Abstract
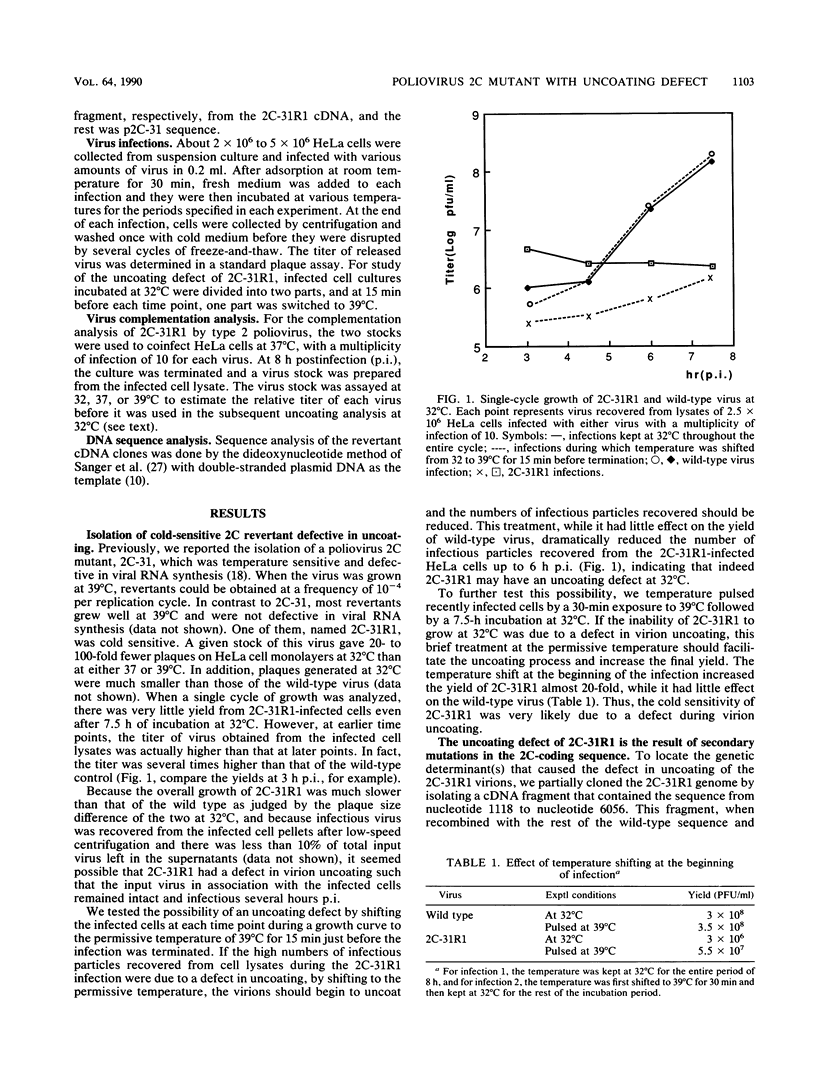
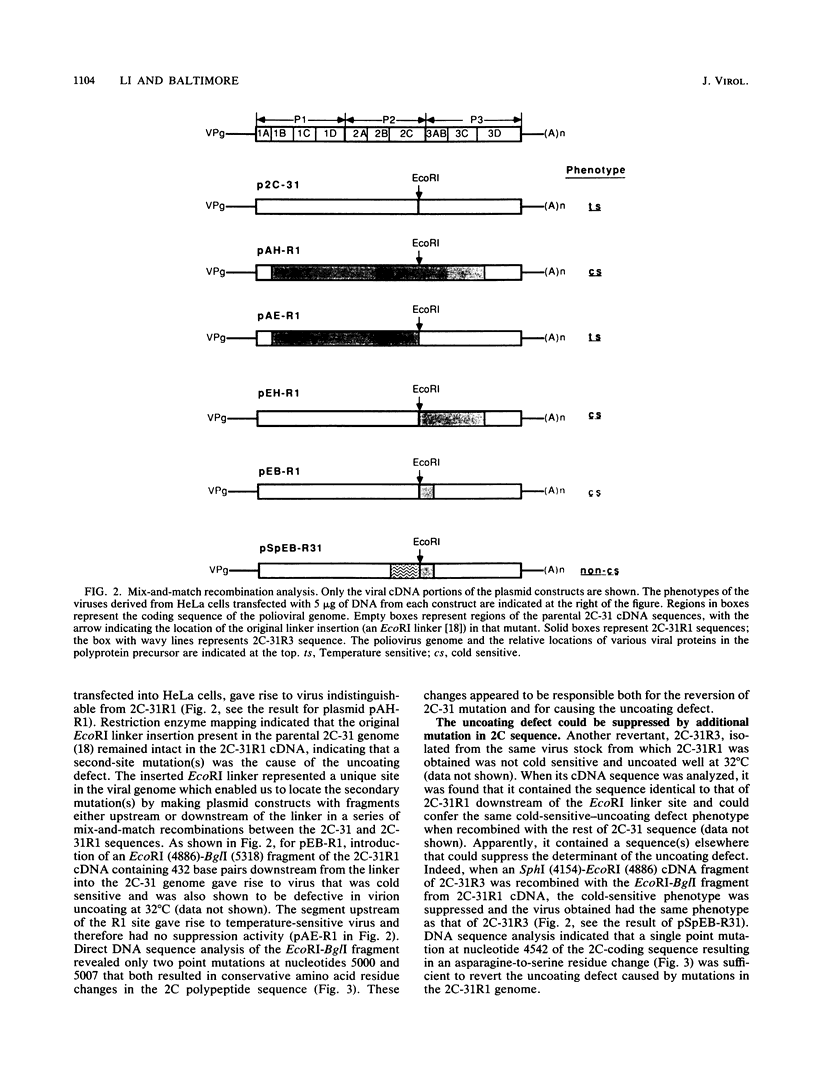
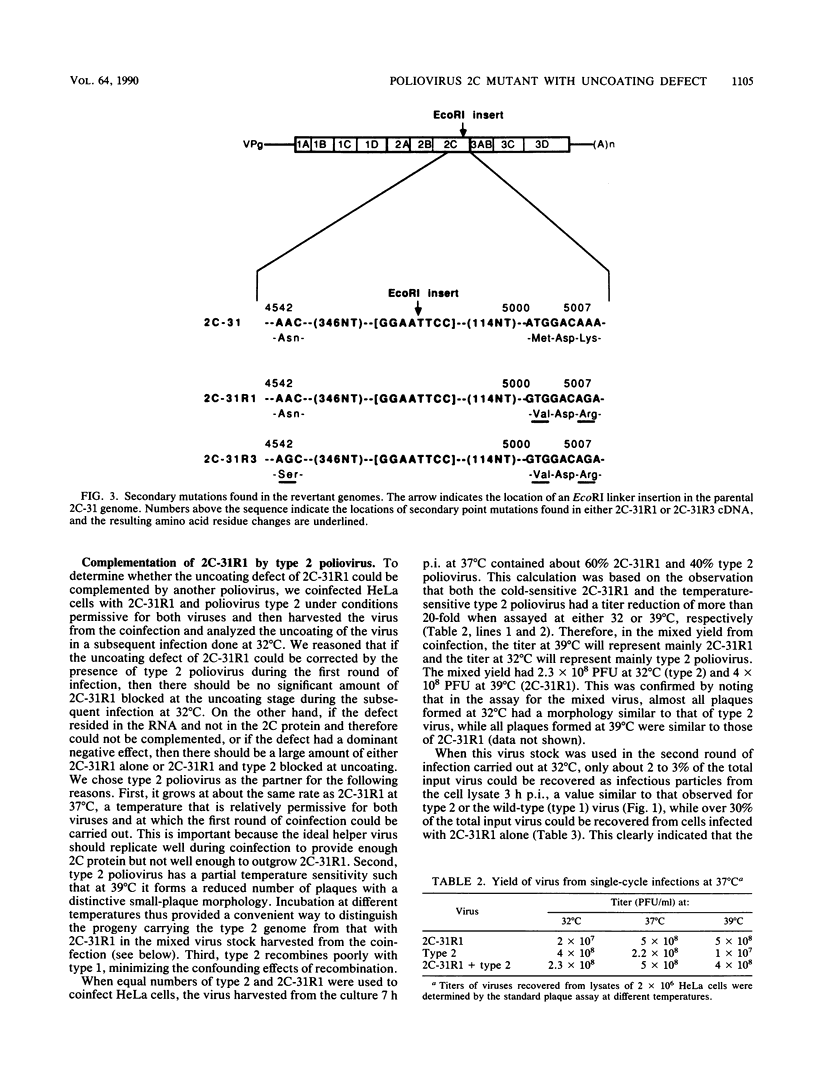
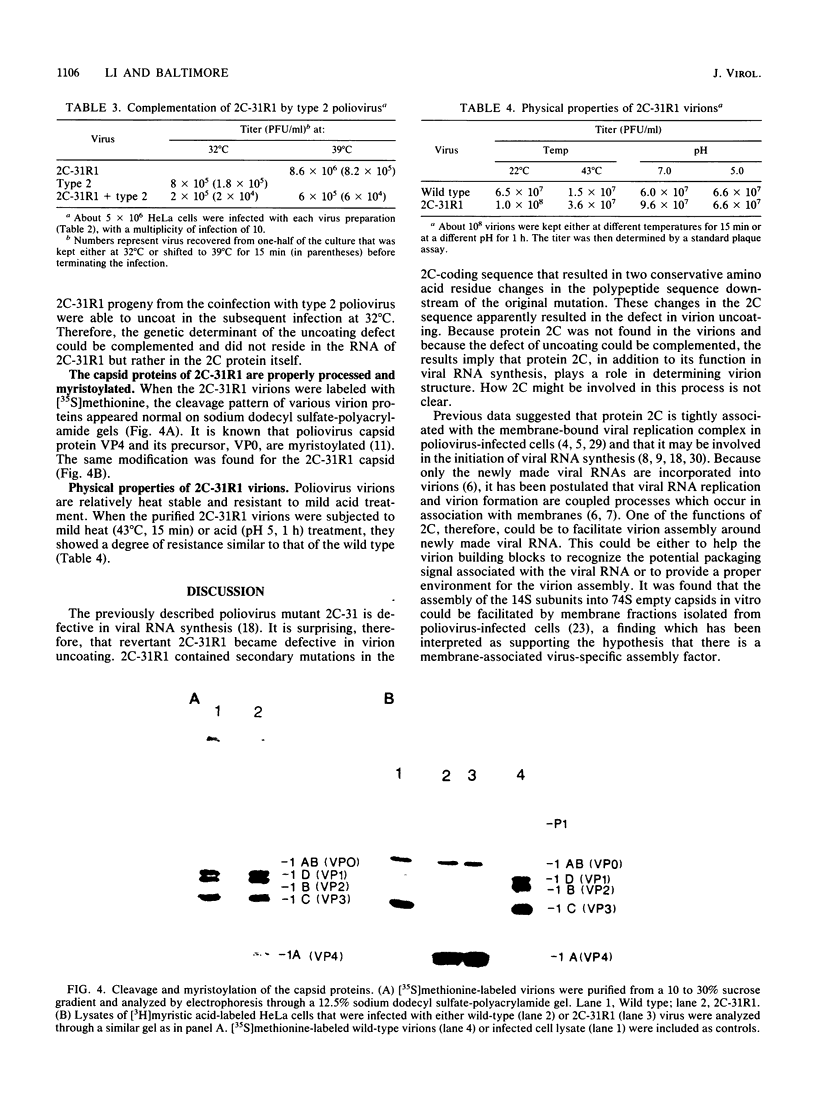
A revertant was isolated from a temperature-sensitive poliovirus 2C mutant, 2C-31, which is defective in viral RNA synthesis. This revertant, called 2C-31R1, grew well at 39 degrees C and was not defective in RNA synthesis. However, in contrast to its parental mutant, 2C-31R1 was cold sensitive and could hardly grow at all at 32 degrees C. Analysis of a single-cycle growth revealed that 2C-31R1 was defective in virion uncoating at 32 degrees C, and a substantial amount (more than 30%) of input viruses could be recovered as infectious particles from an infected cell lysate up to 6 h postinfection. The uncoating defect and the inability to grow at cold temperatures could be overcome by a brief incubation at the permissive temperature (39 degrees C) before the infection was continued at 32 degrees C. cDNA cloning and mix-and-match recombination experiments indicated that the defect in uncoating was the result of two secondary point mutations, seven nucleotides apart, in the 2C-coding sequence downstream of the inserted linker which is the original mutation in the parental 2C-31 genome. Another revertant, 2C-31R3, isolated from the same 2C-31 stock, was not defective in uncoating and appeared to be a secondary revertant that contained an intragenic suppressor for the uncoating defect. The uncoating defect of 2C-31R1 could be complemented by type 2 poliovirus. These results suggested that protein 2C, in addition to its role in viral RNA synthesis, has a function in determining virion structure.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein H. D., Sarnow P., Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986 Dec;60(3):1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. D., Sonenberg N., Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol Cell Biol. 1985 Nov;5(11):2913–2923. doi: 10.1128/mcb.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K., Egger D., Rasser Y., Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983 Nov;131(1):39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- Butterworth B. E., Shimshick E. J., Yin F. H. Association of the polioviral RNA polymerase complex with phospholipid membranes. J Virol. 1976 Aug;19(2):457–466. doi: 10.1128/jvi.19.2.457-466.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliguiri L. A., Compans R. W. The formation of poliovirus particles in association with the RNA replication complexes. J Gen Virol. 1973 Oct;21:99–108. doi: 10.1099/0022-1317-21-1-99. [DOI] [PubMed] [Google Scholar]
- Caliguiri L. A., Mosser A. G. Proteins associated with the poliovirus RNA replication complex. Virology. 1971 Nov;46(2):375–386. doi: 10.1016/0042-6822(71)90039-0. [DOI] [PubMed] [Google Scholar]
- Caliguiri L. A., Tamm I. Action of guanidine on the replication of poliovirus RNA. Virology. 1968 Jul;35(3):408–417. doi: 10.1016/0042-6822(68)90219-5. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chow M., Newman J. F., Filman D., Hogle J. M., Rowlands D. J., Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987 Jun 11;327(6122):482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- Franssen H., Leunissen J., Goldbach R., Lomonossoff G., Zimmern D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984 Apr;3(4):855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman N., Baltimore D. A plasma membrane component able to bind and alter virions of poliovirus type 1: studies on cell-free alteration using a simplified assay. Virology. 1977 Oct 1;82(1):25–36. doi: 10.1016/0042-6822(77)90029-0. [DOI] [PubMed] [Google Scholar]
- Jacobson M. F., Asso J., Baltimore D. Further evidence on the formation of poliovirus proteins. J Mol Biol. 1970 May 14;49(3):657–669. doi: 10.1016/0022-2836(70)90289-5. [DOI] [PubMed] [Google Scholar]
- Jacobson M. F., Baltimore D. Polypeptide cleavages in the formation of poliovirus proteins. Proc Natl Acad Sci U S A. 1968 Sep;61(1):77–84. doi: 10.1073/pnas.61.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P., Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988 Nov;62(11):4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg-Holm K., Gosser L. B., Kauer J. C. Early alteration of poliovirus in infected cells and its specific inhibition. J Gen Virol. 1975 Jun;27(3):329–342. doi: 10.1099/0022-1317-27-3-329. [DOI] [PubMed] [Google Scholar]
- Madshus I. H., Olsnes S., Sandvig K. Mechanism of entry into the cytosol of poliovirus type 1: requirement for low pH. J Cell Biol. 1984 Apr;98(4):1194–1200. doi: 10.1083/jcb.98.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn C. L., Wimmer E., Racaniello V. R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989 Mar 10;56(5):855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- Pallansch M. A., Kew O. M., Semler B. L., Omilianowski D. R., Anderson C. W., Wimmer E., Rueckert R. R. Protein processing map of poliovirus. J Virol. 1984 Mar;49(3):873–880. doi: 10.1128/jvi.49.3.873-880.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin M., Phillips B. A. In vitro assembly of polioviruses. 3. Assembly of 14 S particles into empty capsids by poliovirus-infected HeLa cell membranes. Virology. 1973 May;53(1):107–114. doi: 10.1016/0042-6822(73)90469-8. [DOI] [PubMed] [Google Scholar]
- Pincus S. E., Diamond D. C., Emini E. A., Wimmer E. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J Virol. 1986 Feb;57(2):638–646. doi: 10.1128/jvi.57.2.638-646.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. F., Maizel J. V., Jr Evidence for large precursor proteins in poliovirus synthesis. Proc Natl Acad Sci U S A. 1968 Mar;59(3):966–971. doi: 10.1073/pnas.59.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegami T., Semler B. L., Anderson C. W., Wimmer E. Membrane fractions active in poliovirus RNA replication contain VPg precursor polypeptides. Virology. 1983 Jul 15;128(1):33–47. doi: 10.1016/0042-6822(83)90316-1. [DOI] [PubMed] [Google Scholar]
- Tershak D. R. Inhibition of poliovirus polymerase by guanidine in vitro. J Virol. 1982 Jan;41(1):313–318. doi: 10.1128/jvi.41.1.313-318.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]