Abstract
Growth hormone (GH) binding to its receptor modulates gene transcription by influencing the amount or activity of transcription factors. In the rat, GH exerts sexually dimorphic effects on liver gene transcription through its pattern of secretion which is intermittent in males and continuous in females. The expression of the CYP2C12 gene coding for the female-specific cytochrome P450 2C12 protein is dependent on the continuous exposure to GH. To identify the transcription factor(s) that mediate(s) this sex-dependent GH effect, we studied the interactions of the CYP2C12 promoter with liver nuclear proteins obtained from male and female rats and from hypophysectomized animals treated or not by continuous GH infusion. GH treatment induced the binding of a protein that we identified as hepatocyte nuclear factor (HNF) 6, the prototype of a novel class of homeodomain transcription factors. HNF-6 competed with HNF-3 for binding to the same site in the CYP2C12 promoter. This HNF-6/HNF-3 binding site conveyed both HNF-6- and HNF-3-stimulated transcription of a reporter gene construct in transient cotransfection experiments. Electrophoretic mobility shift assays showed more HNF-6 DNA-binding activity in female than in male liver nuclear extracts. Liver HNF-6 mRNA was barely detectable in the hypophysectomized rats and was restored to normal levels by GH treatment. This work provides an example of a homeodomain-containing transcription factor that is GH-regulated and also reports on the hormonal regulation of HNF-6.
Keywords: CYP2C12, gene transcription, hepatocyte nuclear factor 3, homeoproteins, sexual dimorphism
Growth hormone (GH) is the major regulator of postnatal body growth (1), It also controls lipid and carbohydrate metabolism (2), Binding of GH to its receptor triggers several signaling cascades that account for the short-term effects of GH, which are exerted via the phosphorylation of intracellular proteins, and for its long-term effects, which result from changes in gene transcription (3).
Several genes are considered as primary GH targets. These include the genes for insulin-like growth factor I (IGF-I) (4), which mediates many of the growth-promoting properties of GH, for spi-2.1 (5, 6), a liver-specific serine protease inhibitor, and for the transcription factors c-fos (7), c-jun, and jun B (8), c-myc (9), and C/EBPδ (10). Such GH-induced transcription factors can in turn mediate secondary actions of the hormone by binding to their cognate cis-acting elements in other genes whose rate of transcription is thereby modified.
In adult rodents, the secretion of GH is sexually dimorphic and this leads to a sex-difference in the expression of several genes (11). The male rat secretory profile is characterized by peaks of large amplitude every 3 to 4 h with undetectable interpulse levels, whereas the female profile shows more frequent oscillations of smaller amplitude which result in a continuous presence of GH in the blood (12, 13). The rat CYP2C11 and CYP2C12 genes, which code for steroid hydroxylases expressed in liver, are transcriptionally regulated by GH (11). The CYP2C11 gene is induced by the male GH secretory pattern and is repressed by the female pattern, whereas the CYP2C12 gene is activated by the female pattern (11, 14). We have previously shown that the GH-dependent expression of CYP2C12 requires protein synthesis (15). This result suggested that the GH-dependent transcription factors activating CYP2C12 transcription are themselves regulated at the gene level. We have now identified such a GH-dependent factor as hepatocyte nuclear factor (HNF) 6, a recently described liver-enriched protein that is the prototype of a novel class of homeoproteins (16). We also show that HNF-3, a member of the winged-helix family of transcription factors (17, 18), competes with HNF-6 for binding to the same DNA-binding site on the CYP2C12 promoter and that both HNF-6 and HNF-3 increase the transcriptional activity of this promoter fused to a reporter gene in transfection experiments.
MATERIALS AND METHODS
Animals.
Six-week-old Sprague–Dawley or Wistar rats obtained 7 days after hypophysectomy or sham operation and age-matched control rats (Mollegaards Breeding Centre, Skensved, Denmark, and IFFA-Credo, Lyon, France) were maintained under standardized conditions of light and temperature with free access to standard rat chow and water. Experiments were begun after a 7-day adaptation period. Recombinant bovine (American Cyanamid) or rat (National Institute of Diabetes and Digestive and Kidney Diseases rat pituitary hormone distribution program) GH was delivered to hypophysectomized animals by continuous infusion from an osmotic minipump (model 2001; Alza) so as to mimic the female GH secretory pattern, at a daily dose of 50 or 250 μg/100 g body weight for 7 days. Alternatively, hypophysectomized rats were treated for 7 days with four daily subcutaneous injections of 12.5 μg of GH. These were given at 2 a.m., 8 a.m., 2 p.m., and 8 p.m. The last injection was given on day 8 at 8 a.m. and the rats were killed by decapitation 1, 3, or 6 h afterward. The efficiency of GH treatment was verified by measuring the P450 2C11 and P450 2C12 mRNA levels (19). Nuclear extracts were prepared from individual and from pooled rat livers as described (11). Total RNA was isolated from individual livers by the guanidine thiocyanate/cesium chloride method (20).
Plasmids and Antibodies.
The −131 2C12-Luc construct was identical to the −138 2C12-Luc construct (19) whose coordinates were corrected. Site-directed mutagenesis of the HNF-6/HNF-3 DNA-binding site was done as described (19) by overlap extension from the −131 2C12-Luc construct using PCR (21) and the following oligonucleotide (mutated bases are underlined): CYP2C12 mut (−52 to −30) GCAAAAGATGGTTTTTTATGGTG. To obtain the −131 2C12-Luc mut vector, the PCR product was cleaved with restriction enzymes, subcloned into the corresponding sites of the promoterless luciferase pGL2-basic, and sequenced. The pRL-CMV vector (Promega) coding for Renilla luciferase was used as internal control in transfection assays. The pCMV-HNF-3β (22) and pECE-HNF-6 (16) expression plasmids contain cDNAs coding for HNF-3β and HNF-6, respectively. To obtain anti-HNF-6 antibodies, rabbits were injected at 2-week intervals with recombinant HNF-6 coupled to keyhole limpet hemocyanin and mixed with Freund’s adjuvant. Recombinant HNF-6 was obtained by expressing in Escherichia coli a chimeric protein consisting of glutathione S-transferase fused to residues 111–465 of rat HNF-6. Igs were precipitated from preimmune and immune serum by ammonium sulfate addition. Antisera against HNF-3α, β, or γ were generous gifts from J. E. Darnell (The Rockefeller University, New York).
Electrophoretic Mobility Shift Assay (EMSA).
The following oligonucleotides synthesized by Cybergene (Huddinge, Sweden) were used as probes: CYP2C12 (−52 to −30) GCAAAATATTGATTTTTATGGTG, CYP2C12 mut (see above), and mAP1/HNF-3 GTCTGCTAAGTCAATAATCAGAAT (23). They were labeled with [γ-32P]ATP (Amersham) by T4 polynucleotide kinase (United States Biochemical) and purified on a nondenaturing 15% polyacrylamide gel. Binding reactions involved mixing 10 fmols of probe (5 × 104 cpm) and 1 μg of poly(dI-dC) (Pharmacia) with 2 μg of liver nuclear protein in binding buffer (Hepes, pH 7.9/40 mM KCl/2 mM MgCl2/0.1 mM EGTA/0.5 mM DTT/4% Ficoll 400) adjusted to a volume of 20 μl. After a 30-min incubation at room temperature the resulting DNA–protein complexes were separated at room temperature on preelectrophoresed 5% polyacrylamide gels in a buffer containing 25 mM Tris-borate and 0.5 mM EDTA. DNA-bound factors were identified with immune sera or nonlabeled oligonucleotides added to liver nuclear extracts, on ice, 45 min prior to addition of the labeled probe. Incubation with the labeled probe was then allowed to proceed for 45 min on ice before electrophoresis. HNF-6, HNF-3α, HNF-3β, and HNF-3γ proteins were generated in the presence of 14C-leucine (Amersham) via in vitro transcription and translation of the full-length cDNAs cloned into transcription vectors (16, 22) with the TnT wheat germ extract system (Promega) and were used directly in EMSA. For comparing relative DNA-binding activities, the relative amount of synthesized protein was quantified with a PhosphorImager (Molecular Dynamics) after SDS/PAGE.
RNase Protection Assays.
These experiments were performed as described (16) with 20 μg of total RNA. The HNF-6 probe [290 bases (b)] enables detection of the two HNF-6 isoforms (16). A rat glyceraldehyde-3-phosphate dehyrogenase (GAPDH) antisense RNA probe (Ambion, Austin, TX) was cohybridized (5 × 104 cpm) with the HNF-6 probe as an internal control to correct for variations in RNA concentration. After RNase digestion and separation of the protected fragments on a 6% polyacrylamide denaturing gel, the GAPDH and HNF-6 mRNAs were identified by the 316-b and 215/254-b protected fragments, respectively, and quantified with a PhosphorImager.
Transient Transfections.
The human hepatoma cell line HepG2, obtained from the ATCC repository (National Institutes of Health), was grown in DMEM containing 10% fetal calf serum, 50 units/ml penicillin, and 50 units/ml streptomycin. Cells were seeded at half confluency prior to transfection using the DOTAP reagent (Boehringer Mannheim). The amount of transfected plasmid was kept constant by adding empty expression vector. Plasmids were purified with a Qiagen kit (Qiagen, Chatsworth, CA). Luciferase activities were determined 40 h after the onset of transfection with the Dual-Luciferase Reporter Assay System (Promega) in a luminometer Biocounter M2000 (Lumac, Landgraaf, The Netherlands). Firefly luciferase values were normalized with the Renilla luciferase activity to correct for variations in transfection efficiency. Transfections (three dishes per point) were repeated three times with different plasmid preparations.
RESULTS
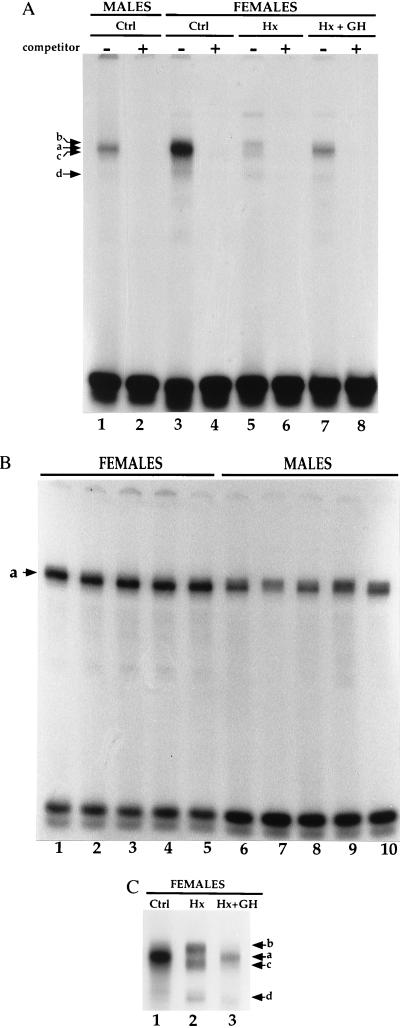
In vitro footprinting of the CYP2C12 promoter with liver nuclear proteins prepared from rats differing in GH status showed GH-dependent differences in the DNase I cleavage pattern within 100 bp from the transcription initiation site (unpublished results). Therefore, using EMSA we analyzed the putative DNA-binding sites present in this region. The band-shift pattern obtained with a CYP2C12 probe extending from −52 to −30 relative to the initiation site showed qualitative and quantitative protein binding differences that were sex- and GH-dependent (Fig. 1). First, the major complex (a) detected using pools of liver extracts was about 2-fold more abundant for female than for male rats (Fig. 1A, lane 3 vs. 1). This sexual dimorphism was confirmed with extracts prepared from individual rats, male extracts yielding complexes of weaker intensity, and of greater interindividual variation than female extracts (Fig. 1B, lanes 6–10 vs. 1–5). Second, hypophysectomy led to the disappearance of the complex seen with control extracts (Complex a) and to the appearance of three other complexes (b–d) (Fig. 1A, lane 5 and Fig. 1C, lane 2). Continuous infusion (female pattern) of GH in hypophysectomized rats restored the pattern seen in control animals (Fig. 1A, lane 7 and Fig. 1C lane 3) although the GH-dependent complex (a) was less abundant than in normal females. None of these experimental conditions altered the band-shift pattern seen with a probe that recognizes the Sp1 transcription factor (data not shown). We concluded that the abundance or DNA-binding capacity of the factor responsible for complex a was not only sex-dependent, but also GH-dependent.
Figure 1.
Sex- and GH-dependent binding of liver nuclear proteins to the CYP2C12 probe. (A) EMSA with the CYP2C12 probe incubated with pooled rat liver nuclear extracts (2 μg of protein per lane) prepared from control (Ctrl) males or females, from hypophysectomized (Hx) females or from Hx females infused with GH for 7 days according to a female-like pattern (Hx + GH). Complexes a, b, c, and d refer to the specific complexes detected in the absence (−), but not in the presence (+), of a 100-fold molar excess of the unlabeled probe. (B) EMSA with the CYP2C12 probe incubated with liver nuclear extracts (2 μg of protein per lane) prepared from five individual males or females. (C) To improve the resolution of the complexes observed in A, the running time of the female samples shown in A was doubled.
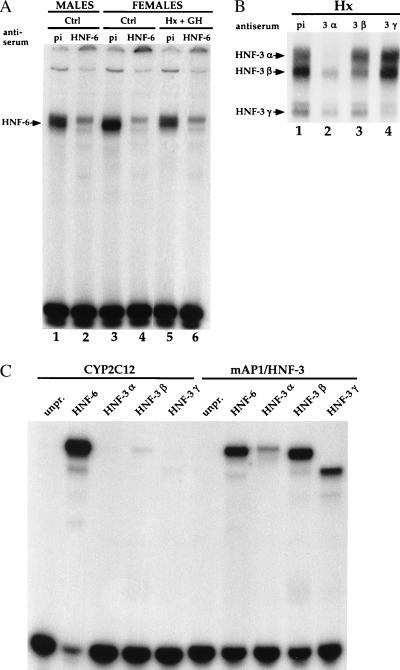
The CYP2C12 probe contains the sequence ATATTGATTT (−47 to −38), which fits with the DNA-binding consensus sequence for HNF-6 [(A/T)(A/T)AT(T/G)GPYTT] (16, 24) and for HNF-3 [(A/T)TRTT(T/G)RYTY] (25, 26). We therefore repeated the EMSA with the different liver extracts in the presence of an anti-HNF-6 immune serum. This specifically prevented the formation of complex a with extracts from normal males and females and from hypophysectomized females treated with GH (Fig. 2A, lanes 2, 4, and 6). These results showed that the only factor responsible for the GH-dependent complex a was HNF-6 and that HNF-6 is expressed in both sexes with a sexual dimorphism.
Figure 2.
Binding of HNF-6 and HNF-3 to the CYP2C12 and mAP1/HNF-3 probes. (A) EMSA performed with the CYP2C12 probe using the same nuclear extracts as those used for the experiment described in Fig. 1A, after preincubation with preimmune serum (pi) or anti-HNF-6 immune serum (HNF-6). (B) EMSA performed with the CYP2C12 probe using nuclear extracts prepared from Hx females. The extract (2 μg of protein per lane) was preincubated with preimmune serum (pi) or with immune serum directed against HNF-3α, β, or γ. (C) EMSA performed with identical amounts of in vitro synthesized proteins. Wheat germ extracts (6 μl per lane), either unprogrammed (unpr.) or programmed with expression vectors for HNF-6, HNF-3α, HNF-3β, or HNF-3γ, were incubated with the CYP2C12 or mAP1/HNF-3 probes before migration.
The complete overlap of the HNF-6 and HNF-3 consensus sequences in the CYP2C12 DNA binding site studied here suggested that complexes b, c, and d, best seen in hypophysectomized females (see Fig. 1C, lane 2), result from the binding of HNF-3. Therefore, we preincubated liver extracts from such animals with immune serum against HNF-3α, β or γ prior to EMSA. This decreased the intensity of complexes b, c, and d, respectively (Fig. 2B). Thus, both HNF-6 and HNF-3 bind to the site detected in the CYP2C12 promoter. To determine their relative affinities, we studied by EMSA the binding of recombinant HNF-6 and HNF-3 to the CYP2C12 probe and to a probe (mAP1/HNF-3) that corresponds to the strong affinity HNF-3 binding site found in the rat transthyretin promoter (27) and which also binds HNF-6 (16, 24). Indeed, the mAP1/HNF-3 probe showed, using EMSA with rat liver nuclear extracts, a GH-dependent binding activity in addition to the HNF-3α, β, and γ complexes usually observed (data not shown). Using identical amounts of recombinant HNF-6 and HNF-3 synthesized in vitro, we observed with the CYP2C12 probe a much greater abundance of the HNF-6 complex than of the HNF-3β or γ complexes (Fig. 2C). A specific HNF-3α complex was observed when a higher amount of protein was used (data not shown). Such a difference in affinity between HNF-6 and HNF-3 results from the nature of the CYP2C12 binding site itself and not from a defective binding of the synthesized HNF-3 proteins. Indeed, when the mAP1/HNF-3 probe was used, HNF-3α, β, and γ complexes were clearly observed (Fig. 2C). HNF-6 and HNF-3β had a similar binding affinity for this probe, stressing that the nucleotide sequence of the site determines the respective affinities of HNF-6 and HNF-3. The experiments with the recombinant proteins (Fig. 2C) and with the liver nuclear extracts (Fig. 1C) also showed that the complexes between DNA and HNF-6 (51 kDa, ref. 16), HNF-3α (50 kDa, ref. 22), and HNF-3β (47 kDa, ref. 22) migrated with similar velocities. Thus, HNF-6 binds as a monomer to the CYP2C12 and mAP1/HNF3 binding sites. Taken together, these results make us conclude that HNF-6 and HNF-3 compete for the same binding site on the CYP2C12 probe but that the affinity of HNF-3 is much lower than that of HNF-6.
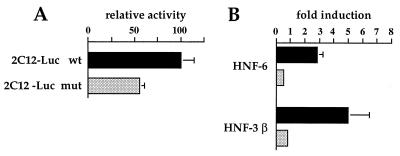
To study the functional consequences of HNF-6 and HNF-3 binding to the CYP2C12 promoter, we cotransfected these factors in hepatoma cells together with the 154 bp (−131 to +23) CYP2C12 promoter fused to a luciferase reporter gene (−131 2C12-Luc). These experiments were also conducted with a reporter construct in which the HNF-6/HNF-3 site was disrupted (−131 2C12-Luc mut). EMSA with the CYP2C12 mut probe demonstrated that the mutation abolished HNF-6 and HNF-3 binding (data not shown). As shown in Fig. 3A, this mutation decreased basal promoter activity by about 50%, showing that the site studied here contributes to the activity of the CYP2C12 promoter. This decrease is consistent with the fact that HNF-6 is present in untransfected HepG2 cells (ref. 24 and unpublished data). The relative luciferase activity of the wild-type construct was specifically increased about 5-fold with HNF-3β and about 3-fold with HNF-6 (Fig. 3B). HNF-6 and HNF-3β had no effect on the mutated construct (Fig. 3B). The transcriptional stimulation by HNF-3β is higher than that observed with HNF-6. This is apparently at odds with the lower affinity of HNF-3β for the binding site (Fig. 2C). As HNF-3β and HNF-6 are structurally unrelated, we cannot exclude a cooperation of HNF-3β with other factors that bind to the reporter construct used. Altogether, our data show that HNF-6 and HNF-3 can stimulate the transcription of the CYP2C12 gene.
Figure 3.
Stimulation of CYP2C12 promoter activity by HNF-6 and HNF-3β. HepG2 cells were transiently transfected with 1 μg of a firefly luciferase reporter gene linked to the wild-type (filled boxes) or mutated (shaded boxes) 2C12-Luc promoter (−131 to +23) in absence (A) or presence (B) of 60 ng of HNF-6 or HNF-3β expression vectors. A Renilla luciferase reporter plasmid (25 ng) was included as internal control. The data are means ± SEM for three independent experiments.
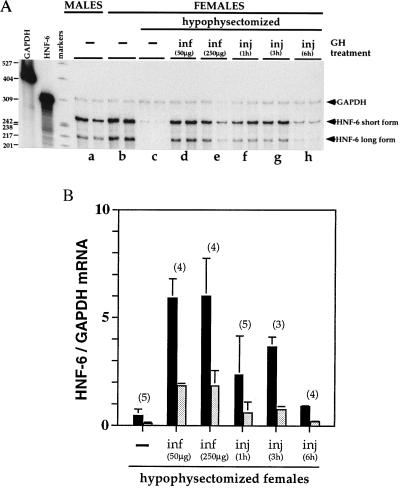
The GH-dependence of the DNA-binding activity of HNF-6 suggested that GH might influence HNF-6 mRNA levels. We therefore quantified HNF-6 mRNA in liver RNA samples prepared from rats with different GH status (Fig. 4 A and B). As expected, two protected fragments were detected, which correspond to the short and long forms of HNF-6 mRNA (16). HNF-6 mRNA was about 2-fold more abundant in female than in male rats (Fig. 4A, lanes b vs. a) and was barely detectable in hypophysectomized animals (Fig. 4A, lanes c). Administration of GH according to a female-like pattern—i.e., continuous infusion—restored both forms of mRNA back to female control levels (Fig. 4A, lanes d and e, and Fig. 4B). Intermittent administration of GH, mimicking the male GH secretory pattern, restored HNF-6 mRNA to the level seen in normal males (Fig. 4A, lanes f and g, and Fig. 4B). Moreover, the latter protocol showed that the effect of GH on HNF-6 mRNA is short-lived. Indeed, this effect peaked between 1 and 3 h and was no longer seen 6 h after the last injection of GH (Fig. 4A, lanes h, and Fig. 4B). Taken together, these results suggest that the constant presence of GH is required to maintain liver HNF-6 mRNA and DNA-binding activity at its physiological levels.
Figure 4.
Influence of GH on the concentration of HNF-6 mRNA detected in rat liver by RNase protection assay. (A) Total RNA (20 μg) was incubated with an HNF-6 and a GAPDH riboprobe. After digestion with RNase, the protected fragments were separated on a 6% denaturing polyacrylamide gel. Each lane corresponds to an individual sample (two animals per experimental condition). Arrows point to the protected fragments expected for GAPDH mRNA (316 b), and for the short (254 b) and long (215 b) forms of HNF-6 mRNA (16). GH was administered for 7 days either by minipump infusion to mimic the female secretory pattern (lane d, 50 μg GH; lane e, 250 μg GH per 100 g body weight) or by daily injections of 12.5 μg GH to mimic the male secretory pattern and sacrificed 1 h (lane f), 3 h (lane g), or 6 h (lane h) after the last injection. (B) Relative concentration of HNF-6 mRNA short form (filled boxes) or long form (shaded boxes) in the liver of hypophysectomized female rats left untreated or treated with GH according to the corresponding protocol described under A above. Data are means ± SEM for the number of animals indicated above the bars.
DISCUSSION
We have shown here that HNF-6, the prototype of a novel class of transcription factors conserved from nematode to mammals (16), is regulated by GH and is therefore a potential mediator of GH action on gene transcription in rat liver.
We previously demonstrated that GH delivered continuously for 6 days (female pattern) to hypophysectomized female rats, in which the CYP2C12 gene was no longer expressed, restored both P450 2C12 mRNA level and CYP2C12 gene transcriptional activity back to control female levels, whereas GH administered intermittently (male pattern) only minimally increased P450 2C12 mRNA (11). Using the same in vivo model, we have now identified in the CYP2C12 promoter a GH-regulated cis-acting element located 10 nucleotides upstream of the TATA box (28). We also show that (i) HNF-6 and HNF-3 compete for the same binding site on this element; (ii) HNF-6 binding activity and mRNA depend on GH and are present in both sexes, being 2-fold higher in females than in males; and (iii) in hypophysectomized animals, the two GH secretory patterns induce liver HNF-6 mRNA, the female pattern being about 2-fold more efficient than the male.
Hypophysectomy dramatically decreased HNF-6 gene expression in liver but it did not abolish it, suggesting there is constitutive expression of HNF-6 that is independent of GH. On the other hand, we cannot exclude an effect of GH synthesized from ectopic sites (29). The rapid decay of HNF-6 mRNA levels observed during the 3- to 6-h period following the last GH injection in the hypophysectomized rats treated intermittently suggests that this mRNA has a half-life shorter than 3 h and that the constant presence of GH in the blood is required for maintenance of adequate HNF-6 mRNA levels and binding activity. The rapid decrease in HNF-6 mRNA is not compatible with an indirect GH action through IGF-I, because, following an injection of GH as done here, the blood level of GH is no longer detectable after 3 h whereas the blood level of IGF-I is still high after 6 h (30). The short half-life of HNF-6 mRNA can also explain why a male pattern of GH secretion is 2-fold less efficient than a female pattern to accumulate HNF-6 mRNA, because the time period between each GH-peak is 3–4 h in the male (13). In females, the higher level of HNF-6 gene expression would result from sustained activation by the constant presence of GH.
We have shown that HNF-6 activates transcription of the CYP2C12 promoter through a site that also binds HNF-3. The much higher affinity of HNF-6 than of HNF-3 for this binding site suggests that it is occupied predominantly by HNF-6 in normal female rats and therefore mediates the GH-dependent transactivating properties of HNF-6. Because HNF-6 is expressed in both sexes, the female-specific expression of the CYP2C12 gene requires additional control mechanisms. GH-regulation of CYP2C12 may result from a cooperativity or synergism between HNF-6 and other GH-regulated transcription factors. Indeed, five other regions of the CYP2C12 promoter are bound by a female-enriched GH-dependent complex called GHNF (31). The binding of these transcription factors, which might be rate-limiting in males, would initiate a GH-dependent transcriptional regulation only in females. In addition to its direct action on the CYP2C12 promoter, HNF-6 may also regulate other genes involved in the sexual regulation of the CYP2C12 gene.
Binding sites for HNF-6 can be identified by computer search in the promoter of several genes—i.e., the mouse Mup genes (32, 33) and the rat CYP2C13 (11, 34), CYP2A1 (35, 36), and CYP2A2 (35–37) genes—whose expression in liver is regulated in a sexually dimorphic way by GH. The present work therefore suggests that HNF-6 might be involved in the sex-dependent control of several GH-regulated genes. Moreover, many binding sites for HNF-6 also bind HNF-3 (refs. 16 and 24, and this report). It is therefore likely that in the liver GH affects through HNF-6 the transcriptional activity of many of the genes regulated by HNF-3, the net effect depending on the relative affinities of HNF-6 and HNF-3 for the site and on the promoter context. Finally, as HNF-6 expression is tissue-specific, with the highest level found in the liver (16), HNF-6 is a crucial determinant of the tissue-specificity of the effects of GH.
Acknowledgments
We thank N. Aidant and S. Neou for technical help, J. E. Darnell for the gifts of HNF-3 expression and in vitro transcription vectors, and S. Muller for the gift of the Sp1 probe. F.P.L. is Research Associate of the National Fund for Scientific Research (Belgium). This work was supported by grants from The Swedish Medical Research Council (03X-06807), the Novo Nordisk Foundation and the Délégation Générale Higher Education and Scientific Research-French Community of Belgium, The Fund for Scientific Medical Research (Belgium), the Belgian Programme on Interuniversity Poles of Attraction, and the D. G. U. I. del Gobierno de Canarias and Fundacion Universitaria de Las Palmas (Spain).
ABBREVIATIONS
- GH
growth hormone
- HNF
hepatocyte nuclear factor
- EMSA
electrophoretic mobility shift assay
- b
base(s)
- IGF-I
insulin-like growth factor I
- GAPDH
glyceraldehyde-3-phosphate dehyrogenase
References
- 1.Isaksson O G, Edén S, Jansson J O. Annu Rev Physiol. 1985;47:483–499. doi: 10.1146/annurev.ph.47.030185.002411. [DOI] [PubMed] [Google Scholar]
- 2.Davidson M B. Endocr Rev. 1987;8:115–131. doi: 10.1210/edrv-8-2-115. [DOI] [PubMed] [Google Scholar]
- 3.Carter-Su C, Schwartz J, Smit L S. Annu Rev Physiol. 1996;58:187–207. doi: 10.1146/annurev.ph.58.030196.001155. [DOI] [PubMed] [Google Scholar]
- 4.Mathews L S, Norstedt G, Palmiter R D. Proc Natl Acad Sci USA. 1986;83:9343–9347. doi: 10.1073/pnas.83.24.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Cam A, Pages G, Auberger P, Le Cam G, Leopold P, Benarous R, Glaichenhaus N. EMBO J. 1987;6:1225–1232. doi: 10.1002/j.1460-2075.1987.tb02358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon J B, Towle H C, Seelig S. J Biol Chem. 1987;262:4284–4289. [PubMed] [Google Scholar]
- 7.Doglio A, Dani C, Grimaldi P, Ailhaud G. Proc Natl Acad Sci USA. 1989;86:1148–1152. doi: 10.1073/pnas.86.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slootweg M C, De Groot R P, Herrmann-Erlee M P, Koornneef I, Kruijer W, Kramer Y M. J Mol Endocrinol. 1991;6:179–188. doi: 10.1677/jme.0.0060179. [DOI] [PubMed] [Google Scholar]
- 9.Murphy L J, Bell G I, Friesen H G. Endocrinology. 1987;120:1806–1812. doi: 10.1210/endo-120-5-1806. [DOI] [PubMed] [Google Scholar]
- 10.Clarkson R W E, Chen C M, Harrison S, Wells C, Muscat G E O, Waters M J. Mol Endocrinol. 1995;9:108–120. doi: 10.1210/mend.9.1.7760844. [DOI] [PubMed] [Google Scholar]
- 11.Legraverend C, Mode A, Westin S, Ström A, Eguchi H, Zaphiropoulos P G, Gustafsson J-Å. Mol Endocrinol. 1992;6:259–266. doi: 10.1210/mend.6.2.1569969. [DOI] [PubMed] [Google Scholar]
- 12.Tannenbaum G S, Martin J B. Endocrinology. 1976;98:562–570. doi: 10.1210/endo-98-3-562. [DOI] [PubMed] [Google Scholar]
- 13.Edén S. Endocrinology. 1979;105:555–560. doi: 10.1210/endo-105-2-555. [DOI] [PubMed] [Google Scholar]
- 14.Liddle C, Mode A, Legraverend C, Gustafsson J-Å. Arch Biochem Biophys. 1992;298:159–166. doi: 10.1016/0003-9861(92)90107-8. [DOI] [PubMed] [Google Scholar]
- 15.Tollet P, Enberg B, Mode A. Mol Endocrinol. 1990;4:1934–1942. doi: 10.1210/mend-4-12-1934. [DOI] [PubMed] [Google Scholar]
- 16.Lemaigre F P, Durviaux S M, Truong O, Lannoy V J, Hsuan J J, Rousseau G G. Proc Natl Acad Sci USA. 1996;93:9460–9464. doi: 10.1073/pnas.93.18.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hromas R, Costa R. Crit Rev Oncol Hematol. 1995;20:129–140. doi: 10.1016/1040-8428(94)00151-i. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann E, Knochel W. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 19.Tollet P, Lahuna O, Ahlgren R, Mode A, Gustafsson J-Å. Mol Endocrinol. 1995;9:1771–1781. doi: 10.1210/mend.9.12.8614413. [DOI] [PubMed] [Google Scholar]
- 20.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 21.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.Lai E, Prezioso V R, Tao W F, Chen W S, Darnell J E., Jr Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 23.Qian X, Samadani U, Porcella A, Costa R H. Mol Cell Biol. 1995;15:1364–1376. doi: 10.1128/mcb.15.3.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samadani U, Costa R H. Mol Cell Biol. 1996;16:6273–6284. doi: 10.1128/mcb.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overdier D G, Porcella A, Costa R H. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux J, Pictet R, Grange T. DNA Cell Biol. 1995;14:385–396. doi: 10.1089/dna.1995.14.385. [DOI] [PubMed] [Google Scholar]
- 27.Costa R H, Grayson D R, Darnell J E., Jr Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaphiropoulos P G, Westin S, Ström A, Mode A, Gustafsson J-Å. DNA Cell Biol. 1990;9:49–56. doi: 10.1089/dna.1990.9.49. [DOI] [PubMed] [Google Scholar]
- 29.Weigent D A, Blalock J E. Int Rev Immunol. 1989;4:193–211. doi: 10.3109/08830188909054418. [DOI] [PubMed] [Google Scholar]
- 30.Maiter D, Underwood L E, Maes M, Davenport M L, Ketelslegers J M. Endocrinology. 1988;123:1053–1059. doi: 10.1210/endo-123-2-1053. [DOI] [PubMed] [Google Scholar]
- 31.Waxman D J, Zhao S, Choi H K. J Biol Chem. 1996;271:29978–29987. doi: 10.1074/jbc.271.47.29978. [DOI] [PubMed] [Google Scholar]
- 32.Norstedt G, Palmiter R. Cell. 1984;36:805–812. doi: 10.1016/0092-8674(84)90030-8. [DOI] [PubMed] [Google Scholar]
- 33.Johnson D, Harrison S, Pineda N, Heinlein C, al-Shawi R, Bishop J O. J Mol Endocrinol. 1995;14:35–49. doi: 10.1677/jme.0.0140035. [DOI] [PubMed] [Google Scholar]
- 34.Legraverend C, Eguchi H, Ström A, Lahuna O, Mode A, Tollet P, Westin S, Gustafsson J-A. Biochemistry. 1994;33:9889–9897. doi: 10.1021/bi00199a010. [DOI] [PubMed] [Google Scholar]
- 35.Matsunaga T, Nagata K, Holsztynska E J, Lapenson D P, Smith A, Kato R, Gelboin H V, Waxman D J, Gonzalez F J. J Biol Chem. 1988;263:17995–18002. [PubMed] [Google Scholar]
- 36.Matsunaga T, Nomoto M, Kozak C A, Gonzalez F J. Biochemistry. 1990;29:1329–1341. doi: 10.1021/bi00457a032. [DOI] [PubMed] [Google Scholar]
- 37.Sundseth S S, Alberta J A, Waxman D J. J Biol Chem. 1992;267:3907–3914. [PubMed] [Google Scholar]