Abstract
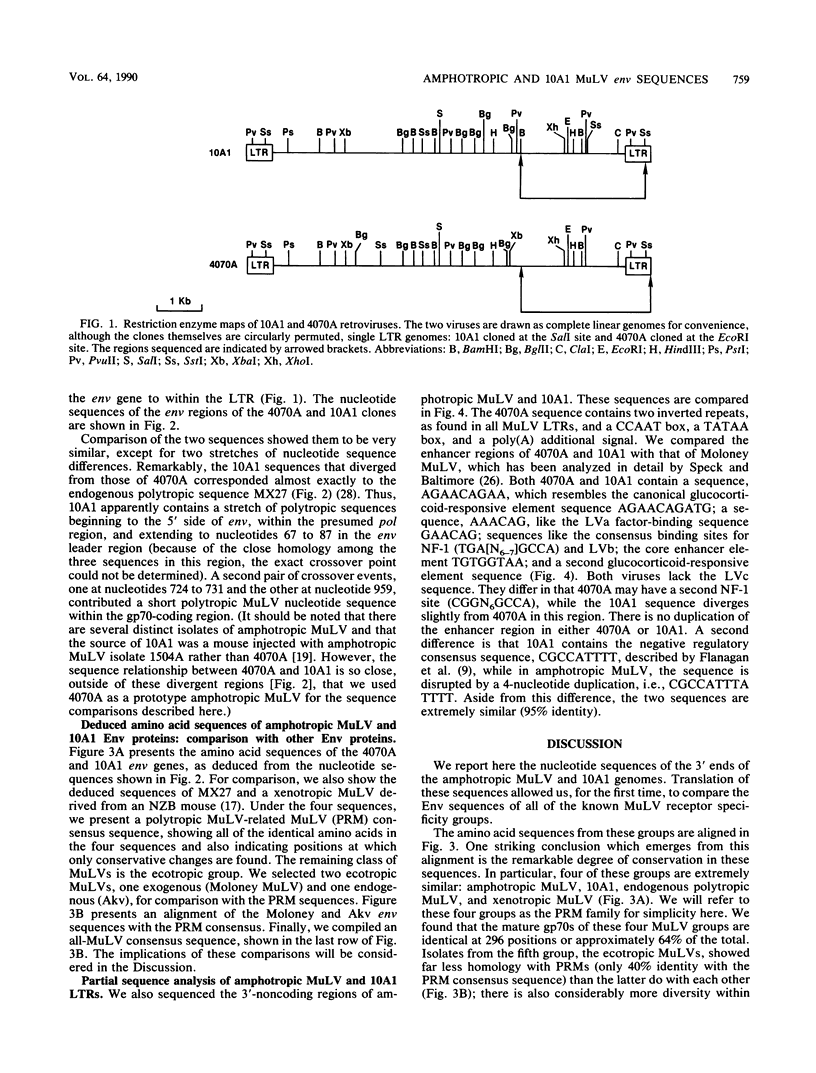
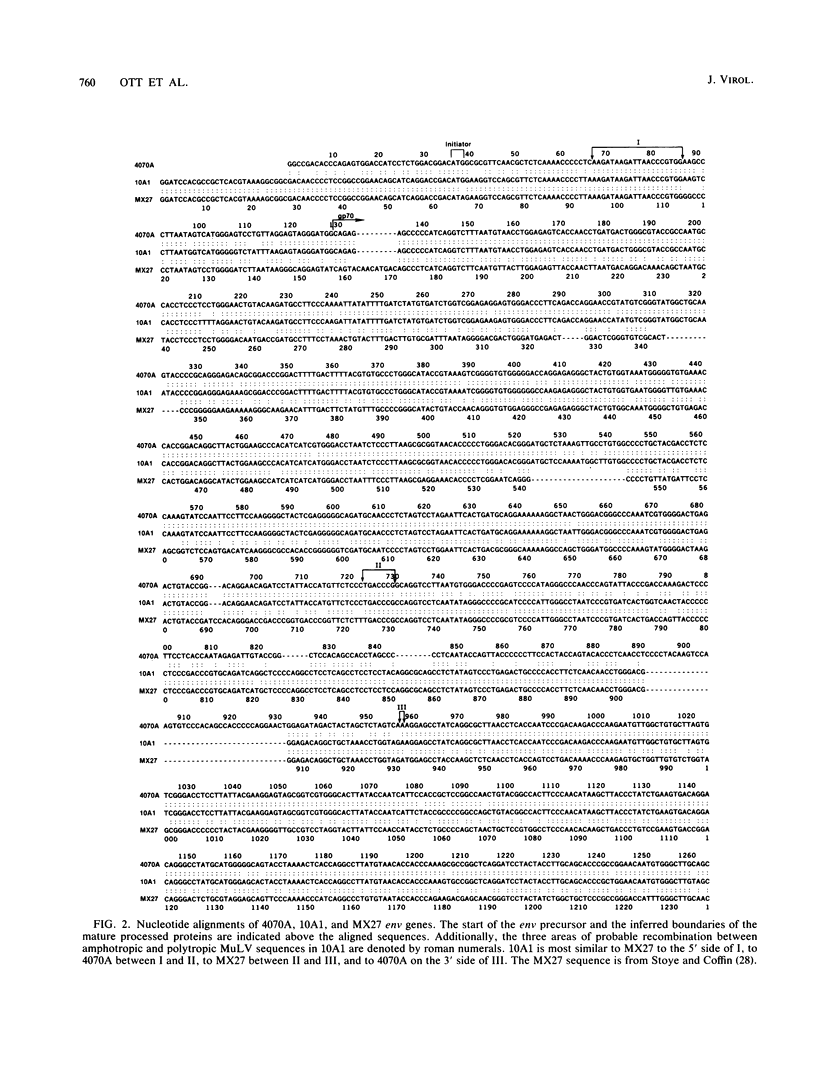
Viral interference studies have demonstrated the existence of four distinct murine leukemia virus (MuLV) receptors on NIH 3T3 mouse cells. The four viral interference groups are ecotropic MuLV; mink cell focus inducing virus (MCF); amphotropic MuLV; and 10A1, a recombinant derivative of amphotropic MuLV that uses a unique receptor but also retains affinity for the amphotropic MuLV receptor. We report here that 10A1 infects rat and hamster cells, unlike its amphotropic parent. We isolated an infectious molecular clone of 10A1 and present here the sequences of the env genes and enhancer regions of amphotropic MuLV and 10A1. The deduced amino acid sequences of amphotropic MuLV and 10A1 gp70su are remarkably similar to those of MCF and xenotropic MuLV (for which mouse cells lack receptors), with 64% amino acids identical in the four groups. We generated a consensus from these comparisons. Further, the differences are largely localized to a few discrete regions: (i) amphotropic MuLV has two short insertions relative to MCF, at residues 87 to 92 and 163 to 169, and (ii) amphotropic MuLV and MCF are totally different in a hypervariable region, which is greater than 30% proline, at residues approximately 253 to 304. 10A1 closely resembles amphotropic MuLV in its N terminus but contains an MCF-type hypervariable region. These results suggest the possibility that receptor specificity is localized in these short variable regions and further that the unique receptor specificity of 10A1 is due to the novel combination of amphotropic MuLV and MCF sequences rather than to the presence of any novel sequences. The Env proteins of ecotropic MuLV are far more distantly related to those of the other four groups than the latter are to each other. We also found that the enhancer regions of amphotropic MuLV and 10A1 are nearly identical, although 10A1 is far more leukemogenic than amphotropic MuLV.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bassin R. H., Tuttle N., Fischinger P. J. Rapid cell culture assay technic for murine leukaemia viruses. Nature. 1971 Feb 19;229(5286):564–566. doi: 10.1038/229564b0. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Besmer P., Baltimore D. Mechanism of restriction of ecotropic and xenotropic murine leukemia viruses and formation of pseudotypes between the two viruses. J Virol. 1977 Mar;21(3):965–973. doi: 10.1128/jvi.21.3.965-973.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bova C. A., Manfredi J. P., Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986 Jul 30;152(2):343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Oliff A. I., Linemeyer D. L., Lander M. R., Lowy D. R. Genomes of murine leukemia viruses isolated from wild mice. J Virol. 1981 Sep;39(3):777–791. doi: 10.1128/jvi.39.3.777-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B., Wehrly K. Different murine cell lines manifest unique patterns of interference to superinfection by murine leukemia viruses. Virology. 1985 Feb;141(1):119–129. doi: 10.1016/0042-6822(85)90188-6. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner A. J., Coffin J. M. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986 May 9;45(3):365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- Flanagan J. R., Krieg A. M., Max E. E., Khan A. S. Negative control region at the 5' end of murine leukemia virus long terminal repeats. Mol Cell Biol. 1989 Feb;9(2):739–746. doi: 10.1128/mcb.9.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984 Feb;49(2):471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. At least four viral genes contribute to the leukemogenicity of murine retrovirus MCF 247 in AKR mice. J Virol. 1985 Jan;53(1):158–165. doi: 10.1128/jvi.53.1.158-165.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Hunsmann G., Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983 Jan;45(1):1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Rasheed S., Shimizu C. S., Gardner M. B. Genomic characterization of a highly oncogenic env gene recombinant between amphotropic retrovirus of wild mouse and endogenous xenotropic virus of NIH swiss mouse. Virology. 1982 Feb;117(1):262–266. doi: 10.1016/0042-6822(82)90526-8. [DOI] [PubMed] [Google Scholar]
- O'Neill R. R., Hartley J. W., Repaske R., Kozak C. A. Amphotropic proviral envelope sequences are absent from the Mus germ line. J Virol. 1987 Jul;61(7):2225–2231. doi: 10.1128/jvi.61.7.2225-2231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Ruscetti S. A 2.4-kilobase-pair fragment of the Friend murine leukemia virus genome contains the sequences responsible for friend murine leukemia virus-induced erythroleukemia. J Virol. 1983 Jun;46(3):718–725. doi: 10.1128/jvi.46.3.718-725.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed S., Pal B. K., Gardner M. B. Characterization of a highly oncogenic murine leukemia virus from wild mice. Int J Cancer. 1982 Mar 15;29(3):345–350. doi: 10.1002/ijc.2910290319. [DOI] [PubMed] [Google Scholar]
- Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982 Jul 15;120(1):251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- Rein A., Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984 Jul 15;136(1):144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. P., Coffin J. M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987 Sep;61(9):2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



