Abstract
The murine B29 (Igβ) promoter is B cell specific and contains essential SP1, ETS, OCT, and Ikaros motifs. Flanking 5′ DNA sequences inhibit B29 promoter activity, suggesting this region contains silencer elements. Two adjacent 5′ DNA segments repress transcription by the murine B29 promoter in a position- and orientation-independent manner, analogous to known silencers. Both these 5′ segments also inhibit transcription by several heterologous promoters in B cells, including mb-1, c-fos, and human B29. These 5′ segments also inhibit transcription by the c-fos promoter in T cells suggesting they are not B cell-specific elements. DNase I footprint analyses show an approximately 70-bp protected region overlapping the boundary between the two negative regulatory DNA segments and corresponding to binding sites for at least two different DNA-binding proteins. Within this footprint, two unrelated 30-bp cis-acting DNA motifs (designated TOAD and FROG) function as position- and orientation-independent silencers when located directly 5′ of the murine B29 promoter. These two silencer motifs act cooperatively to restrict the transcriptional activity of the B29 promoter. Neither of these motifs resembles any known silencers. Mutagenesis of the TOAD and FROG motifs in their respective 5′ DNA segments eliminates the silencing activity of these upstream regions, indicating these two motifs as the principal B29 silencer elements within these regions.
The B29 gene is strictly B cell-specific and expressed at all stages of B cell differentiation (1). The B29 gene product, also called Igβ, is disulfide-linked to the mb-1 gene product Igα and this heterodimeric complex is associated with all Ig isotypes to form the B cell-receptor complex (2–5). The B29–mb-1 heterodimer plays a central and critical role in B cell development. Specific events in B cell development that are controlled by B29–mb-1 heterodimers include allelic exclusion (6, 7) and the pre-B cell transition marked by cell surface translocation of IgM (3, 4, 6, 8). Signal transduction through cross-linked IgM is also controlled by B29–mb-1 heterodimers (for review, see ref. 9). In addition, B29 knock-out mice (i.e., Igβ−/−) produce pro-B cells that fail to progress beyond the DJH stage of rearrangement, suggesting B29 is essential for VDJH recombination resulting in functional μ chain production (10).
We have previously identified and characterized the B29 minimal promoter to resolve the features controlling its B cell specificity (11). The B29 promoter is a TATA-less promoter containing essential cis-acting OCT, ETS, SP1, and Ikaros motifs (11). A similar spectrum of motifs is present in the promoters of other TATA-less genes expressed in lymphocytes including ETS-1 (12), bcl-2 (13), mb-1 (14), TdT (15), VpreB (16), λ5 (17), CD19 (18), and pp52 (19). The strict B cell specificity of the B29 minimal promoter is determined by the combinatorial activities of this cassette of different transcription factors (11, 20).
The current study focuses on the B29 regulatory region 5′ of the murine minimal B29 promoter. Deletion analyses of the B29 gene 5′ flanking region revealed two adjacent regions with significantly lower transcriptional activity than the minimal promoter, suggesting the presence of silencer elements. Silencers are functionally defined cis-acting regulatory DNA elements that down-regulate gene transcription. They generally exhibit activity in either orientation, may be either position-dependent or -independent, and may or may not affect heterologous promoters (13, 21, 22). When tested in transient transfections, two unique DNA segments isolated from the inhibitory region 5′ of the minimal promoter, designated −354/−165 and −565/−355 segments, inhibited transcription of the murine B29 minimal promoter in a position- and orientation-independent manner. Both DNA segments also inhibited transcription from heterologous promoters, further defining them as transcriptional silencers. DNA footprinting showed a 70-bp protected region overlapping the −354/−355 boundary of these two segments. Within this protected region, two unrelated DNA sequences, designated the TOAD and FROG motifs, exhibited both position- and orientation-independent silencing of the B29 promoter. Electrophoretic mobility shift assays (EMSA) indicated that the TOAD and FROG motifs interact with distinct non-tissue-specific nuclear DNA-binding proteins. Site-directed mutagenesis of these two motifs in the context of their respective 5′ DNA segments alleviated transcriptional silencing equivalent to successive deletion of the complete segments, thereby affirming that TOAD and FROG are the principle 5′ silencer motifs in this region acting on the B29 promoter.
MATERIALS AND METHODS
Plasmid Construction and Mutagenesis.
The B29 minimal (−164) promoter and 5′ deletion constructs with endpoints at positions −1230, −890, −719, −565, −354, and −293 were cloned in pCAT Basic (Promega). In all constructs, the B29 endogenous ATG was destroyed, leaving the translation start site as the first methionine codon of the chloramphenicol acetyltransferase (CAT) gene. B29 5′ segments (−354/−165 and −565/−355) were generated by PCR and cloned 5′ of the following promoters: murine B29 (−164), c-fos (−71) (23), human B29 (−193) (24), and murine mb-1 (−-252; provided by Rudolph Grosschedl, University of California at San Francisco) in pCAT Basic (Promega). B29 5′ segments (−354/−165 and −565/−355) were also cloned downstream of CAT in vectors containing either the B29 (−164) or the (−354) promoter segments. The synthetic DNA oligonucleotides [5′-CTCTTCCAGAGCAAGGCAACCACAGGAGACC-3′ (−349/−321, i.e., TOAD) and 5′-GGTGTGCATTTAGCTAAATTCCCCA-3′ (−381/−356, i.e., FROG)] were cloned in single and multiple copies and in both orientations 5′ of B29 promoter constructs (positions −164, −354, and −565). Mutagenized constructs were generated by using the Quik-Change mutagenesis kit (Stratagene) and the following oligonucleotides: mTOAD (positions −335, −336), 5′-CTCTTCCAGAGCAAGttAACCACAGGAGACCA-3′, and mFROG (positions −369, −373), 5′-GGTGTGCAgTTAaCTAAATTCCCCA-3′. Other TOAD motif mutations at bases −329, −331, −333, and −338 had no effect on silencing activity (data not shown).
DNA Transfections and CAT Assays.
M12 B cell line was transfected by the DEAE-dextran method (25). Cells were cotransfected with 5 μg of CAT reporter plasmid and 1 μg of pRSV-luciferase. Extracts were prepared and assayed as in ref. 11 with the exception of quantification by PhosphorImager (Molecular Dynamics) analysis. Results were normalized to luciferase activity and are the average ± SD of at least four transfections using at least two preparations of DNA. Absolute fold inductions for each promoter construct ranged from 8- to 40-fold (muB29), 8- to 30-fold (huB29), 6- to 20-fold (mb-1), 45- to 200-fold (c-fos M12), and 12- to 50-fold (c-fos BW5147) over vector alone (pCAT basic).
Nuclear Extracts and DNA-Binding Assays.
Preparation of crude nuclear extracts from cell lines and DNase I footprinting were performed as described (26). The DNase I footprinting probe was generated by 5′ end labeling with [γ-32P]ATP at the ClaI site of the pCR-Script B29 −411/−293 PCR fragment construct, followed by digestion with SacI. EMSA was performed as described (11). EMSA probes were the identical double-stranded oligonucleotides used in the silencer constructs labeled at both 5′ ends with [γ-32P]ATP.
RESULTS
B29 Minimal Promoter Activity Is Modulated by 5′ Flanking DNA Sequences.
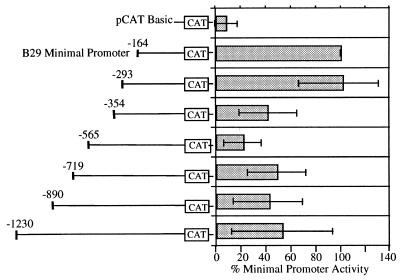
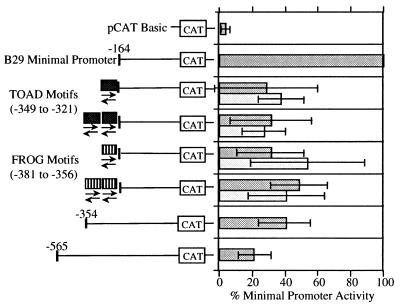
The B29 minimal (i.e., positions −164 to +32) promoter has been previously characterized as a TATA-less promoter controlled by OCT, ETS, Ikaros, and SP1 cis-acting elements (11). Herein, 5′ flanking region deletions of the B29 (−164) promoter revealed multiple segments of negative regulation (Fig. 1). Specifically, deletions ending at positions −354 and −565 exhibited only 40 and 20%, respectively, of the maximal activity of the B29 (−164) promoter. Interestingly, the activity of the human B29 minimal promoter is also strongly inhibited by its 5′ flanking sequences (24), although no significant sequence homology exists between the murine and human B29 gene inhibitory regions. The murine B29 negative DNA −354/−165 and −565/−355 segments were analyzed for their capacity to act as silencer elements.
Figure 1.
B29 minimal promoter activity is modulated by 5′ flanking sequences. Transient transfections to detect transcriptional expression of sequential B29 promoter deletion constructs were carried out in the M12 B cell line. Deletion constructs are identified by nucleotide numbers with respect to the major start site of transcription (+1). The activity of each deletion construct is expressed as a percentage of the CAT activity obtained with the B29 minimal (−164) promoter construct (100%).
Two B29 5′ Gene Segments Function as Position- and Orientation-Independent Silencers.
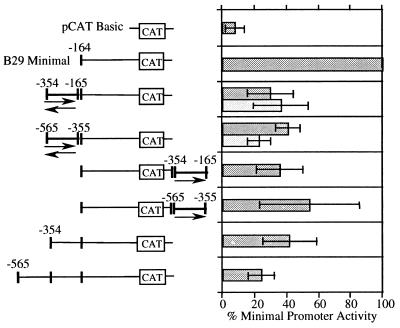
Many silencers, like enhancers, can function in a position- and orientation-independent manner (20, 21, 27, 28). The B29 −354/−165 and −565/−355 segments were individually cloned in forward and reverse orientations directly 5′ of the B29 (−164) promoter and tested for silencer activity. Both segments were also cloned in the forward orientation downstream of the CAT reporter gene in the B29 (−164) promoter construct. These silencer constructs were transiently transfected into the M12 B cell line analyzed in comparison to the B29 position −164, −354, and −565 5′ deletion constructs (Fig. 2). Both the B29 −354/−165 and −565/−355 segments reduced CAT reporter activity by 60–70% in both orientations when located directly 5′ of the B29 (−164) promoter. Both also reduced CAT activity 40–60% when located downstream of the CAT reporter gene. Overall, these silencer constructs exhibited activities comparable to the B29 (−354) promoter. Neither the B29 −354/−165 nor the −565/−355 segment showed further inhibitory effects on CAT reporter gene activity when placed upstream of the B29 position −354 and −565 promoters (data not shown). Thus these data suggest the B29 −354/−165 and −565/−355 segments function independently but may cooperate incrementally in overall silencing activity.
Figure 2.
B29 gene 5′ regions act as position- and orientation-independent silencers. Transient transfections to detect transcriptional expression of B29 promoter/silencer region constructs were carried out in the M12 B cell line. The silencer fragments and their position and orientation relative to the B29 minimal (−164) promoter are as indicated. The activity of each construct is expressed as a percentage of the CAT activity obtained with the B29 minimal (−164) promoter construct (100%). Deletion constructs −164, −354, and −565 are identified by nucleotide numbers with respect to the major start site of transcription (+1) and are shown for comparison.
B29 5′ Segments Silence Heterologous Promoters.
Another feature of many silencer elements is their ability to inhibit transcription from heterologous promoters (12, 13, 20, 21, 27–29). Constructs were generated to assess the silencing activity of the B29 −354/−165 and −565/−355 segments on promoters other than B29. The −354/−165 and −565/−355 segments were tested for silencer activity in both orientations directly 5′ of the human B29 minimal (−193) promoter and the mb-1 minimal (−252) promoter. Regardless of orientation, both segments inhibited the mb-1 (−252) promoter to an extent comparable to the B29 (−164) promoter (Table 1). Both segments also inhibited the human B29 (−193) promoter similarly to the murine B29 (−164) promoter only when in the wild-type orientation. The inhibitory activities of the B29 −354/−165 and −565/−355 segments were not unexpected since the spectrum of active motifs in the mb-1 and human B29 promoters closely resembles that in the murine B29 (−164) promoter (11, 14, 24). To test their activities on a more disparate promoter, the B29 −354/−165 and −565/−355 segments were analyzed for silencing activity on the c-fos (−71) promoter, which is regulated by a different array of cis-acting motifs and contains a TATA box (23). When placed directly 5′ of the c-fos (-71) promoter, both segments inhibited activity to the same extent as the B29 (−164) promoter (Table 1). These data indicate that the silencing activities of the B29 −354/−165 and −565/−355 segments are not restricted to the murine B29 promoter.
Table 1.
Silencer elements are neither promoter- nor cell-type-specific
| Construct generated with each promoter | % promoter activity
|
||||
|---|---|---|---|---|---|
| B cell
|
T cell
|
||||
| muB29 (−164) | huB29 (−193) | mb − 1 (−252) | c-fos (−71) | c-fos (−71) | |
| 8 ± 6 | 6 ± 3 | 13 ± 3 | 1 ± 2 | 4 ± 3 | |
| 100 | 100 | 100 | 100 | 100 | |
| 30 ± 14 | 41 ± 15 | 58 ± 13 | 42 ± 22 | 50 ± 29 | |
| 36 ± 17 | 83 ± 27 | 60 ± 23 | 52 ± 8 | ND | |
| 41 ± 8 | 42 ± 15 | 37 ± 6 | 34 ± 13 | 49 ± 29 | |
| 23 ± 7 | 110 ± 41 | 61 ± 14 | ND | ND | |
ND, not determined.
B29 5′ Silencer Segments Function in T Cells.
The c-fos silencer constructs used above were tested in a T cell line where the c-fos promoter is also active. The B29 −354/−165 and −565/−355 segments inhibited c-fos promoter activity in BW5147 T cells by 50% (Table 1), indicating these silencer elements are not B cell specific. These data, in combination with the earlier heterologous promoter results, strongly suggest the B29 −354/−165 and −565/−355 segments function via a silencing mechanism that is neither promoter- nor cell-type-specific. The B29 −354/−165 and the −565/−355 segments do not contain any sequences with significant homology to known cis-acting silencer motifs or negative regulatory elements (30).
B29 Silencer Elements Interact with DNA-Binding Proteins.
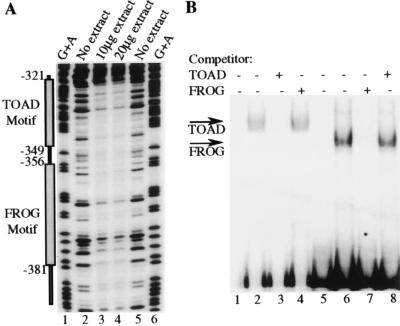
Although some silencer elements do not interact with DNA binding proteins (20), others appear to function through DNA–protein interactions (20, 31–33). DNase I footprinting was performed over the B29 −565 to −164 promoter region to locate nuclear protein binding sites. One extended footprint was observed from −320 to approximately −389 spanning the boundary between the B29 −354/−165 and −565/−355 silencer segments analyzed in Fig. 2. Two oligonucleotides were designed to cover the majority of the footprinted regions (i.e., 60/69 bp) on each side of the B29 (−354) boundary. These two 30-mers were designated the TOAD (−349/−321) and FROG (−381/−356) motifs. The TOAD and FROG motifs each formed specific nuclear protein complexes of differing mobility in EMSA (Fig. 3B). Neither motif competed protein binding to the other, indicating that their protein–DNA complexes differ (Fig. 3B). Protein–DNA complexes with the same mobility were formed with these motifs and nuclear extracts from B cells, T cells, myeloid cells, and fibroblasts (data not shown), suggesting these binding factors may be ubiquitously expressed. These results are not unexpected as the B29 −354/−165 and −565/−355 segments were shown to be neither promoter- nor cell-type-specific silencer elements in Table 1.
Figure 3.
B29 5′ silencer elements interact with DNA binding proteins. (A) The endpoint of the DNase I footprints are identified by nucleotide numbers with respect to the start site of transcription (+1). Lanes containing reactions incubated with either 10 μg or 20 μg M12 B cell crude nuclear extract are indicated (lanes 3 and 4). Lanes containing reactions incubated without extract are indicated (lanes 2 and 5). Reactions were electrophoresed alongside a G+A ladder of the probe (lanes 1 and 6). (B) Double-stranded oligonucleotides corresponding to the sequence from positions −349 to −321 (TOAD motif, lanes 1–4) and −381 to −356 (FROG motif, lanes 5–8) were end-labeled. Lanes 1 and 5 contain TOAD and FROG motif probes alone, respectively. The TOAD motif probe was incubated with 5 μg of M12 B cell nuclear extract (lanes 2–4) and the FROG motif probe was incubated with 5 μg of M12 B cell nuclear extract (lanes 6–8). Probes were incubated in the presence of a 1,000-fold excess of unlabeled TOAD motif competitor (lanes 3 and 8) or were incubated in the presence of a 1,000-fold excess of unlabeled FROG motif competitor (lanes 4 and 7). The specifically formed TOAD and FROG complexes are indicated as such with arrows.
Silencing Activity of the B29 5′ Segments Corresponds to the Two Motifs Shown to Interact with DNA-Binding Proteins.
To determine whether the TOAD and FROG motifs exhibited independent silencing activity without the flanking sequences of the B29 −354/−165 and −565/−355 segments, both 30-mers were inserted in several configurations directly 5′ of the B29 (−164) promoter construct. Single copies of the TOAD or FROG motifs individually produced 60–70% reductions in activity of the B29 (−164) promoter in the wild-type orientation and 50–60% reductions in the opposite orientation (Fig. 4), showing orientation-independent silencing activity out of the context of their 5′ DNA segments. The presence of multiple copies (i.e., two to four) of either the TOAD or FROG motifs did not inhibit the B29 (−164) promoter beyond the levels obtained with single motifs (Fig. 4 and data not shown). Hence, the properties of the isolated TOAD or FROG motifs are identical to those obtained with their respective 5′ B29 DNA segments (i.e., −354/−165 and −565/−355), strongly suggesting that TOAD and FROG are the primary active inhibitory motifs in this region.
Figure 4.
Silencing activity in the B29 5′ gene regions corresponds to TOAD and FROG DNA binding motifs. Transient transfections to detect transcriptional expression of B29 promoter/silencer motif constructs were carried out in the M12 B cell line. The TOAD and FROG silencer motifs, positions −349 to −321 and −381 to −356, respectively, and their positions and orientations relative to the B29 minimal (−164) promoter are as indicated. The activity of each construct is expressed as a percentage of the CAT activity obtained with the B29 minimal (−164) promoter construct (100%). Deletion constructs −164, −354, and −565 are identified by nucleotide numbers with respect to the major start site of transcription (+1) and are shown for comparison.
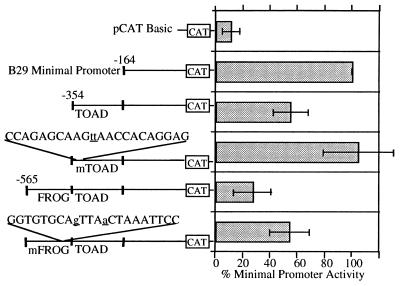
To test this prediction, mutations were introduced into FROG and TOAD motif sequences in the B29 (−565) and (−354) deletion constructs. Base changes disrupting the FROG motif palindrome and one mutant of a series of six scanning mutations of the TOAD motif eliminated silencing activity in a stepwise manner duplicating the effects of deleting the wild-type FROG and TOAD motifs in the B29 (−565) and (−354) deletion constructs (Fig. 5). Specifically, mutation of the TOAD motif restored activity of the B29 (−354) construct to that of the minimal promoter, whereas mutation of the FROG motif raised activity of the B29 (−565) construct to that of the B29 (−354) construct, reflecting the presence of the wild-type TOAD motif within this fragment. These mutations reduced, but did not completely abbrogate, protein binding in EMSA (data not shown). The role of DNA-binding proteins in TOAD and FROG silencer activity remains to be resolved.
Figure 5.
Site-directed mutagenesis of the TOAD and FROG motifs disrupts silencing activity. Transient transfections to detect transcriptional expression of deletion constructs −354 and −565 with introduced point mutations in the TOAD and FROG motifs, respectively, were carried out in the M12 B cell line. Mutagenized nucleotides are indicated by underlined lowercase type. The activity of each construct is expressed as a percentage of the CAT activity obtained with the B29 minimal (−164) promoter construct (100%). Deletion constructs −164, −354, and −565 are identified by nucleotide numbers with respect to the major start site of transcription (+1) and are shown for comparison.
DISCUSSION
We have shown that B29 minimal promoter activity is controlled by dual silencer elements located in the B29 −354/−165 and −565/−355 segments. Both of these 5′ segments exhibit inhibitory activity and the two active motifs (designated TOAD and FROG) identified within them were shown to function as position- and orientation-independent silencers. The B29 −354/−165 and −565/−355 segments were found to inhibit transcription from heterologous promoters including the murine mb-1 and c-fos promoters and the human B29 promoter. In addition to lacking promoter specificity, these silencer elements also lacked cell-type specificity in that they inhibited transcription from the c-fos promoter in both B and T cell lines. These data are not surprising given that DNA–protein complexes formed with TOAD and FROG motif oligonucleotides were found in nuclear extracts from all cell types analyzed by EMSA. The TOAD and FROG motifs exactly mimicked the silencing properties of the B29 −354/−165 and −565/−355 segments. Additionally, mutations in the TOAD and FROG silencer motifs abolished the silencing activity of their respective 5′ segments. These findings strongly implicate the TOAD and FROG motifs as the primary silencer elements in these regions upstream of the B29 promoter.
These two silencer motifs were identified by DNase I protection analyses over the B29 −354/−165 and −565/−355 segments. The TOAD and FROG motif oligonucleotides corresponding to the region protected from DNase I digestion bound distinct protein complexes in EMSA that were not competed by each other. These data suggest the B29 silencing activity is due to at least two separate motifs that bind different proteins. This does not, however, eliminate the possibility that these two motifs and their respective protein complexes work together to provide maximal silencing activity of this region. In fact, only partial alleviation of silencing was observed when only one of the motifs was mutated in a construct where both were present (Fig. 5), suggesting that these motifs act cooperatively.
Neither the TOAD nor the FROG motif harbors any significant sequence homology to other known cis-acting silencer elements (30). Interestingly, the TOAD motif is 50% homologous with the consensus early B cell factor (EBF) motif (34) and 78% homologous to the mb-1 promoter EBF motif (35). EBF is known to function as a positive trans-activator (35–37). Based on EMSA competition experiments, the B29 TOAD motif does not appear to interact with EBF (unpublished data). In addition, TOAD motif DNA–protein complexes are formed with extracts from cell types where B cell-specific EBF is not expressed (36), further negating its possible interaction with the TOAD silencer motif. The FROG motif contains a central 12-bp palindromic sequence. The mutations in the FROG motif, designed to disrupt this palindrome, abolished its silencing activity. Palindromes appear to be necessary for trans-acting factor binding and silencing activity in several silencers including: the huCYP1A1 gene negative regulatory element (NRE) (38), the lox gene NRE (39), and the Igκ gene silencer (40, 41).
Our findings indicate the B29 silencers are non-cell-type-specific elements that modulate the activity of the B cell-specific minimal (-164) promoter but have the ability to silence other promoters as well. Other silencers mimic these features of the B29 silencers by contributing to the transcriptional regulation of the gene but not by controlling tissue specificity. These include bcl-2 (13), ETS-1 (12), and λ5 (42). Interestingly, like B29, all of these genes have TATA-less promoters and initiate transcription at multiple start sites (17, 43, 44), like B29 (11). The bcl-2 NRE is neither promoter- nor cell-type-specific (13). This NRE contains multiple regions contributing to its silencing activity, but it differs from the B29 silencers in that the bcl-2 NRE is both position- and orientation-dependent (13). The human ETS-1 gene contains two NREs that, like the B29 silencers, are orientation-independent and are not promoter-specific (12). The λ5 silencer is less well defined but does not impart tissue or stage specificity as it is active in all stages of B and T cell development (42).
The B29, bcl-2, ETS-1, and λ5 regulatory regions present an interesting conundrum by having silencer elements that are active in cell types where the genes are expressed (12, 13, 42). These silencer elements may serve to modulate the level of transcription of their respective genes rather than to control cell-type-specific gene expression. Such silencers would function as governors restricting fluctuations in gene activity, thereby preventing deleterious consequences of overexpression. Overexpression of bcl-2 is postulated to underlie the prolonged survival in neoplastic B cell disorders of follicular non-Hodgkin lymphoma and chronic lymphocytic leukemia (45, 46). CD3ɛ overexpression blocks T cell development at a very early stage preceding T cell receptor (TCR) rearrangement (47). CD3ɛ also acts as an oncogene when overexpressed in prothymocytes (48). These findings are especially relevant to the possible consequences of B29 overexpression as CD3ɛ fulfills the same role in the TCR as B29 does in the B cell receptor.
The finding that multiple copies of either TOAD or FROG did not inhibit the activity of the B29 promoter beyond the combined effects of single FROG and TOAD motifs may be a reflection of a fundamental property of the class of silencers that function as gene expression governors. Namely, their function is to limit but not completely repress gene transcription. Accordingly, the maintenance of B29 gene expression at a particular level by the TOAD and FROG motifs may be essential for normal B cell development or function.
Acknowledgments
This research was supported by National Institutes of Health Grants CA12800 and GM40185. C.S.M. was supported by Public Health Service National Research Service Award AI07126, the Dr. Ursula Mandel Scholarship, the UC Dissertation Year Fellowship, and a Sigma Xi Grant-In-Aid. S.A.O. was supported by Public Health Service National Research Service Award GM07104.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- EBF
early B cell factor
- EMSA
electrophoretic mobility-shift assay
- NRE
negative regulatory element
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF002279).
References
- 1.Hermanson G G, Briskin M, Sigman D, Wall R. Proc Natl Acad Sci USA. 1989;86:7341–7345. doi: 10.1073/pnas.86.19.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hombach J, Tsubata T, Leclercq L, Stappert H, Reth M. Nature (London) 1990;343:760–762. doi: 10.1038/343760a0. [DOI] [PubMed] [Google Scholar]
- 3.Campbell K S, Cambier J C. EMBO J. 1990;9:441–448. doi: 10.1002/j.1460-2075.1990.tb08129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkitaraman A R, Williams G T, Dariavach P, Neuberger M S. Nature (London) 1991;352:777–781. doi: 10.1038/352777a0. [DOI] [PubMed] [Google Scholar]
- 5.Ishihara K, Wood W J, Jr, Damore M, Hermanson G G, Wall R, Kincade P W. Proc Natl Acad Sci USA. 1992;89:633–637. doi: 10.1073/pnas.89.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papavasiliou F, Misulovin Z, Suh H, Nussenzweig M C. Science. 1995;268:408–411. doi: 10.1126/science.7716544. [DOI] [PubMed] [Google Scholar]
- 7.Manz J, Denis K, Witte O, Brinster R, Storb U. J Exp Med. 1988;168:1363–1381. doi: 10.1084/jem.168.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hombach J, Sablitzky F, Rajewsky K, Reth M. J Exp Med. 1988;167:652–657. doi: 10.1084/jem.167.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cambier J C. Curr Opin Immunol. 1992;4:257–264. doi: 10.1016/0952-7915(92)90074-o. [DOI] [PubMed] [Google Scholar]
- 10.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 11.Omori S A, Wall R. Proc Natl Acad Sci USA. 1993;90:11723–11727. doi: 10.1073/pnas.90.24.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J H, Jeha S, Oka T. Oncogene. 1993;8:133–139. [PubMed] [Google Scholar]
- 13.Young R L, Korsmeyer S J. Mol Cell Biol. 1993;13:3686–3697. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Travis A, Hagman J, Grosschedl R. Mol Cell Biol. 1991;11:5756–5766. doi: 10.1128/mcb.11.11.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landau N R, St. John T P, Weissman I L, Wolf S C, Silverstone A E, Baltimore D. Proc Natl Acad Sci USA. 1984;81:5836–5840. doi: 10.1073/pnas.81.18.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okabe T, Bauer S R, Kudo A. Eur J Immunol. 1992;22:31–36. doi: 10.1002/eji.1830220106. [DOI] [PubMed] [Google Scholar]
- 17.Kudo A, Sakaguchi N, Melchers F. EMBO J. 1987;6:103–107. doi: 10.1002/j.1460-2075.1987.tb04725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omori S A, Smale S, O’Shea-Greenfield A, Wall R. J Immunol. 1997;159:1800–1808. [PubMed] [Google Scholar]
- 20.Ernst P, Smale S T. Immunity. 1995;2:311–319. doi: 10.1016/1074-7613(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 21.Sawada S, Scarborough J D, Killeen N, Littman D R. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 22.Ye J, Ghosh P, Cippitelli M, Subleski J, Hardy K J, Ortaldo J R, Young H A. J Biol Chem. 1994;269:25728–25734. [PubMed] [Google Scholar]
- 23.Gilman M Z, Wilson R N, Weinberg R A. Mol Cell Biol. 1986;6:4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson A A, Wood W J, Jr, Gilly M J, Damore M A, Omori S A, Wall R. Blood. 1996;87:666–673. [PubMed] [Google Scholar]
- 25.Grosschedl R, Baltimore D. Cell. 1985;41:885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- 26.Lo K, Landau N R, Smale S T. Mol Cell Biol. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanuma Y, Nakabayashi H, Esumi M, Endo H. Mol Cell Biol. 1995;15:517–523. doi: 10.1128/mcb.15.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart M J, Cox G, Reifel-Miller A, Kim S Y, Westbrook C A, Leibowitz D S. J Biol Chem. 1994;269:10820–10829. [PubMed] [Google Scholar]
- 29.Ye J, Cippitelli M, Dorman L, Ortaldo J R, Young H A. Mol Cell Biol. 1996;16:4744–4753. doi: 10.1128/mcb.16.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quandt K, Frech K, Karas H, Wingender E, Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raich N, Clegg C H, Grofti J, Romeo P H, Stamatoyannopoulos G. EMBO J. 1995;14:801–809. doi: 10.1002/j.1460-2075.1995.tb07058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boucher P D, Ruch R J, Hines R N. J Biol Chem. 1993;268:17384–17391. [PubMed] [Google Scholar]
- 33.Szabo P, Moitra J, Rencendorj A, Rakhely G, Rauch T, Kiss I. J Biol Chem. 1995;270:10212–10221. doi: 10.1074/jbc.270.17.10212. [DOI] [PubMed] [Google Scholar]
- 34.Travis A, Hagman J, Hwang L, Grosschedl R. Mol Cell Biol. 1993;13:3392–3400. doi: 10.1128/mcb.13.6.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldhaus A L, Mbangkollo D, Arvin K L, Klug C A, Singh H. Mol Cell Biol. 1992;12:1126–1133. doi: 10.1128/mcb.12.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagman J, Belanger C, Travis A, Turck C W, Grosschedl R. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 37.Sigvardsson M, Bemark M, Leanderson T. Eur J Immunol. 1995;25:298–301. doi: 10.1002/eji.1830250150. [DOI] [PubMed] [Google Scholar]
- 38.Boucher P D, Hines R N. Carcinogenesis. 1995;16:383–392. doi: 10.1093/carcin/16.2.383. [DOI] [PubMed] [Google Scholar]
- 39.O’Prey J, Harrison P R. Nucleic Acids Res. 1995;23:3664–3672. doi: 10.1093/nar/23.18.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierce J W, Gifford A M, Baltimore D. Mol Cell Biol. 1991;11:1431–1437. doi: 10.1128/mcb.11.3.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saksela K, Baltimore D. Mol Cell Biol. 1993;13:3698–3705. doi: 10.1128/mcb.13.6.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Glozak M A, Blomberg B B. J Immunol. 1995;155:2498–2514. [PubMed] [Google Scholar]
- 43.Oka T, Rairkar A, Chen J H. Oncogene. 1991;6:2077–2083. [PubMed] [Google Scholar]
- 44.Dynan W S, Tjian R. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 45.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed J C. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 46.Korsmeyer S J. Blood. 1992;80:879–886. [PubMed] [Google Scholar]
- 47.Wang B, Levelt C, Salio M, Zheng D, Sancho J, Liu C P, She J, Huang M, Higgins K, Sunshine M J, Eichmann K, Lacy E, Lonberg N, Terhorst C. Int Immunol. 1995;7:435–448. doi: 10.1093/intimm/7.3.435. [DOI] [PubMed] [Google Scholar]
- 48.Wang B, She J, Salio M, Allen D, Lacy E, Lonberg N, Terhorst C. Mol Med. 1997;3:72–81. [PMC free article] [PubMed] [Google Scholar]