Abstract
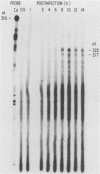
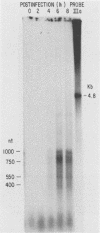
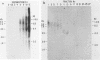
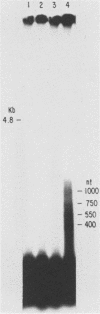
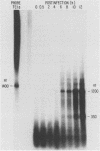
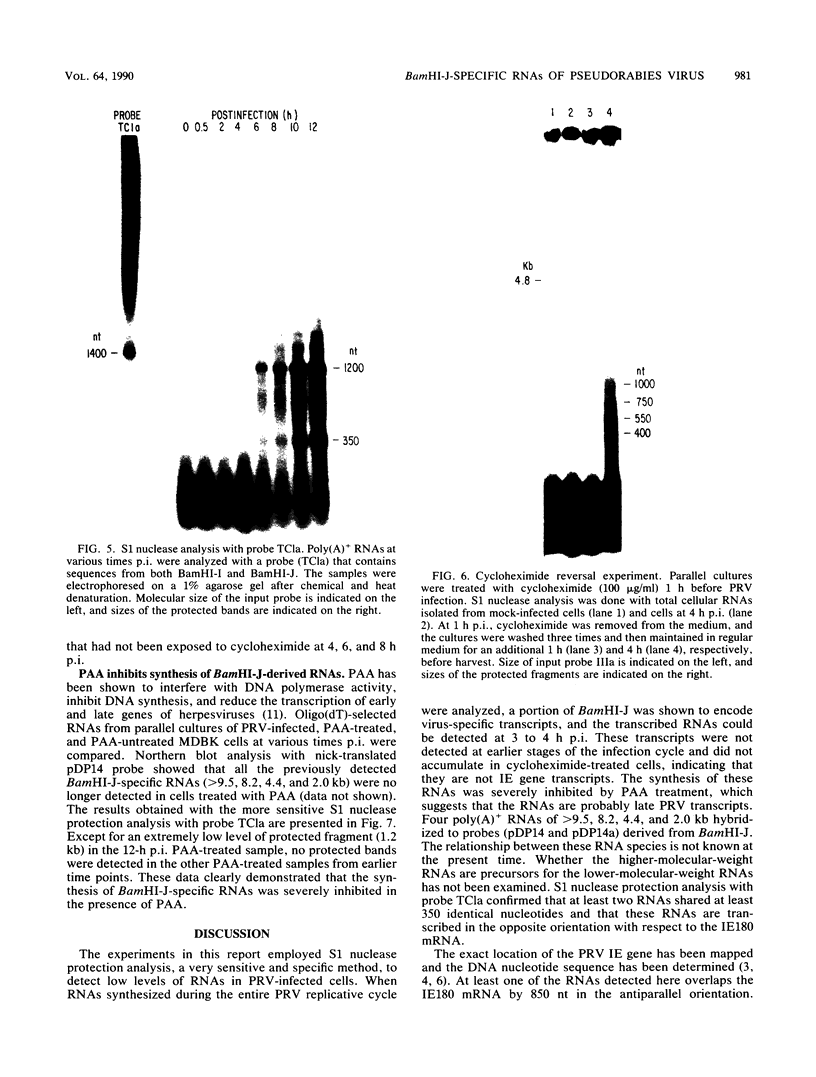
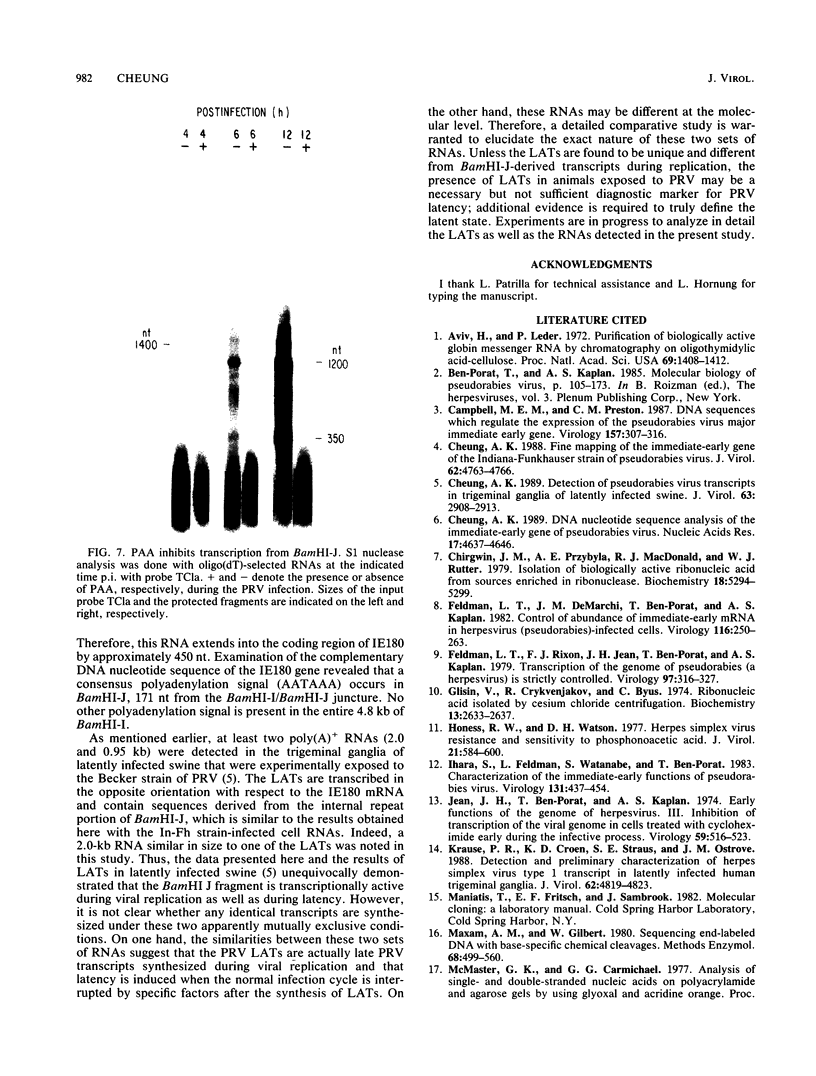
The BamHI J fragment of the pseudorabies virus (PRV) genome has not been associated with any viral transcripts during viral replication. In this report, data are presented to show that a portion of BamHI-J is transcribed during a productive infection. Four oligo(dT)-cellulose-selected RNA species were detected by hybridization with probes derived from BamHI-J. These RNAs were partially colinear, and they were transcribed in the opposite orientation with respect to the immediate-early gene (IE180) of PRV. At least one of the transcripts overlapped (antisense) the coding sequence of IE180 by 450 nucleotides. Expression of these RNAs was sensitive to phosphonoacetic acid, indicating that they are transcripts of PRV late genes. There were several similarities between these RNAs and the latency-associated transcripts detected in the trigeminal ganglia of swine latently infected with PRV (A. K. Cheung, J. Virol. 63: 2908-2913, 1989).
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. E., Preston C. M. DNA sequences which regulate the expression of the pseudorabies virus major immediate early gene. Virology. 1987 Apr;157(2):307–316. doi: 10.1016/0042-6822(87)90273-x. [DOI] [PubMed] [Google Scholar]
- Cheung A. K. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 1989 Jun 26;17(12):4637–4646. doi: 10.1093/nar/17.12.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. K. Detection of pseudorabies virus transcripts in trigeminal ganglia of latently infected swine. J Virol. 1989 Jul;63(7):2908–2913. doi: 10.1128/jvi.63.7.2908-2913.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. K. Fine mapping of the immediate-early gene of the Indiana-Funkhauser strain of pseudorabies virus. J Virol. 1988 Dec;62(12):4763–4766. doi: 10.1128/jvi.62.12.4763-4766.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Feldman L. T., Demarchi J. M., Ben-Porat T., Kaplan A. S. Control of abundance of immediate-early mRNA in herpesvirus (pseudorabies)-infected cells. Virology. 1982 Jan 15;116(1):250–263. doi: 10.1016/0042-6822(82)90417-2. [DOI] [PubMed] [Google Scholar]
- Feldman L., Rixon F. J., Jean J. H., Ben-Porat T., Kaplan A. S. Transcription of the genome of pseudorabies virus (A herpesvirus) is strictly controlled. Virology. 1979 Sep;97(2):316–327. doi: 10.1016/0042-6822(79)90343-x. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Herpes simplex virus resistance and sensitivity to phosphonoacetic acid. J Virol. 1977 Feb;21(2):584–600. doi: 10.1128/jvi.21.2.584-600.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S., Feldman L., Watanabe S., Ben-Porat T. Characterization of the immediate-early functions of pseudorabies virus. Virology. 1983 Dec;131(2):437–454. doi: 10.1016/0042-6822(83)90510-x. [DOI] [PubMed] [Google Scholar]
- Jean J. H., Ben-Porat T., Kaplan A. S. Early functions of the genome of herpesvirus. 3. Inhibition of the transcription of the viral genome in cells treated with cycloheximide early during the infective process. Virology. 1974 Jun;59(2):516–523. doi: 10.1016/0042-6822(74)90461-9. [DOI] [PubMed] [Google Scholar]
- Krause P. R., Croen K. D., Straus S. E., Ostrove J. M. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J Virol. 1988 Dec;62(12):4819–4823. doi: 10.1128/jvi.62.12.4819-4823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirtle E. C., Wathen M. W., Paul P. S., Mengeling W. L., Sacks J. M. Evaluation of field isolates of pseudorabies (Aujeszky's disease) virus as determined by restriction endonuclease analysis and hybridization. Am J Vet Res. 1984 Oct;45(10):1906–1912. [PubMed] [Google Scholar]
- Puga A., Notkins A. L. Continued expression of a poly(A)+ transcript of herpes simplex virus type 1 in trigeminal ganglia of latently infected mice. J Virol. 1987 May;61(5):1700–1703. doi: 10.1128/jvi.61.5.1700-1703.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusanova T., Ben-Porat T., Himeno M., Kaplan A. S. Early functions of the genome of herpesvirus. I. Characterization of the RNA synthesized in cycloheximide-treated, infected cells. Virology. 1971 Dec;46(3):877–889. doi: 10.1016/0042-6822(71)90088-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rock D. L., Nesburn A. B., Ghiasi H., Ong J., Lewis T. L., Lokensgard J. R., Wechsler S. L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987 Dec;61(12):3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivack J. G., Fraser N. W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987 Dec;61(12):3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. G., Wagner E. K., Devi-Rao G. B., Cook M. L., Feldman L. T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987 Feb 27;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- Wagner E. K., Devi-Rao G., Feldman L. T., Dobson A. T., Zhang Y. F., Flanagan W. M., Stevens J. G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988 Apr;62(4):1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler S. L., Nesburn A. B., Watson R., Slanina S. M., Ghiasi H. Fine mapping of the latency-related gene of herpes simplex virus type 1: alternative splicing produces distinct latency-related RNAs containing open reading frames. J Virol. 1988 Nov;62(11):4051–4058. doi: 10.1128/jvi.62.11.4051-4058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler S. L., Nesburn A. B., Zwaagstra J., Ghiasi H. Sequence of the latency-related gene of herpes simplex virus type 1. Virology. 1989 Jan;168(1):168–172. doi: 10.1016/0042-6822(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]