Abstract
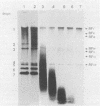
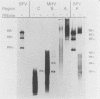
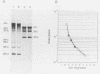
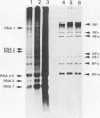
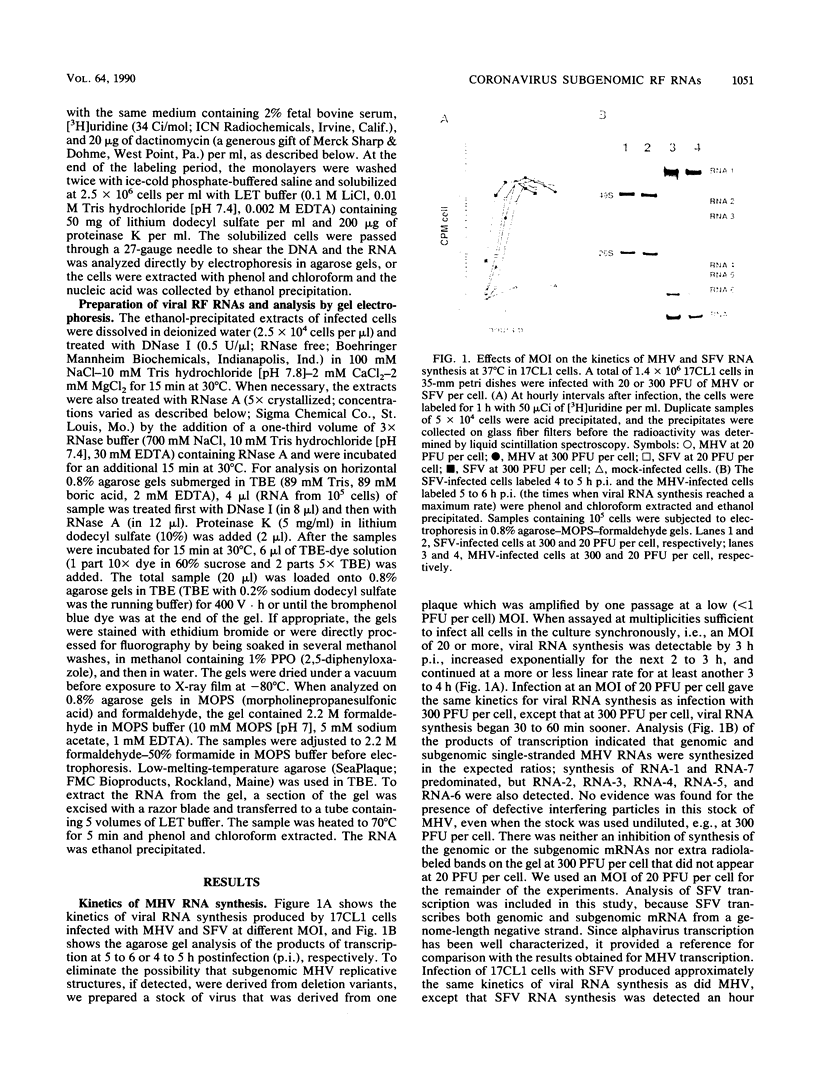
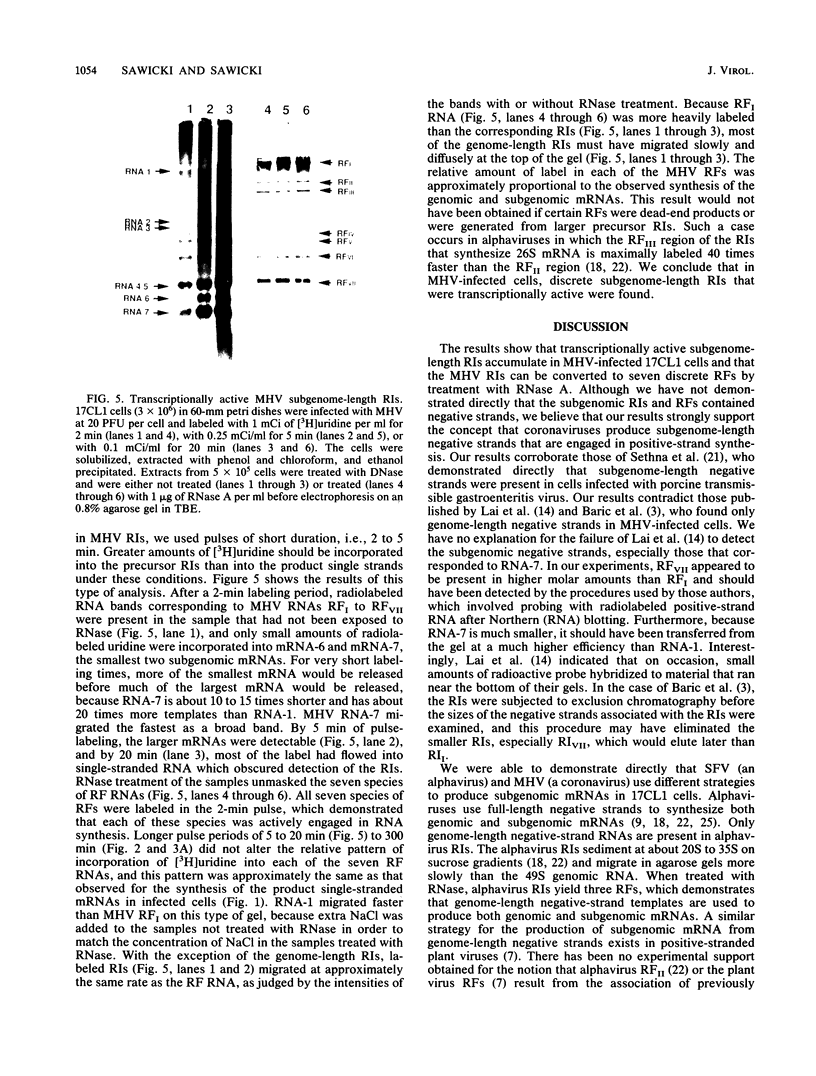
Both genomic and subgenomic replicative intermediates (RIs) and replicative-form (RF) structures were found in 17CL1 mouse cells that had been infected with the A59 strain of mouse hepatitis virus (MHV), a prototypic coronavirus. Seven species of RNase-resistant RF RNAs, whose sizes were consistent with the fact that each was derived from an RI that was engaged in the synthesis of one of the seven MHV positive-strand RNAs, were produced by treatment with RNase A. Because the radiolabeling of the seven RF RNAs was proportional to that of the corresponding seven positive-strand RNAs, the relative rate of synthesis of each of the MHV positive-strand RNAs may be controlled by the relative number of each of the size classes of RIs that are produced. In contrast to alphavirus, which produced its subgenome-length RF RNAs from genome-length RIs, MHV RF RNAs were derived from genome- and subgenome-length RIs. Only the three largest MHV RF RNAs (RFI, RFII, and RFIII) were derived from the RIs that migrated slowest on agarose gels. The four smallest RF RNAs (RFIV, RFV, RFVI, and RFVII) were derived from RIs that migrated in a broad region of the gel that extended from the position of 28S rRNA to the position of the viral single-stranded MHV mRNA-3. Because all seven RIs were labeled during very short pulses with [3H]uridine, we concluded that the subgenome-length RIs are transcriptionally active. These findings, with the recent report of the presence of subgenome-length negative-strand RNAs in cells infected with porcine transmissible gastroenteritis virus (P. B. Sethna, S.-L. Hung, and D. A. Brian, Proc. Natl. Acad. Sci. USA 86: 5626-5630, 1989), strongly suggest that coronaviruses utilize a novel replication strategy that employs the synthesis of subgenomic negative strands to produce subgenomic mRNAs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984 Apr 19;308(5961):751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J., Smeekens S., Rottier P. Sequence of the nucleocapsid gene from murine coronavirus MHV-A59. Nucleic Acids Res. 1983 Feb 11;11(3):883–891. doi: 10.1093/nar/11.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Lai M. M. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983 Dec;48(3):633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M. E., Brown T. D., Foulds I. J., Green P. F., Tomley F. M., Binns M. M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987 Jan;68(Pt 1):57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bozarth R. F., Harley E. H. The electrophoretic mobility of double-stranded RNA in polyacrylamide gels as a function of molecular weight. Biochim Biophys Acta. 1976 May 19;432(3):329–335. doi: 10.1016/0005-2787(76)90142-8. [DOI] [PubMed] [Google Scholar]
- Budzilowicz C. J., Wilczynski S. P., Weiss S. R. Three intergenic regions of coronavirus mouse hepatitis virus strain A59 genome RNA contain a common nucleotide sequence that is homologous to the 3' end of the viral mRNA leader sequence. J Virol. 1985 Mar;53(3):834–840. doi: 10.1128/jvi.53.3.834-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargouri R., Joshi R. L., Bol J. F., Astier-Manifacier S., Haenni A. L. Mechanism of synthesis of turnip yellow mosaic virus coat protein subgenomic RNA in vivo. Virology. 1989 Aug;171(2):386–393. doi: 10.1016/0042-6822(89)90606-5. [DOI] [PubMed] [Google Scholar]
- Jacobs L., Spaan W. J., Horzinek M. C., van der Zeijst B. A. Synthesis of subgenomic mRNA's of mouse hepatitis virus is initiated independently: evidence from UV transcription mapping. J Virol. 1981 Aug;39(2):401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings D. A., Bredenbeek P. J., Noten J. F., Hogeweg P., Spaan W. J. Differential premature termination of transcription as a proposed mechanism for the regulation of coronavirus gene expression. Nucleic Acids Res. 1988 Nov 25;16(22):10849–10860. doi: 10.1093/nar/16.22.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R. M. The role of RNA priming in viral and trypanosomal mRNA synthesis. Cell. 1985 Jul;41(3):651–652. doi: 10.1016/S0092-8674(85)80041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käriäinen L., Takkinen K., Keränen S., Söderlund H. Replication of the genome of alphaviruses. J Cell Sci Suppl. 1987;7:231–250. [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Patton C. D., Baric R. S., Stohlman S. A. Presence of leader sequences in the mRNA of mouse hepatitis virus. J Virol. 1983 Jun;46(3):1027–1033. doi: 10.1128/jvi.46.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Patton C. D., Stohlman S. A. Replication of mouse hepatitis virus: negative-stranded RNA and replicative form RNA are of genome length. J Virol. 1982 Nov;44(2):487–492. doi: 10.1128/jvi.44.2.487-492.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P. J., Noten A. F., Horzinek M. C., Spaan W. J. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988 Oct;166(2):415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D. L., Kaariainen L., Lambek C., Gomatos P. J. Mechanism for control of synthesis of Semliki Forest virus 26S and 42s RNA. J Virol. 1978 Jan;25(1):19–27. doi: 10.1128/jvi.25.1.19-27.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986 Jan;57(1):328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. The effect of overproduction of nonstructural proteins on alphavirus plus-strand and minus-strand RNA synthesis. Virology. 1986 Jul 30;152(2):507–512. doi: 10.1016/0042-6822(86)90157-1. [DOI] [PubMed] [Google Scholar]
- Sethna P. B., Hung S. L., Brian D. A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. T., Strauss J. H. Replication of Sindbis virus. II. Multiple forms of double-stranded RNA isolated from infected cells. J Mol Biol. 1972 Nov 28;71(3):615–631. doi: 10.1016/s0022-2836(72)80027-5. [DOI] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. C. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B. A., Siddell S. G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]