Abstract
Cryo-electron tomography (cryo-ET) allows the visualization of cellular structures under close-to-life conditions and at molecular resolution. While it is inherently a static approach, yielding structural information about supramolecular organization at a certain time point, it can nevertheless provide insights into function of the structures imaged, in particular, when supplemented by other approaches. Here, we review the use of experimental methods that supplement cryo-ET imaging of whole cells. These include genetic and pharmacological manipulations, as well as correlative light microscopy and cryo-ET. While these methods have mostly been used to detect and identify structures visualized in cryo-ET or to assist the search for a feature of interest, we expect that in the future they will play a more important role in the functional interpretation of cryo-tomograms.
Keywords: Cryo-electron tomography, Correlative light microscopy, Electron microscopy
Introduction
Among techniques used in cell biology, cryo-electron tomography (cryo-ET) is a relatively recent one. It combines the advantages of 3D imaging with a close-to-life preservation and it allows studying biological material at molecular resolution (rev. in Lucic et al. 2005; Nicastro et al. 2005). Rapid freezing followed by the investigation of the frozen-hydrated samples avoids artifacts notorious to chemical fixation and dehydration procedures (Dubochet et al. 1988). Furthermore, the biological material is observed directly, without heavy metal staining, avoiding problems in interpretation caused by an unpredictable accumulation of staining material. Consequently, cryo-ET of whole cells has the advantage that the supramolecular architecture can be studied in unperturbed cellular environments (Baumeister 2002).
We can distinguish two types of cryo-preparation. Thin samples (typically below 0.5 μm in thickness) can be rapidly frozen by plunging into liquid ethane (Dubochet et al. 1988) and imaged in an electron microscope without further processing. While plunge-freezing is limited to small cells and thin regions of cells, thicker samples are frozen by high-pressure freezing (Moor 1987) and then cryo-sectioned, that is sectioned to typically 50–200 nm thick sections at cryogenic temperatures (Al-Amoudi et al. 2004). In both cases the high freezing rates ensure that a sample is vitrified, i.e. formation of ice crystals is avoided. Also, samples are kept and imaged in EM at cryogenic temperatures to avoid devitrification.
Electron microscopy (EM) in general, and cryo-ET being no exception, delivers static images of a biological system, as does X-ray crystallography. In spite of this apparent limitation, both methods can provide a structural framework for a mechanistic understanding of molecular and cellular functions. Cryo-ET has already pushed the resolution limits and increased the fidelity of the images, and we can expect this trend to continue. In this review, we focus on the experimental methods that supplement cryo-ET imaging of whole cells. Various approaches were used to assist the detection or identification of imaged structures and also to allow cryo-ET imaging of intracellular structures arrested in a particular functional state. This is a very young field, and consequently the current literature is not extensive. We paid particular attention to the methods used and to the development of methods that might be relevant for the future. The methods were separated in two main categories. On one side are genetic and pharmacological manipulations that are used along the lines of the well-proven basic paradigm that involves investigations of untreated, treated and (when applicable) control samples. When applied to cryo-ET, a number of tomograms are recorded for samples in each group and a value is associated with each tomogram in order to assess the effect of a treatment. Correlative light microscopy (LM) and EM methods fall in the second category. The advantage of this approach is that LM provides information about the larger environment and the functional states of structures visualized in cryo-ET, or alternatively, cryo-ET effectively extends the resolution achievable in LM.
Genetic, pharmacological and related treatments integrated with cryo-ET
Finding and identifying a structure of interest in tomograms recorded under low-dose conditions having a poor signal-to-noise ratio is one of the basic problems in cryo-ET. It is also a prerequisite for providing insights to its function. Although a large number of observations obtained from conventional EM are certainly very useful in this respect, more sophisticated detection methods are needed if cryo-ET is expected to reach beyond ultrastructure to the molecular architecture level. In straightforward cases, when features of interest possess strong intrinsic contrast, they can be easily identified. For example, the location and spatial arrangement of magnetosomes, small organelles containing magnetite crystals present in magnetotactic bacteria, were compared in wild-type and mutant cells lacking proteins that were expected to be relevant for magnetosome organization and function (Komeili et al. 2006; Scheffel et al. 2006). In both studies, the wild-type organization of magnetosomes was restored after overexpression of the deleted proteins. Cryo-preparation was necessary for the preservation of cytoskeletal filaments of Spiroplasma melliferum (Kurner et al. 2005) and of cell surface filaments of Flavobacterium johnsoniae (Liu et al. 2007) and allowed a relatively straightforward detection. This in turn made an integrative approach possible, whereby the genetic deletion of GldF, one of the envelope-associated gliding proteins, led to a complete loss of filaments (Liu et al. 2007). The reintroduction of the deleted protein rescued the filaments, thus, providing new evidence for the involvement of GldF in filament formation. Another challenge inherent to cryo-ET is to obtain enough cryo-structures so that they can be statistically analyzed at a reasonable confidence level. Difficulties in sample preparation, as well as the time needed to record, reconstruct and analyze a cryo-tomogram limit throughput. For example, the investigation of actin networks in filopodia of slime mold Dictyostelium cells was facilitated by using genetically modified cells that overexpress small GTPase Rac1A and show an abundance of filopodia (Medalia et al. 2007). Tomograms of wild-type filopodia were used as a control, to make sure that the overexpression of Rac1A did not alter actin networks.
Cytoskeletal filament bundles observed in cryo-tomograms of wild-type Caulobacter crescentus were classified in four types based on their dimensions and cellular localization (Briegel et al. 2006). In an attempt to identify at least some of the filaments, the authors also imaged cells lacking crescentin, one of the previously identified cytoskeletal proteins. Additionally, they imaged both wild-type and crescentin knockout cells that were treated with a drug that is known to depolymerize filaments composed of another cytoskeletal protein, MreB. While crescentin and MreB could be ruled out as constituents of some filament types, the interpretation of the results was complicated by ambiguous results. For example, the lack of one type of filament in the MreB depolymerizing-drug-treated crescentin knockout cells, but not in the non-treated knockout and treated wild-type cells, points to a particular interaction between crescentin, MreB and an as yet unidentified molecule forming these filaments. Also, although the authors analyzed a fair number of tomograms, another type of filament could not be identified because its abundance in wild-type cells was too low. Consequently, this example shows how even a moderately complicated experimental system (four conditions, four filament types) can go beyond current limits of the interpretability of cellular cryo-ET. An even more elaborate version of the same wild-type—mutant—rescue scheme was applied in the investigation of Escherichia coli chemotaxis receptor arrays (Zhang et al. 2004, 2007). It involved genetic deletion of chemoreceptors and/or another two proteins of the chemosensory system, CheA and CheW, and their subsequent expression at different relative levels. As a result, it was confirmed that the periodic structures located at cell poles represent chemoreceptors, illustrating how a genetic approach can assist the identification of structures observed in cryo-tomograms.
In all examples presented so far, various experimental assays were used to assist detection and identification of structures of interest in cryo-tomograms, but not to investigate cells in a well-defined functional or developmental stage. The latter approach was implemented by partially synchronizing the growth phase of the unicellular eukaryote Ostreococcus tauri by light–dark cycles and revealed the detailed ultrastructure of whole cells, tightly packed with organelles, even though the resolution was effectively limited by the thickness of this sample (Henderson et al. 2007).
Rapid freezing techniques currently allow EM imaging of the same process at well-defined time points. This method, called time-resolved cryo-EM, was used to observe growth and shrinkage in reconstituted microtubules (Mandelkow et al. 1991) and liposome formation under different conditions (Frederik and Sommerdijk 2005). A very precise control of reaction time can be achieved by spraying an interacting component on a sample just milliseconds before vitrification (Berriman and Unwin 1994), or by flash-photolysis (Subramaniam et al. 1993). While it may not be straightforward to achieve such precise timings for cryo-ET of whole cells, its application for investigations of cellular processes in vitro may complement cellular studies.
Correlative light microscopy and cryo-ET
In cell biology journals, green pictures abound. The vast array of currently available fluorescent probes as well as the pervasive use of fluorescence LM contributed to our knowledge about the identity and the spatial distribution of molecular players involved in many cellular processes, but left many open questions regarding the molecular and supramolecular architecture of these players. Resolving these questions may help us understand the principles underlying a large number of cellular processes. Correlative LM and EM methods allow imaging of the same feature over multiple length scales and can combine the advantages of both methods, namely live cell imaging and the ability to obtain high-resolution information. Approaches that combine LM with other EM preservation methods are already showing their importance for the integration of structure and function and are beginning to become important tools in cell biology (rev. in Koster and Klumperman 2003). We first review the current state of this topic and then outline some future directions.
Lessons from correlative approaches involving other EM preparations
Conventional sample preparation in EM involves a sequence of steps: a chemical fixation, dehydration, embedding, staining and sectioning. Better preservation can be obtained if instead of a chemical fixation and dehydration a sample is rapidly frozen using a high-pressure freezer and then freeze substituted. In this type of EM, labeling by electron-dense markers is typically achieved using the immunolabeling methods and it is a trade-off between obtaining good structural preservation and good labeling. In general, pre-embedding labeling methods (typically a sample is mildly fixed, permeabilized and labeled by primary reagents before it is embedded and sectioned) can achieve more efficient labeling; post-embedding methods provide better preservation, while the Tokuyasu method often provides the best compromise (Griffiths 1993).
For correlative EM–LM, one way of introducing fluorescent markers is by immunolabeling, where one proceeds in a similar fashion to immunolabeling with electron-dense labels. However, postembedding labeling is generally more difficult to achieve and it is incompatible with the use of osmium tetroxide, a common fixative and staining agent (Giepmans et al. 2005; van Driel et al. 2008). The use of genetic manipulations allows non-invasive introduction of fluorescence into living cells and facilitates the monitoring of various cellular processes by time-lapse or laser scan fluorescence imaging. This in turn allows the identification of a time point where a process that is followed reaches a particular stage, so that the process can be arrested in that state and examined in EM. For example, individual tubulovesicular transport compartments in axons of cultured neurons were labeled by the overexpression of GFP-fusion proteins that are known to associate with these organelles and their movement was observed by recovery after photobleaching of small areas of axons (Nakata et al. 1998).
The movement was arrested by chemical fixation and the area where the fluorescence was observed was imaged by EM. Similarly, the simultaneous labeling by a GFP-fusion protein and a membrane dye allowed the authors to detect the exact time point when individual synapses were formed in neuronal cultures and to arrest further development by chemical fixation in order to image the nascent synapses in conventional EM (Ahmari et al. 2000). Thanks to the clearly identifiable features of neurons in culture and with the help of external orientation marks in both studies the same areas were imaged in both LM and EM thus revealing the cellular environment at the ultrastructural level of the fluorescently labeled tubulovesicular structures involved in the transport of presynaptic components and the synaptogenesis.
The ability of horseradish peroxidase to catalyze the oxidation of diaminobenzidine (DAB) and induce formation of diffusible osmiophilic precipitate has been extensively exploited for immunolabeling in conventional EM. DAB is introduced to an osmium treated and immunolabeled sample, where horseradish peroxidase is typically conjugated to the secondary antibodies, to form an electron-dense reaction product visible in EM. This approach was used to investigate the ultrastructure and identify components of the secretory pathway involved in the transport of cargo from the Golgi apparatus towards the plasma membrane (Polishchuk et al. 2000), within Golgi apparatus (Mironov et al. 2001), and between endoplasmic reticulum and Golgi (Mironov et al. 2003). Pre-embedding fluorescence labeling of structures involved in the transport allowed the authors to follow the transport by time-lapse fluorescence microscopy and to stop it at a desired moment. Furthermore, the immunolabeling was directed against the lumenal part of the transport structures, and so the precipitate formed by the subsequent oxidation of DAB delineated these structures and allowed their detection in EM.
The photooxidation of DAB obtained upon strong illumination of a fluorescent molecule was also used for labeling in conventional EM, and was first shown for Lucifer yellow (Maranto 1982). It brings the additional benefit of observing first a fluorescent label directly in LM and then the product of the photoconversion in EM. FM dyes, widely used labels for studies of synaptic vesicle endocytosis and exocytosis, can be applied to live cells and, under suitable experimental conditions, provide electron-dense labeling of internalized synaptic vesicles, thus functionally distinguishing synaptic vesicle pools (Henkel et al. 1996). Also, fluorescence from a membrane-permeable biarsenical dye ReAsH, induced by its binding to any of the specially designed short genetically encoded tetra-cysteine motives as well as from GFP can induce photooxidation of DAB; these compounds were used for correlative LM and EM (Gaietta et al. 2002; Grabenbauer et al. 2005).
Rapid freezing arrests intracellular processes within milliseconds, and therefore avoids structural changes associated with chemical fixation. For example, the development of Caenorhabditis elegans embryos was followed in LM and stopped at specific stages of early mitosis using high-pressure freezing (Muller-Reichert et al. 2007). In some of the previously mentioned publications on the secretory pathway, tomograms of high pressure frozen, freeze-substituted samples were obtained (Mironov et al. 2001, 2003). However, because pre-embedding immunolabeling methods require chemical fixation and permeabilization and consequently are not compatible with rapid freezing, it was not possible to obtain direct correlation between fluorescent labeling and structures observed in tomograms. Another possible approach for fluorescent labeling of high-pressure frozen samples is to add a fluorescent dye during freeze-substitution (Biel et al. 2003). This type of fluorescence labeling allows imaging of the sample in LM before sectioning, but unlike the pre-embedding methods does not interfere with the sample structure.
Examples of correlation between LM and cryo-ET
At first glance, the correlative LM and cryo-ET procedure appears quite straightforward: a feature of interest is first identified on an LM image, and then traced through a series of EM images of increasing magnification until the feature of interest is resolved at the required level of detail. Nevertheless, its application to samples vitrified in a thin layer of ice or cryo-sectioned brings new challenges and constraints. These samples are quite sensitive to electron irradiation, and imaging in cryo-ET has to be done in the low-dose mode. Consequently, the electron dose spent searching for a feature of interest has to be minimized in order to leave enough room for the recording of a tomographic series. In addition, it is not always possible to move between lower and higher magnification in EM without incurring a positional shift that would leave a feature of interest outside of the field of view. Sometimes, large morphological features are clear enough so that a limited number of low-dose images suffice to establish a correlation based solely on visual cues. Cultured neurons in early developmental stages are sparse enough so that cryo-EM images of microtubules in intact neurons can be correlated to LM images in a straightforward manner (Garvalov et al. 2006). In another study, thin fenestrated areas of cultured endothelial cells not only proved to be thin enough to be investigated by cryo-ET but also provided convenient orientation so that tomograms could be correlated to LM images without resorting to special information or software (Braet et al. 2007). In the previous two examples, LM images were recorded in live cultures before they were frozen, and consequently the ability to make a correlation relied on the adherent nature of these cells. However, thanks to important new developments it is now possible to investigate frozen samples in LM. Namely, cryo-holders that allow LM imaging of EM-grids at liquid nitrogen temperatures were developed (Sartori et al. 2005, 2007; Schwartz et al. 2007). Their designs ensure that a grid in the holder is kept at the liquid nitrogen temperature, so that a sample remains in the vitrified state during imaging under LM. Currently, these devices are limited to long working distance objectives, effectively limiting the resolution that can be achieved. In the design by Schwartz et al. (2007), the rate of cooling is automatically controlled and does not require manual handling. However, loading of EM grids is difficult and the imaging is restricted to episcopic illumination. Liquid nitrogen for cooling needs to be added manually in the design by Sartori et al. (2007) (Fig. 1), but the overall handling is rather easy and transmission illumination (as needed for phase-contrast, for example) can be used for imaging. Both designs are expected to be improved in the forthcoming versions.
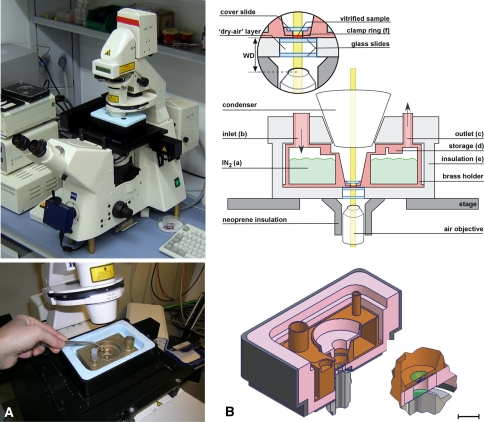
Fig. 1.
Cryo-holder. a Cryo-holder mounted on an LM, general view (above) and a close-up showing the interior of the cryo-holder. b Schematic of the whole cryo-holder assembly. Section of the cryo-holder in operational mode on the stage of the optical microscope (above) and a view in perspective. Insets show magnified views of the central part (scale bar 24 μm). (Modified from Sartori et al. 2007 with permission.)
The development of the cryo-holder for LM opened up the possibility to correlate tomograms obtained from cryo-sections and LM. Ribbons of cryo-sectioned cardiomyocytes that were labeled live with a mitochondrion-specific fluorescent dye were placed on EM grids and imaged in cryo-fluorescence (Gruska et al. 2008) (Fig. 2). Even though longer exposure times were required, fluorophore bleaching did not affect imaging, possibly due to the reduced bleaching rate at cryogenic temperatures (Schwartz et al. 2007; Gruska et al. 2008). This demonstration provides an important “proof of concept” for correlative, low-dose screening capabilities in the electron microscope. Low copy-number features that might even evade detection in 2D electron microscope projection images can be pinpointed with sufficient accuracy to enable subsequent tomography and even serial tomography. Another advantage of LM imaging of frozen material is that features of interest are in the same position during both LM and EM imaging, a prerequisite for the establishment of a direct correlation. This is important not only for highly motile cells but also for cellular processes that change their cellular location rapidly. In an alternative approach, one type of chemosensory receptors present in C. crescentus was fluorescently labeled using genetic methods, allowing the authors to establish a direct correlation between the fluorescence and the periodic structures visualized in tomograms of chemically fixed and immobilized cells (Briegel et al. 2008). This allowed them to identify the periodic structures in non-immobilized cells, thus making an indirect correlation between LM and cryo-EM, and to strengthen conclusions reached earlier (Zhang et al. 2007, discussed earlier). In the earlier examples, correlation between LM and cryo-ET was established based on visual cues, with the help of automated EM grid-scanning procedures (Mastronarde 2005; Sartori et al. 2007). In other cases, the low contrast of the material on the LM level, or technical aspects of the zero-loss energy-filtered EM (the most advantageous for cellular tomography) preclude the effective use of the low magnifications in EM for a precise establishment of correlation. While grid-scanning procedures are still useful to establish a rough correlation, a more precise scheme is needed, especially when the correlation is used to navigate the search in cryo-EM to the position of a given fluorescent spot. A purely coordinate transformation-based software correlation scheme was developed and applied to mature neurons grown in culture, yielding a correlation between synaptic vesicles visualized in cryo-ET and labeled by FM fluorescent dye (Lucic et al. 2007), (Fig. 3). The error of the software-established correlation was determined in the test examples to be between 1 and 3 μm, probably just about the level that would allow a reliable EM search for fluorescent spots. An additional benefit of this approach is that it further reduces the electron dose spent on the feature of interest. The labeling method used here is expected to label functional presynaptic terminals that are capable of synaptic vesicle exocytosis and endocytosis. Therefore, a functional state of a particular presynaptic terminal identified by fluorescent labeling is directly linked to its structural features observed in cryo-tomograms.
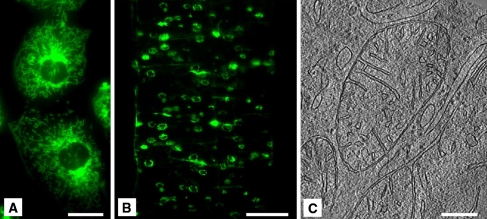
Fig. 2.
Application of LM and cryo-ET correlation on cryo-sections. a Adherent murine cardiomyocytes viewed by epifluorescence microscopy at 37°C after staining with Mitotracker Green FM. b Vitreously sectioned, detached cardiomyocytes as shown in a, viewed using cryo-fluorescence microscopy. Nominal section thickness, 150 nm. c Tomographic slice from a vitreously sectioned cardiomyocyte, showing several mitochondria in cross-section. Nominal section thickness, 85 nm. Scale bars: a 20 μm, b 100 μm, c = 200 nm. (Adapted from Gruska et al. 2008 with permission.)
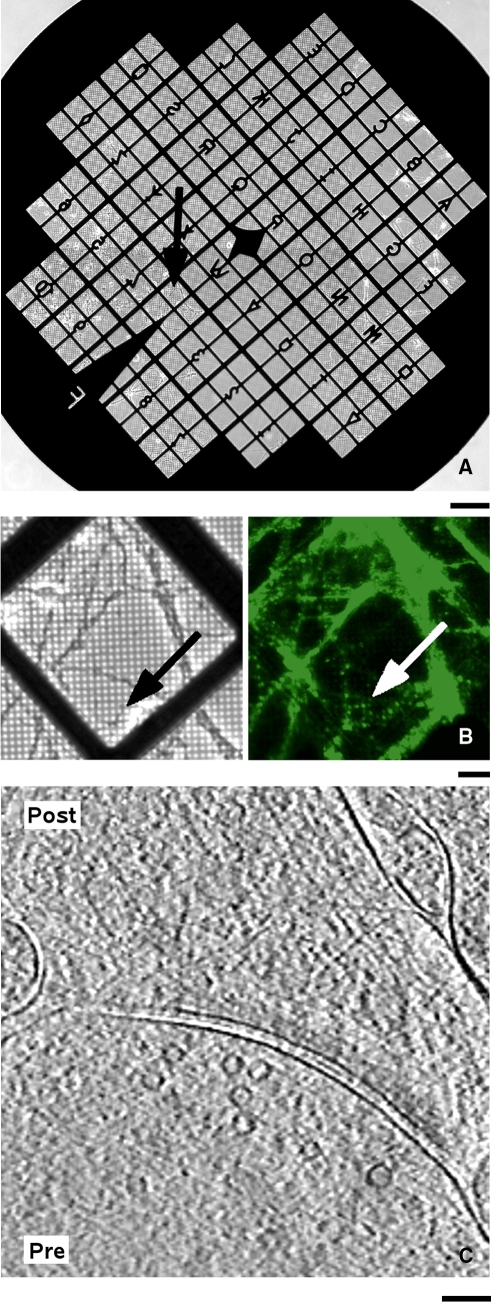
Fig. 3.
Application of LM and cryo-ET correlation on cultured neurons. a The whole grid imaged in phase-contrast LM. The arrow points to a grid square shown in b. Scale bar 200 μm. b Live phase contrast (left) and FM1-43 labeled cryo-fluorescence (right) images. The arrows point to the spot that was correlated to the image shown in c. Scale bar 20 μm. c Tomographic slices showing the correlated synapse. Presynaptic and postsynaptic terminals are labeled by “Pre” and “Post”, respectively. Scale bar 100 nm. (Modified from Lucic et al. 2007 with permission.)
Strategies for correlative LM and cryo-ET
In order to combine the advantages of cryo-ET with LM, fluorescent labeling methods for the correlative approach should be non-invasive, and the labeling has to be done before freezing. Immunolabeling of intracellular epitopes requires membrane permeabilization, and considering that the pore-forming methods for living cells appear to induce significant structural changes (Humbel et al. 1998), immunolabeling methods for living cells seem to be largely restricted to labeling of proteins with extracellular epitopes. This approach was successfully applied to investigate the lateral movement of AMPA type ionic channels/receptors present at the postsynaptic side (Tardin et al. 2003). Of particular interest is labeling with quantum dots (Dahan et al. 2003; Nisman et al. 2004) because they possess strong fluorescence and they can be observed in cryo-tomograms, allowing the detection of a molecule of interest the structure of which is not known or cannot be detected in individual cryo-EM images. However, given the typical distance between the probe and the molecule of interest (around 10 nm), the detection of the molecule in a crowded environment would likely require additional information. In addition, considering that the size of quantum dots is above 10 nm, steric hindrance may prohibit the access of quantum dots to regions with high protein densities, such as cellular junctions. Nevertheless, immunolabeling methods, together with related approaches where a fluorescent or an electron-dense label is conjugated to a protein of interest, may have a wider application to isolated cellular components, as shown in a study where nuclear pore complexes and the positions of gold-labeled proteins involved in nuclear import were visualized in cryo-tomograms of isolated nuclei, and the tomograms were superimposed to obtain the spatial distribution of the labeled protein (Beck et al. 2007). The integration of cryo-tomography with computer simulations of the biochemical reactions underlying nuclear import allowed the authors to obtain a dynamic picture of the cargo as it traverses the nuclear pore (Fig. 4), exemplifying another strategy for providing insights into dynamic processes.
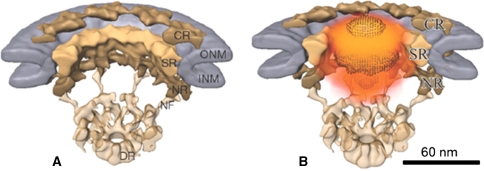
Fig. 4.
From a static to the dynamic picture of nuclear import. a Cut-away view of the nuclear pore complex structure with subjective segmentation for the cytoplasmic (CR), spoke (SR) and nuclear rings (NR) in shades of yellow, and the inner (INM) and outer nuclear membrane (ONM) in bluish-grey. b The same as in a, except that the localization of the labeled cargo at the nuclear pore is shown in orange. (Modified from Beck et al. 2007 with permission.)
Many genetically encoded or non-toxic fluorescent indicators can be introduced into living cells. These include different membrane dyes, probes for various organelles, and metal-ion or pH indicators that can be used for correlations to detect and identify a cellular compartment of interest. Promising candidates for this approach might be smaller membrane-enclosed compartments, the identity and function of which cannot be readily inferred from their pleomorphic structures. This approach can also be useful in cases where the cellular localization or a larger environment of a structure of interest (molecule, membranous structure or an organelle) needs to be determined, or to capture the functional state of a structure of interest and its environment in the case where the fluorescent probe indeed provides such information. In addition, fluorescent fusion proteins the overexpression of which causes functional changes in a cell offer numerous possibilities to identify structures involved in these functional changes.
Considering the observation that imaging in EM rapidly quenches fluorescence (Lucic et al. 2007; van Driel et al. 2008), it follows that fluorescence imaging needs to precede acquisition of a cryotomogram, but the decision between live and cryo fluorescent imaging, or both, often depends on other considerations. We are particularly interested in the approach where correlation is used to navigate the search in cryo-EM to the position of a given fluorescent spot, essentially replacing the search for a feature of interest in EM. However, sample vitrification, especially in cellular tomography, tends to be an unreliable process, yielding ice of heterogenous thickness and not always providing the best preservation, so it is advisable to image a large portion of, if not a whole grid, in order to increase chances that a region where a tomogram can be recorded was imaged in LM. Obviously, the higher the resolution required, the more LM images are needed. Considering that grids are usually slightly bent, z-stacks have to be recorded at each position in order to obtain in-focus images, which greatly increases the time needed for the imaging and handling of the images. On the other hand, imaging with a cryo-stage for LM ensures that the location of imaged structures remains the same in both LM and cryo-EM. Cryo-phase contrast images have poor resolution, but they can be used to estimate ice thickness (Sartori et al. 2007) and to direct (higher resolution) fluorescence imaging only to regions where ice thickness is appropriate for cryo-ET, thus greatly reducing the number of images taken.
As for the correlative approaches involving cryo-sections, additional complications arising from the 3D nature of the frozen samples argue for cryo-fluorescence imaging. Development of special devices and protocols that would allow easier manipulation and confocal imaging of frozen samples prior to cryo-sectioning, perhaps along the lines as for the conventionally prepared or freeze-substituted material (Pfeiffer et al. 2003; Biel et al. 2003), may be quite useful to assist cryo-sectioning. A clonable label for EM would represent one way of localizing supramolecular structures within the cellular context. At first glance, such an approach would be analogous to the use of green fluorescent protein in live cell imaging. Small metal-binding proteins such as metallothionein can be fused to cellular proteins, thus acting as nucleation sites for (electron-dense) gold atoms taken up from the surrounding medium (Mercogliano and DeRosier 2007). This approach is not completely straightforward, however, since it is difficult to distinguish between non-specific uptake/precipitation and bona fide labeling. Metallothioneins occur naturally in many organisms, thus relegating the use of this and similar strategies to organisms where endogenous expression has been suppressed. Furthermore, the need for relatively high concentrations of gold salts in the growth medium and their usefulness as chemotherapeutic agents raises the issue of cytotoxicity or other physiological effects. One must also bear in mind that strategies aimed at pinpointing certain proteins indirectly can only hope to visualize a very small subset of the proteome simultaneously, as opposed to a pattern recognition-based approach with a theoretically comprehensive/complete database of unique templates. On the other hand, information on the 3D subcellular distribution of one particular protein may be a valuable piece of information if morphology is well preserved. Classical immunolabeling of sectioned material grossly underestimates numbers of a particular protein in a 3D volume because high-affinity binding with gold-conjugated antibodies relies on accessibility of epitopes. Hence, labeling of sectioned cells is essentially a 2D (surface) phenomenon. The insertion of a clonable label provides possibilities for 3D labeling but relies on high-affinity binding, or simultaneous nucleation of gold at all target sites, which clearly conflicts with the dynamic nature of protein turnover. Therefore, neither of these approaches can be regarded as quantitative.
Summary and outlook
Cryo-ET is an emerging technique in cell biology that aims to provide faithful 3D images of cellular structures at molecular resolution. The first category of experimental methods integrated with cryo-ET that we considered are genetic and pharmacological manipulations of cells. In the literature we reviewed, these manipulations influenced structures that could be observed directly in tomograms, in most cases cytoskeletal filaments. In some cases, the manipulations increased the frequency of the formation of these structures, thus aiding search routines in cryo, while in other cases, they assisted the detection and molecular identification of structures. While the template matching approaches have a prominent role in the detection and identification of molecular complexes in cryo-ET (Ortiz et al. 2006) (Fig. 5), further technical developments leading to an improved resolution of cryo-tomograms, are likely needed to assure their pervasive applicability.
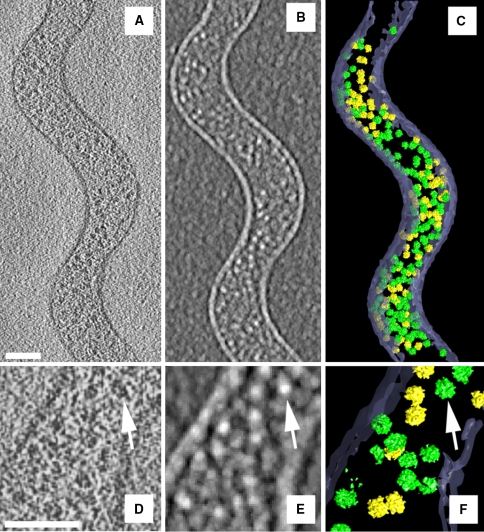
Fig. 5.
Identification by pattern recognition. Scale bars 100 nm. a, d Tomographic slice of an intact S. melliferum cell, b, e Pattern recognition using a 70S ribosome structure as a template, bright spots correspond to the detection peaks. c, f Final ribosome atlas after removal of putative false positives. Green denotes high, while yellow denotes medium detection fidelity. d–f Insets from (a–c). (Modified from Ortiz et al. 2006 with permission.)
Furthermore, we expect that genetic or pharmacological manipulations that can bring a cell or a cellular process into a well-defined state will be more prominent in future cryo-ET work. These investigations might benefit from hybrid approaches involving complementary techniques that can provide information about the molecular composition of a complete cell or some of its organelles, interaction networks, formation of transient molecular complexes, or spatial proximity of different molecules. However, these are mostly “bulk” measurements that do not account for the underlying variability that is evident at the level of single molecules. In order to reconcile this with the “individualistic” nature of cryo-ET, where structures are accessed individually and only a limited number of cryo-tomograms can be analyzed, great care should be taken to assure that the manipulations used are reliable and show high efficiency.
It is somewhat of a paradox that increasing magnification and the concomitant increase in information at a certain structural level also carries the risk of losing the overall picture. In other words, so-called ‘representative images’ may indeed represent the exception rather than the rule. This issue is very much related to specimen quality, since aesthetically acceptable regions of a specimen (suitable ice thickness in a plunge-frozen specimen, for example) will naturally bias a microscopist’s data collection strategy. Thinner ice may, however, simply be the result of smaller cells that in turn may not be representative of a population of cells. Pre-selection of live cells for a particular phenotype by flow cytometry may increase the chances or even ensure that any imaged cell is representative of that phenotype. Also, selection of small cells and their subsequent vitrification, for example, can lead to more efficient data collection, provided that this phenotype is not an artifact, such as a starved or dormant cell phenotype.
The correlative LM and cryo-ET approaches, where the same feature is imaged in both LM (particularly in fluorescence) and cryo-ET, fall in the second category of experimental methods that supplement cryo-ET with information about the dynamics or a functional state of a cell under investigation. This approach was used to help navigate in cryo-EM to a structure of interest, provide information about the wider environment, and to associate dynamic information with structures visualized in cryo-tomograms.
Although correlating LM and cryo-ET is still time-consuming, there are several improvements that are expected to make the correlation approach more productive. Straightforward improvements focusing on flexibility are needed to streamline the software aspect of correlative work. Typically, different computers and operating systems are used, while different samples and microscopes require different correlative approaches. It would be useful to develop quick and flexible computer routines that cover several correlation scenarios. It is crucial to choose open file formats to allow easy exchange of data (such as coordinates of correlated spots) between a program that controls EM and other correlation-related programs, allowing users to develop their own correlation scripts that best fit their needs. If a correlation is precise, reliable procedures for automated recording of multiple tomographic series and procedures for automated tomographic reconstruction may then allow us to obtain several tomograms correlated to features of interest in LM, with minimal user interference.
One can envisage correlative approaches based on advanced LM techniques, such as time-lapse microscopy, or FRET-based methods that allow monitoring of different functional states of molecules in living cells. Although these methods can provide precise information about complex formation and translocation, they do not reveal structural components. Consequently, their correlation with cryo-ET has great potential in identifying structural correlates of many functional states. Furthermore, recent fluorescence imaging techniques that can reach resolutions above the diffraction limit, such as stimulated emission depletion (Hell 2003), photo-activated localization microscopy (Betzig et al. 2006), stochastic optical reconstruction microscopy (Rust et al. 2006), or nonlinear structured illumination (Gustafsson 2005), may in principle allow detection of individual fluorescently labeled molecules in correlated cryo-tomograms. While in principle any information from LM can be used for a correlation, difficulties associated with cellular cryo-ET might seriously limit such an approach. The diffraction limit breaking methods typically require high illumination intensities, which may limit their applicability to intact cells and make the correlative approach even more involved. In any case, one would need to ensure that a fair number of fluorescently labeled structures exist on a given sample, so that the chances of recording a cryo-tomogram correlated to one of these structures are reasonably high. Cryo-X-ray tomography provides the enticing possibility to visualize entire cells in 3D at a resolution intermediate between that of LM and EM, and to directly correlate tomograms with 3D LM volumes of the same cell (Schneider 1998). In fact, a light microscope can be integrated into the system to directly screen for features of interest prior to tomography. Interpretation of X-ray microscopy data is straightforward because image formation is based on absorption contrast (note that phase contrast imaging using a Zernicke-type phase stop is also possible). A further attractive feature of this technique is that specimens can be prepared in a manner identical to that used for cryo-ET. Vitreous ice is essentially transparent to X-rays between the K-absorption edges of carbon and oxygen to a depth of 10 µm, thus allowing imaging of relatively large structures at these energies. Ice embedded specimens with suitable size/geometry can be tilted over the full angular range, giving rise to tomograms with isotropic resolution. Resolution in X-ray tomography is largely a function of the outermost ring of the zone plate used to focus the X-rays with the caveat that the use of high-resolution zone plates reduces the amount of the specimen kept in focus: for example, a 40 nm outer ring corresponds to a 1.3 μm depth of focus, and 25 nm zone plates are now being introduced. Fortunately, cryogenically prepared cells can tolerate large doses of X-ray radiation before the onset of visible damage (>1010 Gray), which makes the acquisition of a focus series for each tilt angle a viable proposition.
Arguably, the careful and often tedious optimization of sample preparation remains among the most important objectives and holds the key for the further integration of cryo-ET with other experimental methods, be it for sophisticated treatment schemes or correlations using advanced LM methods. We expect that these, as well as computational and technical developments will lead to attributing further functional aspects to cellular structures visualized in cryo-ET.
Acknowledgments
We thank Gabriela J. Greif for critical reading of the manuscript. This work was supported by DFG SPP 1128 grant and by the EU within the Network of Excellence NoE-3DEM.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ahmari SE, Buchanan J, Smith SJ. Assembly of presynaptic active zones from cytoplasmic transport packets. Nat Neurosci. 2000;3:445–451. doi: 10.1038/74814. [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlen LP, Richter K, Blanc NS, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–684. doi: 10.1016/S0959-440X(02)00378-0. [DOI] [PubMed] [Google Scholar]
- Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- Berriman J, Unwin N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy. 1994;56:241–252. doi: 10.1016/0304-3991(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Biel SS, Kawaschinski K, Wittern KP, Hintze U, Wepf R. From tissue to cellular ultrastructure: closing the gap between micro- and nanostructural imaging. J Microsc. 2003;212:91–99. doi: 10.1046/j.1365-2818.2003.01227.x. [DOI] [PubMed] [Google Scholar]
- Braet F, Wisse E, Bomans P, Frederik P, Geerts W, Koster A, Soon L, Ringer S. Contribution of high-resolution correlative imaging techniques in the study of the liver sieve in three-dimensions. Microsc Res Tech. 2007;70:230–242. doi: 10.1002/jemt.20408. [DOI] [PubMed] [Google Scholar]
- Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Jensen RB, Jensen G. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- van Driel LF, Knoops K, Koster AJ, Valentijn JA. Fluorescent labeling of resin-embedded sections for correlative electron microscopy using tomography-based contrast enhancement. J Struct Biol. 2008;161:372–383. doi: 10.1016/j.jsb.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Frederik PM, Sommerdijk N. Spatial and temporal resolution in cryo-electron microscopy—a scope for nano-chemistry. Curr Op Coll Interface Sci. 2005;10:245–249. doi: 10.1016/j.cocis.2005.09.018. [DOI] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Garvalov BK, Zuber B, Bouchet-Marquis C, Kudryashev M, Gruska M, Beck M, Leis A, Frischknecht F, Bradke F, Baumeister W, Dubochet J, Cyrklaff M. Luminal particles within cellular microtubules. J Cell Biol. 2006;174:759–765. doi: 10.1083/jcb.200606074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Griffiths G (1993) Fine structure immunocytochemistry link. Springer, Berlin
- Gruska M, Medalia O, Baumeister W, Leis A. Electron tomography of vitreous sections from cultured mammalian cells. J Struct Biol. 2008;161:384–392. doi: 10.1016/j.jsb.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell SW. Toward fluorescence nanoscopy. Nat Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- Henderson GP, Gan L, Jensen GJ. 3-D ultrastructure of O. tauri: electron cryotomography of an entire eukaryotic cell. PLoS ONE. 2007;2:e749. doi: 10.1371/journal.pone.0000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel AW, Lubke J, Betz WJ. FM1–43 dye ultrastructural localization in and release from frog motor nerve terminals. Proc Natl Acad Sci USA. 1996;93:1918–1923. doi: 10.1073/pnas.93.5.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbel BM, de Jong MD, Muller WH, Verkleij AJ. Pre-embedding immunolabeling for electron microscopy: an evaluation of permeabilization methods and markers. Microsc Res Tech. 1998;42:43–58. doi: 10.1002/(SICI)1097-0029(19980701)42:1<43::AID-JEMT6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Koster AJ, Klumperman J (2003) Electron microscopy in cell biology: integrating structure and function. Nat Rev Mol Cell Biol (Suppl:S)S6–S10 [PubMed]
- Kurner J, Frangakis AS, Baumeister W. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science. 2005;307:436–438. doi: 10.1126/science.1104031. [DOI] [PubMed] [Google Scholar]
- Liu J, McBride MJ, Subramaniam S. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol. 2007;189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Forster F, Baumeister W. structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Lucic V, Kossel A, Yang T, Bonhoeffer T, Baumeister W, Sartori A. Multiscale imaging of neurons grown in culture: from light microscopy to cryo-electron tomography. J Struct Biol. 2007;160:146–156. doi: 10.1016/j.jsb.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranto AR. Neuronal mapping: a photooxidation reaction makes Lucifer yellow useful for electron microscopy. Science. 1982;217:953–955. doi: 10.1126/science.7112109. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, Gerisch G. Organization of actin networks in intact filopodia. Curr Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV, Nicoziani P, Martella O, Trucco A, Kweon HS, Di Giandomenico D, Polishchuk RS, Fusella A, Lupetti P, Berger EG, Geerts WJ, Koster AJ, Burger KN, Luini A. Small cargo proteins and large aggregates can traverse the golgi by a common mechanism without leaving the lumen of cisternae. J Cell Biol. 2001;155:1225–1238. doi: 10.1083/jcb.200108073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Mironov AA, Jr, Beznoussenko GV, Trucco A, Lupetti P, Smith JD, Geerts WJ, Koster AJ, Burger KN, Martone ME, Deerinck TJ, Ellisman MH, Luini A. ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev Cell. 2003;5:583–594. doi: 10.1016/S1534-5807(03)00294-6. [DOI] [PubMed] [Google Scholar]
- Moor H. Theory and practice of high pressure freezing. In: Steinbrecht RA, Zierold K, editors. Cryotechniques in biological electron microscopy. Berlin: Springer; 1987. pp. 175–191. [Google Scholar]
- Muller-Reichert T, Srayko M, Hyman A, O’Toole ET, McDonald K. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 2007;79:101–119. doi: 10.1016/S0091-679X(06)79004-5. [DOI] [PubMed] [Google Scholar]
- Nakata T, Terada S, Hirokawa N. Visualization of the dynamics of synaptic vesicle and plasma membrane proteins in living axons. J Cell Biol. 1998;140:659–674. doi: 10.1083/jcb.140.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP. Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. J Histochem Cytochem. 2004;52:13–18. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- Ortiz JO, Forster F, Kurner J, Linaroudis AA, Baumeister W. Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol. 2006;156:334–341. doi: 10.1016/j.jsb.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Pfeiffer S, Beese M, Boettcher M, Kawaschinski K, Krupinska K. Combined use of confocal laser scanning microscopy and transmission electron microscopy for visualisation of identical cells processed by cryotechniques. Protoplasma. 2003;222:129–137. doi: 10.1007/s00709-003-0014-6. [DOI] [PubMed] [Google Scholar]
- Polishchuk RS, Polishchuk EV, Marra P, Alberti S, Buccione R, Luini A, Mironov AA. Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. J Cell Biol. 2000;148:45–58. doi: 10.1083/jcb.148.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Kossel A, Leis A, BaumeisterW, Plitzko JM (2005) Correlationmicroscopy: Bridging the gap between light- and cryo-electronmicroscopy. Microsc and Microanal 11(Suppl 2):16–17
- Sartori A, Gatz R, Beck F, Rigort A, BaumeisterW Plitzko J. Correlative microscopy: bridging the gap between light- and cryo-electron microscopy. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Schneider G. Cryo X-ray microscopy with high spatial resolution in amplitude and phase contrast. Ultramicroscopy. 1998;75:85–104. doi: 10.1016/S0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J Microsc. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardin C, Cognet L, Bats C, Lounis B, Choquet D. Direct imaging of lateral movements of ampa receptors inside synapses. EMBO J. 2003;22:4656–4665. doi: 10.1093/emboj/cdg463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Bos E, Heymann J, Gnaegi H, Kessel M, Peters PJ, Subramaniam S. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J Microsc. 2004;216:76–83. doi: 10.1111/j.0022-2720.2004.01395.x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–81. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]