGVLESDMII | - | 131±103 | 37±3 | ||
| 266GPWHLGKLE274 | HTQTAGPWHLGKLE | - | 333±6 | - | |
| 294RGPSLRTTT302 | TRGPSLRTTTVSGKL | - | - | 11±4 | |
| NS3 | 189LTIMDLHPG197 | KKRNLTIMDLHPGSG | - | - | 50±16 |
| 296AARGYISTRV305 | ASIAARGYISTRVGM | 40±14 | - | - | |
| ARGYISTRVGMGEAA | 9±4 | - | - | ||
| 313IFMTATPPG321 | EAAAIFMTATPPGTA | - | - | 474±116 | |
| IFMTATPPGTADAFP | - | - | 323±287 | ||
| 357GKTVWFVPSIK367 | TDFAGKTVWFVPSIK | 48±15 | - | - | |
| GKTVWFVPSIKAGND | 396±14 | - | - | ||
| 383VIQLSRKTFD392 | KKVIQLSRKTFDTEY | - | 21±3 | - | |
| 406VVTTDISEMGANF418 | FVVTTDISEMGANFK | - | - | 408±104 | |
| TDISEMGANFKADRV | - | 152±33 | - | ||
| NS5 | 302TWAYHGSYE310 | DENPYKTWAYHGSYEVK | 126±10* | - | 14±5 |
| TWAYHGSYEVKATGSA | 161±20* | - | 63±17 | ||
| 342AMTDTTPFGQQRVFKEKVDTRT363 | MVTQMAMTDTTPFGQQR | - | - | 28±0* | |
| 450CVYNMMGKREKKLGEFG466 | GSCVYNMMGKREKKLGE | - | - | 13±2 | |
| 505SGVEGEGLH513 | NSYSGVEGEGLHKLGYI | - | - | 184±15 | |
| 531YADDTAGWDTRIT543 | KIPGGAMYADDTAGWDT | - | - | 46±3 | |
| 568IFKLTYQNKVV578 | ANAIFKLTYQNKVVKVQ | 577±384 | - | 24±9* | |
| 597DQRGSGQVGTYGLNTFTNME616 | VMDIISRKDQRGSGQVG | - | 88±1 | - | |
| 658RMAISGDDCVVKP670 | VERLKRMAISGDDCVVK | - | 159±24 | 16±6 | |
| MAISGDDCVVKPIDDRF | - | 249±39 | - | ||
| 707VPFCSHHFH715 | DWQQVPFCSHHFHELIM | 32±8* | 34±11 | - | |
| 765LMYFHRRDLRLA776 | MYFHRRDLRLASNAI | 75±16* | - | 33±9 | |
| 790PTSRTTWSIHA800 | VHWVPTSRTTWSIHAHH | - | - | 83±1 | |
| SRTTWSIHAHHQWMTTE | - | - | 122±46 | ||
Assessed by IFN-γ ELISpot assay in HLA-DR2 (DRB1*1501), HLA-DR3 (DRB1*0301) and HLA-DR4 (DRB1*0401) Tg mice immunized with DENV-3 peptides (see Methods)
Amino acid positions numbered according to the sequence alignments of the 4 DENV types
Sequences present in the pan-DENV sequences are underlined
SFC, spot-forming cells; SD, standard deviation. Representative results from at least two immunized Tg mice are shown, except when indicated by an asterisk
Discussion
In this study, we identified and characterized pan-DENV sequences that were highly conserved in all recorded DENV isolates. The large number of sequences analyzed (12,404 as of December 2007), and their wide distribution in terms of geography and time (1945–2007) (data not shown), offered information for a broad survey of DENV protein diversity in nature. The 44 pan-DENV protein sequences of at least 9 aa, covering 514 aa or about 15% of the complete DENV polyprotein of ∼3390 aa, were conserved in at least 80% of all recorded DENV sequences, and 34 of the 44 (∼77%) were conserved in ≥95% of DENV sequences. All the 44 were in the non-structural proteins except for the two E sequences. These conserved sequences have shown remarkable stability over the entire history of DENV sequences deposited in the NCBI Entrez protein database, as illustrated by their low peptide entropy values and variant frequencies. In addition, 27 of the pan-DENV sequences were conserved in 64 other Flaviviruses, as further evidence of prolonged evolutionary stability within this genus, as previously discussed [59]–[61]. Two are also present in the proteomes of the Aedes albopictus mosquito and the bacteria Chromohalobacter salexigens, possibly in keeping with recent reports of the genetic recombination between phyla [58]. It is likely that these pan-DENV sequences have been under selection pressure to fulfill critical biological and/or structural properties, some of which have been identified for the E (fusion peptide, dimerization domain), NS3 (peptidase S7, DEAD/H domains) and NS5 proteins (MTPase, RdRp domains) [51]–[56]. Hence, these conserved sequences are unlikely to significantly diverge in newly emerging DENV isolates in the future, and represent attractive targets for the development of specific anti-viral compounds and vaccine candidates.
There also is evidence that many of the conserved sequences are immunologically relevant. A majority (26/44) contained at least 9 amino acids overlapping with a total of 45 peptides that have been reported to be immunogenic in humans and/or HLA-DR Tg mice. In addition, putative T-cell determinants for 12 major HLA class I supertypes and for class II DR supertype, with broad application to the immune responses of human population worldwide, were predicted by computational analysis. Some of the putative T-cell determinants were predicted to be promiscuous to multiple HLA supertypes, in addition to multiple alleles of a given HLA supertype. Such a degree of promiscuity has previously been observed for DENV [62] and HIV peptides [63], among others. The existence of conserved T-cell determinants specific for multiple HLA supertypes further supports their evaluation as vaccine targets, since they would provide broader population coverage [63]. Many of the predicted HLA binding nonamers were localized in clusters, as we have also observed in HLA Tg mice immunized with WNV proteins and DNA encoding the SARS coronavirus N protein [64], and has been reported in studies of human immunodeficiency virus (HIV) type 1 proteins [65]–[68], the outer membrane protein of Chlamydia trachomatis [69], and other antigens [64].
The significant sequence variations between the proteins of the 4 DENV types represent a cardinal issue for the development of a tetravalent DENV vaccine that provides robust protection against each DENV type. Subtle amino acid substitutions within T-cell determinants restricted by a given HLA allomorph, such as in the event of sequential heterologous infections, or between a vaccine formulation and a subsequent natural infection [7], can dramatically alter the phenotype of the specific T cells, resulting in a wide range of effects from agonism to antagonism [9], [12]–[15]. Because of the extent of intra-type (1 to 21%) and inter-type (14 to 67%) amino acid variability among DENV isolates [48], many nonamer T-cell determinants contain single or multiple amino acid difference(s). When the 4 DENV types were analyzed together, a majority of the nonamer positions across the full proteome exhibited variants that together were present in ∼60 to ∼85% of all sequences. The frequencies of variant peptides across the 4 DENV types suggest that vaccine strategies incorporating whole DENV immunogens, such as inactivated and recombinant subunit vaccines, live attenuated viruses, or chimeric viruses expressing structural DENV genes, are likely to elicit T-cell responses to altered peptide ligands. This phenomenon is also likely to occur in individuals exposed to several Flaviviruses, such as DENV, JEV and YFV that are co-circulating in regions of Asia, India or South America, or following vaccination [70].
While the immune correlates of DENV protection remain poorly documented, there is evidence that both neutralizing antibody and specific T-cell responses are required [7],[71]. The incorporation of defined HLA-restricted T-cell determinants within DENV vaccine candidates might improve vaccine efficiency by increasing T-cell help to sustain a robust, long-lived immunity, and possibly through direct cytostatic and cytotoxic effects on infected cells. For tetravalent formulations, it may be relevant to focus primarily on sequences that are conserved in all 4 DENV types and to avoid the regions of T-cell immunity that are highly variable, unless they are strictly type-specific [17],[72]. The two pan-DENV E sequences (positions 97–111 and 252–261) and the exposed domain III of the E antigen (positions 300–400) [73],[74], are also candidate sequences for neutralizing antibody responses. An additional criterion for the selection of T-cell targets is the need for determinants with broad HLA representation, as it has been emphasized in the recognition of HLA supertypes [18]–[20]. Further investigations are needed to validate the immunogenicity of the candidate T-cell determinants in human subjects, and to identify sequences associated with deleterious T-cell responses.
The global approach described herein provides a framework and methodology for large-scale and systematic analysis of conserved sequences of other pathogens, in particular for rapidly evolving viruses such as influenza A virus [75] and HIV [63]. These studies will offer insights into their diversity and evolutionary history, together with providing critical data for rational vaccine development, structure-based design of candidate inhibitory compounds, and improvement of the current diagnostic methods.
Supporting Information
Average nonamer peptide entropy for each protein of each DENV type and all the four types combined. The values are shown for the 2005 dataset.
(0.70 MB TIF)
Molecular location of 19 pan-DENV sequences (in red) on the protein's 3-D structure. These sequences were mapped on the available crystallographic models of the E ectodomain (PDB Accession No. 1OAN; 394 out of 493-495 residues), NS3 (1BEF and 2BMF, 181 and 451 out of 618-619 residues, respectively) and NS5 fragments (1R6A, 295 out of 900-904 residues). The major portions of eleven of the 19 pan-DENV sequences were buried (NS3-148GLYGNGVVT156, 256EIVDLMCHATFT267, 284MDEAHFTDP292, 296AARGYISTRV305, 313IFMTATPPG321, 357GKTVWFVPSIK367, 406VVTTDISEMGANF418, and 491EAKMLLDNI499; NS5-79DLGCGRGGWSYY90, 141DTLLCDIGESS151 and 209PLSRNSTHEMYW220), 2 were partially buried/exposed (NS3-46FHTMWHVTRG55 and 537LMRRGDLPVWL547) and the remaining 6 were exposed (E-97VDRGWGNGCGLFGKG111 and 252VLGSQEGAMH261; NS3-189LTIMDLHPG197 and 383VIQLSRKTFD392; NS5-6GETLGEKWK14 and 104TKGGPGHEEP113) at the surface of the corresponding structures.
(9.65 MB DOC)
The intra-type percentage representation of pan-DENV sequences.
(0.10 MB DOC)
Pan-DENV sequences, entropy and representation of variants.
(0.08 MB DOC)
Functional and structural properties of pan-DENV sequences.
(0.06 MB DOC)
Distribution of pan-DENV sequences in nature.
(0.12 MB DOC)
Candidate putative HLA supertype-restricted binding nonamer peptides in pan-DENV sequences, predicted by immunoinformatic algorithms.
(0.20 MB DOC)
Intra-type representation of candidate putative HLA supertype-restricted nonamer peptides predicted by immunoinformatics algorithms.
(0.22 MB DOC)
GI numbers.
(0.86 MB XLS)
Translation of the abstract into Chinese by Guang Lan Zhang.
(0.06 MB PDF)
Acknowledgments
The authors are grateful to Lars Fugger (Weatherall Institute of Molecular Medicine, Oxford, UK) and Arthur Vanderbark (Oregon Health and Science University, Portland), Chella S. David (Mayo Clinic, Rochester), and Grete Sønderstrup (Stanford University School of Medicine) for providing HLA-DR2, -DR3 and -DR4 Tg mice, respectively. Overlapping DENV-3 peptide arrays were obtained through by the NIH Biodefense and Emerging Infectious Disease Research Resources Repository, NIAID, NIH. The authors thank Paul Nordstrom August, T. Jahan, and Aslam Khan for their valuable suggestions and help with the illustrations. The authors are grateful to Yu Jianshi for his help in translating the abstract to Chinese.
Footnotes
The authors have declared that no competing interests exist.
This project has been funded in part with the Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, USA, under Grant No. 5 U19 AI56541. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–341. doi: 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes EC. The evolutionary biology of dengue virus. Novartis Found Symp. 2006;277:177–187; discussion 187–192, 251–173. doi: 10.1002/0470058005.ch13. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 4.Esser MT, Marchese RD, Kierstead LS, Tussey LG, Wang F, et al. Memory T cells and vaccines. Vaccine. 2003;21:419–430. doi: 10.1016/s0264-410x(02)00407-3. [DOI] [PubMed] [Google Scholar]
- 5.Zinkernagel RM, Hengartner H. On immunity against infections and vaccines: credo 2004. Scand J Immunol. 2004;60:9–13. doi: 10.1111/j.0300-9475.2004.01460.x. [DOI] [PubMed] [Google Scholar]
- 6.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livingston PG, Kurane I, Dai LC, Okamoto Y, Lai CJ, et al. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. 1995;154:1287–1295. [PubMed] [Google Scholar]
- 9.Sloan-Lancaster J, Allen PM. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu Rev Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Welsh RM, Rothman AL. Dengue immune response: low affinity, high febrility. Nat Med. 2003;9:820–822. doi: 10.1038/nm0703-820. [DOI] [PubMed] [Google Scholar]
- 11.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 12.Evavold BD, Sloan-Lancaster J, Allen PM. Tickling the TCR: selective T-cell functions stimulated by altered peptide ligands. Immunol Today. 1993;14:602–609. doi: 10.1016/0167-5699(93)90200-5. [DOI] [PubMed] [Google Scholar]
- 13.Madrenas J, Germain RN. Variant TCR ligands: new insights into the molecular basis of antigen-dependent signal transduction and T-cell activation. Semin Immunol. 1996;8:83–101. doi: 10.1006/smim.1996.0011. [DOI] [PubMed] [Google Scholar]
- 14.Kalergis AM, Nathenson SG. Altered peptide ligand-mediated TCR antagonism can be modulated by a change in a single amino acid residue within the CDR3 beta of an MHC class I-restricted TCR. J Immunol. 2000;165:280–285. doi: 10.4049/jimmunol.165.1.280. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura Y, Chen YZ, Uemura Y, Tanaka Y, Tsukamoto H, et al. Degenerate recognition and response of human CD4+ Th cell clones: implications for basic and applied immunology. Mol Immunol. 2004;40:1089–1094. doi: 10.1016/j.molimm.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Loke H, Bethell DB, Phuong CX, Dung M, Schneider J, et al. Strong HLA class I–restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. 2001;184:1369–1373. doi: 10.1086/324320. [DOI] [PubMed] [Google Scholar]
- 17.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 18.Sette A, Livingston B, McKinney D, Appella E, Fikes J, et al. The development of multi-epitope vaccines: epitope identification, vaccine design and clinical evaluation. Biologicals. 2001;29:271–276. doi: 10.1006/biol.2001.0297. [DOI] [PubMed] [Google Scholar]
- 19.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 20.Khan AM, Miotto O, Heiny AT, Salmon J, Srinivasan KN, et al. A systematic bioinformatics approach for selection of epitope-based vaccine targets. Cell Immunol. 2006;244:141–147. doi: 10.1016/j.cellimm.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osatomi K, Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990;176:643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- 23.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, et al. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 27.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423 and 623–656. [Google Scholar]
- 28.Miotto O, Heiny A, Tan TW, August JT, Brusic V. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics. 2008;9(Suppl 1):S18. doi: 10.1186/1471-2105-9-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paninski L. Estimation of entropy and mutual information. Neural Computation. 2003;15:1191–1253. [Google Scholar]
- 30.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters B, Sidney J, Bourne P, Bui HH, Buus S, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi:10.1371/journal.pbio.0030091. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35:2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 36.Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V. MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res. 2005;33:W172–179. doi: 10.1093/nar/gki452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 38.Bian H, Hammer J. Discovery of promiscuous HLA-II-restricted T cell epitopes with TEPITOPE. Methods. 2004;34:468–475. doi: 10.1016/j.ymeth.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 40.Sette A, Sidney J, Livingston BD, Dzuris JL, Crimi C, et al. Class I molecules with similar peptide-binding specificities are the result of both common ancestry and convergent evolution. Immunogenetics. 2003;54:830–841. doi: 10.1007/s00251-002-0530-0. [DOI] [PubMed] [Google Scholar]
- 41.Vandenbark AA, Rich C, Mooney J, Zamora A, Wang C, et al. Recombinant TCR ligand induces tolerance to myelin oligodendrocyte glycoprotein 35-55 peptide and reverses clinical and histological signs of chronic experimental autoimmune encephalomyelitis in HLA-DR2 transgenic mice. J Immunol. 2003;171:127–133. doi: 10.4049/jimmunol.171.1.127. [DOI] [PubMed] [Google Scholar]
- 42.Strauss G, Vignali DA, Schonrich G, Hammerling GJ. Negative and positive selection by HLA-DR3(DRw17) molecules in transgenic mice. Immunogenetics. 1994;40:104–108. [PubMed] [Google Scholar]
- 43.Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, et al. Mice lacking all conventional MHC class II genes. Proc Natl Acad Sci U S A. 1999;96:10338–10343. doi: 10.1073/pnas.96.18.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito K, Bian HJ, Molina M, Han J, Magram J, et al. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J Exp Med. 1996;183:2635–2644. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fugger L, Michie SA, Rulifson I, Lock CB, McDevitt GS. Expression of HLA-DR4 and human CD4 transgenes in mice determines the variable region beta-chain T-cell repertoire and mediates an HLA-DR-restricted immune response. Proc Natl Acad Sci U S A. 1994;91:6151–6155. doi: 10.1073/pnas.91.13.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cope AP, Patel SD, Hall F, Congia M, Hubers HA, et al. T cell responses to a human cartilage autoantigen in the context of rheumatoid arthritis-associated and nonassociated HLA-DR4 alleles. Arthritis Rheum. 1999;42:1497–1507. doi: 10.1002/1529-0131(199907)42:7<1497::AID-ANR25>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 47.Roederer M, Koup RA. Optimized determination of T cell epitope responses. J Immunol Methods. 2003;274:221–228. doi: 10.1016/s0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- 48.Khan AM, Heiny AT, Lee KX, Srinivasan KN, Tan TW, et al. Large-scale analysis of antigenic diversity of T-cell epitopes in dengue virus. BMC Bioinformatics. 2006;7(Suppl 5):S4. doi: 10.1186/1471-2105-7-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8:74–77. doi: 10.1016/s0966-842x(99)01669-8. [DOI] [PubMed] [Google Scholar]
- 50.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 51.Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 53.Murthy HM, Clum S, Padmanabhan R. Dengue virus NS3 serine protease. Crystal structure and insights into interaction of the active site with substrates by molecular modeling and structural analysis of mutational effects. J Biol Chem. 1999;274:5573–5580. doi: 10.1074/jbc.274.9.5573. [DOI] [PubMed] [Google Scholar]
- 54.Xu T, Sampath A, Chao A, Wen D, Nanao M, et al. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, et al. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haydon DT, Woolhouse ME. Immune avoidance strategies in RNA viruses: fitness continuums arising from trade-offs between immunogenicity and antigenic variability. J Theor Biol. 1998;193:601–612. doi: 10.1006/jtbi.1998.0726. [DOI] [PubMed] [Google Scholar]
- 58.Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, et al. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85:1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- 59.Henchal EA, Putnak JR. The dengue viruses. Clin Microbiol Rev. 1990;3:376–396. doi: 10.1128/cmr.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Billoir F, de Chesse R, Tolou H, de Micco P, Gould EA, et al. Phylogeny of the genus flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J Gen Virol. 2000;81:781–790. doi: 10.1099/0022-1317-81-3-781. [DOI] [PubMed] [Google Scholar]
- 62.Gagnon SJ, Zeng W, Kurane I, Ennis FA. Identification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clones. J Virol. 1996;70:141–147. doi: 10.1128/jvi.70.1.141-147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson CC, McKinney D, Anders M, MaWhinney S, Forster J, et al. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. J Immunol. 2003;171:5611–5623. doi: 10.4049/jimmunol.171.10.5611. [DOI] [PubMed] [Google Scholar]
- 64.Gupta V, Tabiin TM, Sun K, Chandrasekaran A, Anwar A, et al. SARS coronavirus nucleocapsid immunodominant T-cell epitope cluster is common to both exogenous recombinant and endogenous DNA-encoded immunogens. Virology. 2006;347:127–139. doi: 10.1016/j.virol.2005.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berzofsky JA, Pendleton CD, Clerici M, Ahlers J, Lucey DR, et al. Construction of peptides encompassing multideterminant clusters of human immunodeficiency virus envelope to induce in vitro T cell responses in mice and humans of multiple MHC types. J Clin Invest. 1991;88:876–884. doi: 10.1172/JCI115389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shankar P, Fabry JA, Fong DM, Lieberman J. Three regions of HIV-1 gp160 contain clusters of immunodominant CTL epitopes. Immunol Lett. 1996;52:23–30. doi: 10.1016/0165-2478(96)02574-6. [DOI] [PubMed] [Google Scholar]
- 67.Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, et al. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci U S A. 2001;98:4587–4592. doi: 10.1073/pnas.071063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown SA, Stambas J, Zhan X, Slobod KS, Coleclough C, et al. Clustering of Th cell epitopes on exposed regions of HIV envelope despite defects in antibody activity. J Immunol. 2003;171:4140–4148. doi: 10.4049/jimmunol.171.8.4140. [DOI] [PubMed] [Google Scholar]
- 69.Kim SK, DeMars R. Epitope clusters in the major outer membrane protein of Chlamydia trachomatis. Curr Opin Immunol. 2001;13:429–436. doi: 10.1016/s0952-7915(00)00237-5. [DOI] [PubMed] [Google Scholar]
- 70.Moran E, Simmons C, Vinh Chau N, Luhn K, Wills B, et al. Preservation of a critical epitope core region is associated with the high degree of flaviviral cross-reactivity exhibited by a dengue-specific CD4(+) T cell clone. Eur J Immunol. 2008;38:1050–1057. doi: 10.1002/eji.200737699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. 2007;5:518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- 72.Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 73.Mota J, Acosta M, Argotte R, Figueroa R, Mendez A, et al. Induction of protective antibodies against dengue virus by tetravalent DNA immunization of mice with domain III of the envelope protein. Vaccine. 2005;23:3469–3476. doi: 10.1016/j.vaccine.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 74.Chin JF, Chu JJ, Ng ML. The envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microbes Infect. 2007;9:1–6. doi: 10.1016/j.micinf.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 75.Heiny AT, Miotto O, Srinivasan KN, Khan AM, Zhang GL, et al. Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets. PLoS ONE. 2007;2:e1190. doi:10.1371/journal.pone.0001190. doi: 10.1371/journal.pone.0001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simmons CP, Dong T, Chau NV, Dung NT, Chau TN, et al. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J Virol. 2005;79:5665–5675. doi: 10.1128/JVI.79.9.5665-5675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurane I, Dai LC, Livingston PG, Reed E, Ennis FA. Definition of an HLA-DPw2-restricted epitope on NS3, recognized by a dengue virus serotype-cross-reactive human CD4+ CD8- cytotoxic T-cell clone. J Virol. 1993;67:6285–6288. doi: 10.1128/jvi.67.10.6285-6288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okamoto Y, Kurane I, Leporati AM, Ennis FA. Definition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4+ T cell clones. J Gen Virol. 1998;79 (Pt 4):697–704. doi: 10.1099/0022-1317-79-4-697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average nonamer peptide entropy for each protein of each DENV type and all the four types combined. The values are shown for the 2005 dataset.
(0.70 MB TIF)
Molecular location of 19 pan-DENV sequences (in red) on the protein's 3-D structure. These sequences were mapped on the available crystallographic models of the E ectodomain (PDB Accession No. 1OAN; 394 out of 493-495 residues), NS3 (1BEF and 2BMF, 181 and 451 out of 618-619 residues, respectively) and NS5 fragments (1R6A, 295 out of 900-904 residues). The major portions of eleven of the 19 pan-DENV sequences were buried (NS3-148GLYGNGVVT156, 256EIVDLMCHATFT267, 284MDEAHFTDP292, 296AARGYISTRV305, 313IFMTATPPG321, 357GKTVWFVPSIK367, 406VVTTDISEMGANF418, and 491EAKMLLDNI499; NS5-79DLGCGRGGWSYY90, 141DTLLCDIGESS151 and 209PLSRNSTHEMYW220), 2 were partially buried/exposed (NS3-46FHTMWHVTRG55 and 537LMRRGDLPVWL547) and the remaining 6 were exposed (E-97VDRGWGNGCGLFGKG111 and 252VLGSQEGAMH261; NS3-189LTIMDLHPG197 and 383VIQLSRKTFD392; NS5-6GETLGEKWK14 and 104TKGGPGHEEP113) at the surface of the corresponding structures.
(9.65 MB DOC)
The intra-type percentage representation of pan-DENV sequences.
(0.10 MB DOC)
Pan-DENV sequences, entropy and representation of variants.
(0.08 MB DOC)
Functional and structural properties of pan-DENV sequences.
(0.06 MB DOC)
Distribution of pan-DENV sequences in nature.
(0.12 MB DOC)
Candidate putative HLA supertype-restricted binding nonamer peptides in pan-DENV sequences, predicted by immunoinformatic algorithms.
(0.20 MB DOC)
Intra-type representation of candidate putative HLA supertype-restricted nonamer peptides predicted by immunoinformatics algorithms.
(0.22 MB DOC)
GI numbers.
(0.86 MB XLS)
Translation of the abstract into Chinese by Guang Lan Zhang.
(0.06 MB PDF)