Abstract
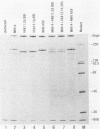
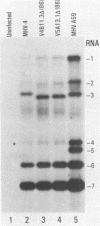
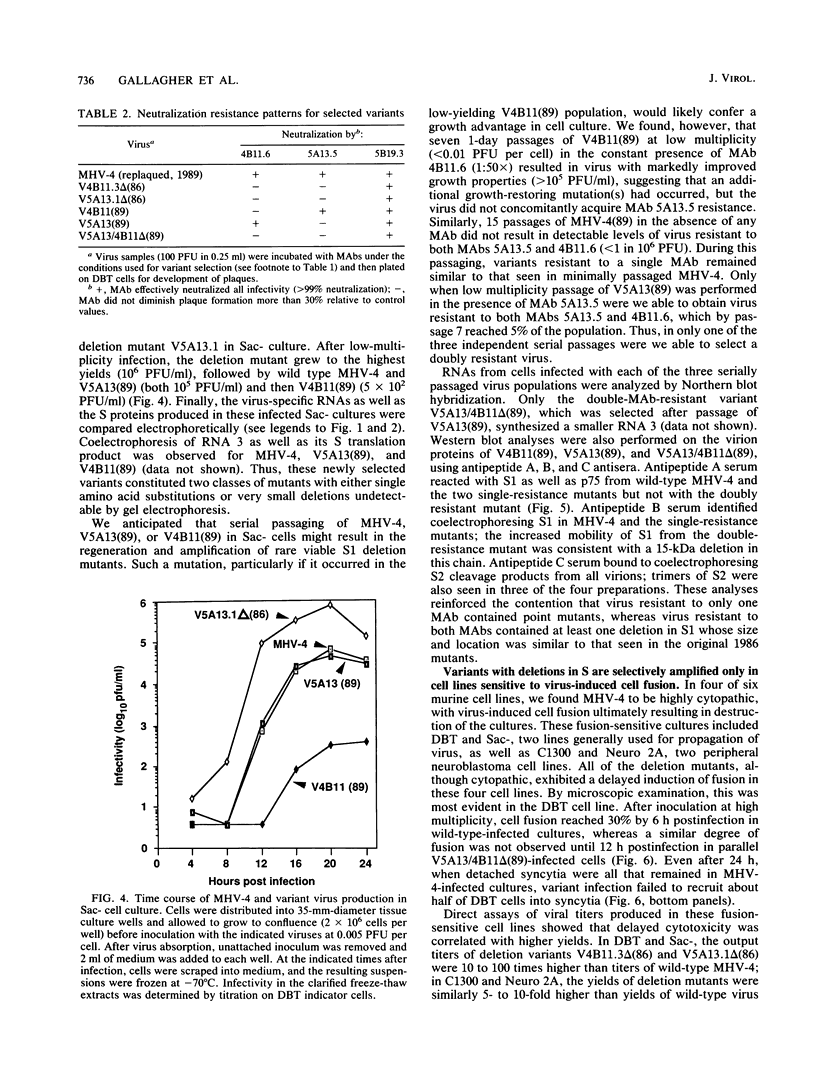
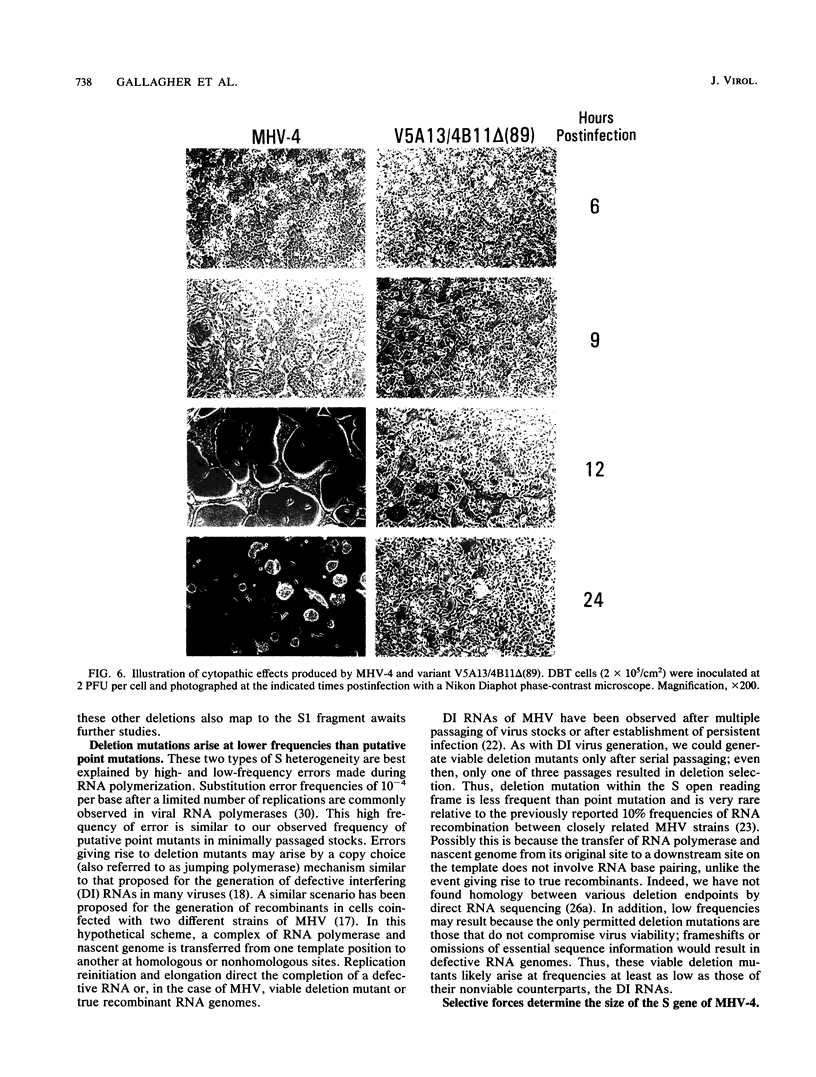
Neuroattenuated variants of mouse hepatitis virus type 4 (MHV-4) selected for resistance to neutralizing monoclonal antibodies (R.G. Dalziel, P.W. Lampert, P. J. Talbot, and M. J. Buchmeier, J. Virol. 59:463-471, 1986) were found to harbor large deletions in both mRNA 3 and its protein product, the 180-kilodalton viron spike (S) glycoprotein. By using antipeptide antibodies directed against selected portions of the chain, deletions were mapped to the middle of the amino-terminal S1 fragment, one of the two posttranslational cleavage products of S, and involved omission of 15 kilodaltons of protein. Deletion mutants could be selected only after multiple passage of virus through cultured cell lines; minimally passaged MHV-4 stocks contained putative point mutants selectable by neutralizing monoclonal antibodies but no deletions. Enhanced growth of deletion mutants relative to wild-type virus was observed in four cell lines used for virus propagation and was attributed to delayed and diminished cytopathic effects that allowed cultures to support virus production for prolonged periods. This hypothesis was reinforced by the finding that no selective advantage for the deletion mutants was observed in two cell lines resistant to virus-induced cytopathic effects. These results indicate that the passaging of MHV-4 in culture generates heterogeneity in S structure and eventually selects for rare neutralization-resistant deletion mutants with decreased virulence properties.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baybutt H. N., Wege H., Carter M. J., ter Meulen V. Adaptation of coronavirus JHM to persistent infection of murine sac(-) cells. J Gen Virol. 1984 May;65(Pt 5):915–924. doi: 10.1099/0022-1317-65-5-915. [DOI] [PubMed] [Google Scholar]
- Buchmeier M. J., Southern P. J., Parekh B. S., Wooddell M. K., Oldstone M. B. Site-specific antibodies define a cleavage site conserved among arenavirus GP-C glycoproteins. J Virol. 1987 Apr;61(4):982–985. doi: 10.1128/jvi.61.4.982-985.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J Gen Virol. 1983 Dec;64(Pt 12):2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- Collins A. R., Knobler R. L., Powell H., Buchmeier M. J. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell--cell fusion. Virology. 1982 Jun;119(2):358–371. doi: 10.1016/0042-6822(82)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel R. G., Lampert P. W., Talbot P. J., Buchmeier M. J. Site-specific alteration of murine hepatitis virus type 4 peplomer glycoprotein E2 results in reduced neurovirulence. J Virol. 1986 Aug;59(2):463–471. doi: 10.1128/jvi.59.2.463-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J. O., Stohlman S. A., Harmon R. C., Lai M. M., Frelinger J. A., Weiner L. P. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology. 1983 Dec;131(2):296–307. doi: 10.1016/0042-6822(83)90498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J. O., Trousdale M. D., el-Zaatari F. A., Stohlman S. A., Weiner L. P. Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies. J Virol. 1986 Jun;58(3):869–875. doi: 10.1128/jvi.58.3.869-875.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frana M. F., Behnke J. N., Sturman L. S., Holmes K. V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985 Dec;56(3):912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel M. V., Lampert P. W., Oldstone M. B. Temperature-sensitive mutants of mouse hepatitis virus produce a high incidence of demyelination. Proc Natl Acad Sci U S A. 1978 Aug;75(8):4033–4036. doi: 10.1073/pnas.75.8.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Goto N., Makino S., Fujiwara K. Persistent infection with mouse hepatitis virus, JHM strain in DBT cell culture. Adv Exp Med Biol. 1981;142:301–308. doi: 10.1007/978-1-4757-0456-3_24. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Knobler R. L., Tunison L. A., Lampert P. W., Oldstone M. B. Selected mutants of mouse hepatitis virus type 4 (JHM strain) induce different CNS diseases. Pathobiology of disease induced by wild type and mutants ts8 and ts15 in BALB/c and SJL/J mice. Am J Pathol. 1982 Nov;109(2):157–168. [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Makino S., Soe L. H., Shieh C. K., Keck J. G., Fleming J. O. Coronavirus: a jumping RNA transcription. Cold Spring Harb Symp Quant Biol. 1987;52:359–365. doi: 10.1101/sqb.1987.052.01.041. [DOI] [PubMed] [Google Scholar]
- Lazzarini R. A., Keene J. D., Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981 Oct;26(2 Pt 2):145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Luytjes W., Geerts D., Posthumus W., Meloen R., Spaan W. Amino acid sequence of a conserved neutralizing epitope of murine coronaviruses. J Virol. 1989 Mar;63(3):1408–1412. doi: 10.1128/jvi.63.3.1408-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Sturman L. S., Bredenbeek P. J., Charite J., van der Zeijst B. A., Horzinek M. C., Spaan W. J. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987 Dec;161(2):479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985 May;54(2):329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J. G., Stohlman S. A., Lai M. M. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris V. L., Tieszer C., Mackinnon J., Percy D. Characterization of coronavirus JHM variants isolated from Wistar Furth rats with a viral-induced demyelinating disease. Virology. 1989 Mar;169(1):127–136. doi: 10.1016/0042-6822(89)90048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S. E., Gallagher T. M., Buchmeier M. J. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein gene of several strains of murine hepatitis virus. Virology. 1989 Dec;173(2):664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt I., Skinner M., Siddell S. Nucleotide sequence of the gene encoding the surface projection glycoprotein of coronavirus MHV-JHM. J Gen Virol. 1987 Jan;68(Pt 1):47–56. doi: 10.1099/0022-1317-68-1-47. [DOI] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. C. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- Stohlman S. A., Brayton P. R., Fleming J. O., Weiner L. P., Lai M. M. Murine coronaviruses: isolation and characterization of two plaque morphology variants of the JHM neurotropic strain. J Gen Virol. 1982 Dec;63(2):265–275. doi: 10.1099/0022-1317-63-2-265. [DOI] [PubMed] [Google Scholar]
- Sturman L. S., Holmes K. V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980 Jan;33(1):449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L. S., Ricard C. S., Holmes K. V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J Virol. 1985 Dec;56(3):904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Fleming J. O. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology. 1989 Mar;169(1):233–235. doi: 10.1016/0042-6822(89)90061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Siddell S. G., Wege H., ter Meulen V. Characterization of a variant virus selected in rat brains after infection by coronavirus mouse hepatitis virus JHM. J Virol. 1985 May;54(2):429–435. doi: 10.1128/jvi.54.2.429-435.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot P. J., Buchmeier M. J. Antigenic variation among murine coronaviruses: evidence for polymorphism on the peplomer glycoprotein, E2. Virus Res. 1985 Jun;2(4):317–328. doi: 10.1016/0168-1702(85)90028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot P. J., Salmi A. A., Knobler R. L., Buchmeier M. J. Topographical mapping of epitopes on the glycoproteins of murine hepatitis virus-4 (strain JHM): correlation with biological activities. Virology. 1984 Jan 30;132(2):250–260. doi: 10.1016/0042-6822(84)90032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Dörries R., Wege H. Hybridoma antibodies to the murine coronavirus JHM: characterization of epitopes on the peplomer protein (E2). J Gen Virol. 1984 Nov;65(Pt 11):1931–1942. doi: 10.1099/0022-1317-65-11-1931. [DOI] [PubMed] [Google Scholar]
- Wege H., Winter J., Meyermann R. The peplomer protein E2 of coronavirus JHM as a determinant of neurovirulence: definition of critical epitopes by variant analysis. J Gen Virol. 1988 Jan;69(Pt 1):87–98. doi: 10.1099/0022-1317-69-1-87. [DOI] [PubMed] [Google Scholar]
- Weiner L. P. Pathogenesis of demyelination induced by a mouse hepatitis. Arch Neurol. 1973 May;28(5):298–303. doi: 10.1001/archneur.1973.00490230034003. [DOI] [PubMed] [Google Scholar]