Abstract
The discovery that peptide nucleic acids (PNA) mimic DNA and RNA by forming complementary duplex structures following Watson–Crick base pairing rules opens fields in biochemistry, diagnostics, and medicine for exploration. Progress requires the development of modified PNA duplexes having unique and well defined properties. We find that anthraquinone groups bound to internal positions of a PNA oligomer intercalate in the PNA–DNA hybrid. Their irradiation with near-UV light leads to electron transfer and oxidative damage at remote GG doublets on the complementary DNA strand. This behavior mimics that observed in related DNA duplexes and provides the first evidence for long range electron (hole) transport in PNA–DNA hybrid. Analysis of the mechanism for electron transport supports hole hopping.
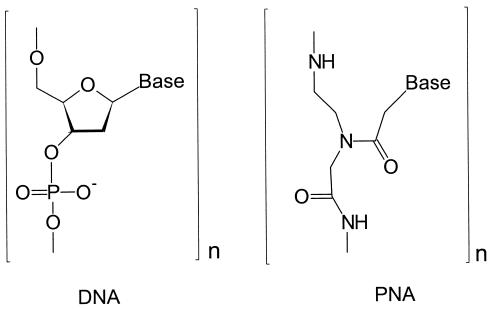
Peptide nucleic acid (PNA) oligomers are DNA/RNA analogs (see Fig. 1) in which the natural sugar–phosphate backbone is replaced by a synthetic peptide backbone (1). PNA oligomers that contain purine and pyrimidine nucleobases hybridize with complementary DNA and RNA strands to form right-handed, double-helical complexes according to the Watson–Crick rules of hydrogen bond-mediated base pair formation (2). Although much has been learned about the structural (3) and thermodynamic (4) factors involved in hybridization, little is known about the chemical reactivity of PNA/DNA hybrids. It is crucial to understand how PNA/DNA hybrids mimic the reactions and functions of duplex DNA. Of immediate importance for their application as clinical diagnostic agents is investigation of the conductivity of DNA and its PNA analogs (5, 6).
Figure 1.
A comparison of DNA and PNA building blocks.
DNA must balance the dual requirements of chemical stability and ease of transcription and replication (7). It is clear that DNA is far from inert toward a variety of different reactive species, particularly oxidizing agents. Oxidative damage to DNA produced by normal metabolism, deep-UV laser irradiation (8), gamma rays (9), or pulse radiolysis (10) accumulates at guanine residues, an effect attributed to one-dimensional migration of a radical cation (“hole”) along the DNA helix (11). Both the low oxidation potential and reactivity of the guanine radical cation contribute to the effectiveness of guanine as a trap for the migrating hole. Because guanine lesions may be the major cause of mutations (12), intense attention is focused on understanding the conductivity properties of DNA to elucidate the mechanisms by which migration of oxidative damage occurs (13). In this regard, the recent reports by Barton and coworkers are particularly important (14, 15). They describe a system consisting of a rhodium complex that is covalently linked to one end of a DNA duplex. Irradiation caused damage to the DNA more than 30 Å from where the complex was presumed to intercalate. This observation opens up exciting opportunities to study the factors that control hole migration in nucleic acids.
Photosensitizers often react with nucleic acids by single electron transfer to oxidize a base. Recent findings reveal that the light-induced reactions of a photosensitizer bound to duplex DNA by intercalation frequently generate alkali-dependent cleavage sites selectively at the 5′-G of G-purine doublets, with a strong preference for GG steps. These photosensitizers include substituted anthraquinones (16, 17), naphthalimides (18, 19), a rhodium metal complex (14), and riboflavin (20). Breslin and Schuster (17) demonstrated unambiguously that GG-selective, photoinduced damage of DNA arises by an electron transfer pathway from an intercalated anthraquinone. Time-resolved spectroscopy reveals that the excited state of the quinone accepts an electron from a base in the DNA within 20 ps of excitation (21). The base radical cation (hole) can either recombine with the electron, be trapped by reaction with water and/or oxygen, or migrate along the DNA helix to the lowest oxidation potential sites that serve as traps (22, 23).
We prepared a series of PNA oligomers with anthraquinone derivatives (AQ) covalently linked to internal positions. The ability of the quinone to photosensitize DNA damage by electron transfer when bound to the duplex by intercalation suggested it could serve a similar role in PNA-containing duplexes. The inability of common intercalators to bind to PNA (24) required that the quinone chromophore be covalently linked to the PNA backbone. The facile modification of PNA at internal residues as well as the superior hybridization properties of PNA oligomers offer distinct advantages relative to synthesis of modified DNA oligomers. Irradiation of the anthraquinone in the hybrid duplex leads to long distance hole migration and damage at GG sites in the DNA strand. Additional experiments reveal the mechanism for hole migration by its directional preference in the stacked base pairs of a PNA–DNA hybrid duplex.
MATERIALS AND METHODS
Radiolabeling of DNA.
DNA oligomers, including those with 8-OxoG and abasic modifications, were purchased from Midland Certified Reagent (Midland, Texas) and were used as received. DNA oligomers were labeled at the 5′ terminus using [γ-32P]-ATP and T4 polynucleotide kinase, according to standard procedures. The end-labeled DNA was purified by electrophoresis through a 20% denaturing polyacrylamide gel. The DNA band was excised from the gel, eluted overnight, and ethanol-precipitated.
Photocleavage Experiments.
In a typical experiment, PNA–DNA hybrids were formed by mixing together in a microcentrifuge tube PNA and unlabeled DNA (5 μM each) with 40,000-cpm labeled DNA in 200 μl of sodium phosphate buffer (10 mM phosphate, pH 7.0). The mixture was heated to 90°C for 2 min, then allowed to cool to room temperature over a period of 1 h. Two 10-μl aliquots were removed from the sample and kept in the dark, while the remainder was irradiated in a Rayonet Photoreactor (Southern New England Ultraviolet Company, Bransford, CT) equipped with eight lamps (λ = 350 nm). The sample tube was suspended from a rotating platform, and cooling air was supplied by a fan in the bottom of the unit. At the desired times, 2 × 10-μl aliquots were removed and kept in the dark. After irradiation, the DNA was precipitated with ethanol and dried. One tube from each set of aliquots was then incubated in 1 M piperidine at 90°C for 30 min. After evaporation of the piperidine and drying, all samples were suspended in denaturing loading buffer, then loaded onto a 20% denaturing polyacrylamide gel. Autoradiography was used to detect cleavage products.
Quantum Yield Determination.
A 40-μM sample of the PNA-2/DNA-3 hybrid duplex (Tm = 51°C) was prepared from stoichiometric amounts of each strand in 10 mM sodium phosphate buffer by heating the solution to 90°C for 5 min and then cooling it slowly (5 h) to room temperature. The sample was irradiated at 350 nm for 1 h in a calibrated Rayonet reactor equipped with eight 15-W lamps (light flux = 2.6 × 10−8 Eins/min⋅cm2). The samples were treated with piperidine (90°C for 30 min) and 5′-dephosphorylated with bacterial alkaline phosphatase. The 5′-AAT-3′ and 5′-GAAT-3′ fragments resulting from cleavage at the GG step of the DNA strand were separated and quantified by HPLC on a Rainin Microsorb-MV C18 column (Rainin Instruments) with an acetonitrile/water/ammonium acetate mobile phase. The fragments were identified by comparison with authentic samples independently prepared.
RESULTS AND DISCUSSION
The Structure of PNA Conjugates.
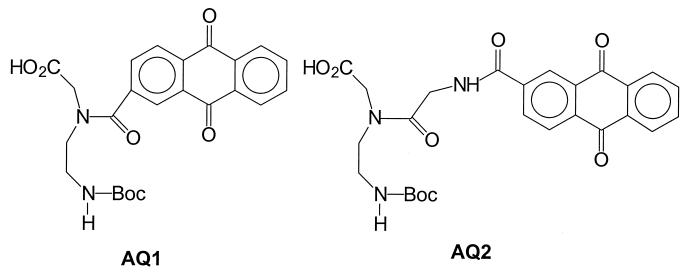
PNA oligomers were synthesized from N-Boc protected (2-aminoethyl)glycine monomers (25). The AQ-containing monomers are shown in Fig. 2. In AQ1, the anthraquinone group is linked to the peptide backbone by a single amide bond whereas for AQ2 there is an intervening glycine group between the quinone and backbone. A third monomer (Ac, used in control experiments) has an acetyl group bound to the backbone nitrogen atom of the (2-aminoethyl)glycine. The synthetic details and characterization of these compounds and the oligomers described below are reported elsewhere (unpublished work).
Figure 2.
PNA monomers covalently linked to AQ.
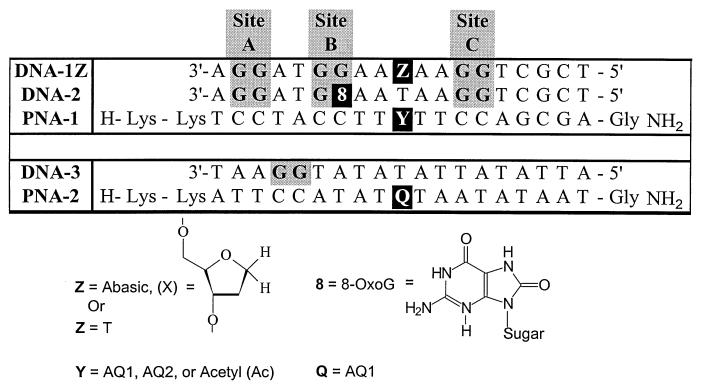
The PNA–AQ monomers were incorporated into 19-base oligomers (Fig. 3). The quinones are at the central position of PNA-1, equidistant from two CC sequences (sites B and C), with a third CC sequence (site A) two bases beyond site B. Hybridization of the PNA-1 with its complementary DNA-1Z oligomers gives duplexes with three GG sites to act as traps of a migrating radical cation. Placement of the quinone at the center of the duplex permits study of both the distance and directional dependence of hole migration.
Figure 3.
PNA–DNA duplexes.
A model of a PNA–DNA duplex containing an intercalated, covalently linked AQ1 group was built in sybyl 6.0 (Tripos Associates, St. Louis) using the coordinates for a PNA–DNA structure determined using NMR spectroscopy by Erikkson and Nielsen (27). One of the internal base pairs of the duplex was removed by excision of the PNA and DNA bases. The AQ carboxamide was then linked to the PNA backbone in place of the nucleobase while the DNA base was replaced by a hydrogen atom to create an abasic site. Even though this structure was not subjected to energy minimization, it clearly showed that there is space within the helix to accommodate the intercalated AQ group. This result and other experiments including thermal denaturation and phosphorescence quenching are consistent with an intercalated conformation of the AQ and are reported in detail elsewhere (unpublished work).
Light Causes Long Range DNA Damage.
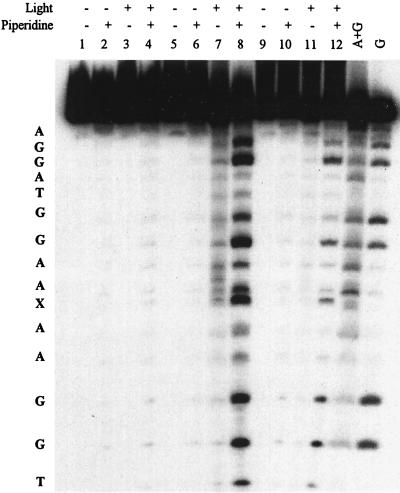
The irradiation of AQs intercalated in duplex DNA gives efficient, piperidine-requiring strand cleavage selectively at GG sites by an electron transfer mechanism (16, 17). The base sequence in PNA-1/DNA-1X was specifically designed to probe for the corresponding reaction in PNA/DNA hybrids and to examine its mechanism. DNA-1X was labeled at its 5′ terminus with 32P by standard methods (28) and hybridized with the complementary PNA oligomers. Irradiation of the PNA-1(AQ1)/DNA-1X hybrid at 350 nm lead to piperidine-requiring DNA strand cleavage at the three GG sites (Fig. 4, lane 8). Cleavage at the 3′-G is favored for site A whereas the 5′-G is favored at site B (Fig. 3); cleavage occurs with equal efficiency at each G of site C. DNA strand cleavage also is observed at the abasic residue directly opposite the AQ. The pattern of cleavage bands is identical for the PNA-1(AQ2) hybrid (Fig. 4, lane 12) although the efficiency is lower. Control experiments with PNA-1(Ac), which lacks the quinone group, show essentially no cleavage of the DNA strand at any site (Fig. 4, lanes 1–4).
Figure 4.
Cleavage of DNA by AQ conjugate within PNA/DNA hybrids. Samples were prepared by mixing 5000 cpm 5′-32P-end-labeled DNA-1X and 5.0 μM each of unlabeled DNA-1X and PNA in 10 mM sodium phosphate buffer (pH 7.0). Samples were irradiated for 60 min in a Rayonet equipped with eight lamps (λmax = 350 nm) in colorless microcentrifuge tubes, divided in two, precipitated with ethanol, washed, and dried. One set was incubated with 100 μl of 1.0 M piperidine at 90°C for 30 min. The samples were suspended in denaturing loading buffer and electrophoresed through a 20% denaturing polyacrylamide gel. The image was obtained by scanning the autoradiogram of the gel. Conditions of irradiation or piperidine treatment are indicated above each lane. Three different PNAs were investigated: PNA(Ac) (lanes 1–4), PNA(AQ1) (lanes 5–8), and PNA(AQ2) (lanes 9–12). The sequence of DNA-1X, determined from the Maxam–Gilbert A+G and G sequencing lanes, is on the left side of the image.
Additional control experiments show that cleavage of the DNA strand results from an intramolecular reaction initiated by excitation of the quinone in the hybrid duplex. In the photocleavage experiment described above, the PNA and labeled DNA were hybridized in the presence of a stoichiometric amount of unlabeled DNA-1X (relative to PNA-1). This ensures that all of the PNA will be hybridized during the experiment, with some fraction of the hybrids containing labeled DNA-1X strands. Hybridization of PNA-1(AQ1) with labeled DNA-1X and with a 10-fold excess of unlabeled DNA-1X strongly inhibits cleavage of the labeled DNA because the fraction of PNA-1(AQ1) that is hybridized to labeled DNA-1X is 10-fold lower. However, hybridization in the presence of a stoichiometric amount of unlabeled complementary DNA and a 9-fold excess of noncomplementary single-stranded DNA has no effect on the cleavage of the labeled DNA. (The presence of a stoichiometric amount of complementary, unlabeled DNA in the latter case ensures that the PNA is hybridized while maintaining the concentration of unhybridized DNA.) The lack of inhibition in this experiment demonstrates that cleavage is not mediated by a freely diffusing intermediate because the excess of single-stranded, unlabeled DNA should effectively inhibit such a process. Finally, cleavage is still observed when excess unlabeled DNA-1X is added after hybridization with labeled DNA, demonstrating the kinetic stability of the PNA(AQ)–DNA hybrids studied in these experiments.
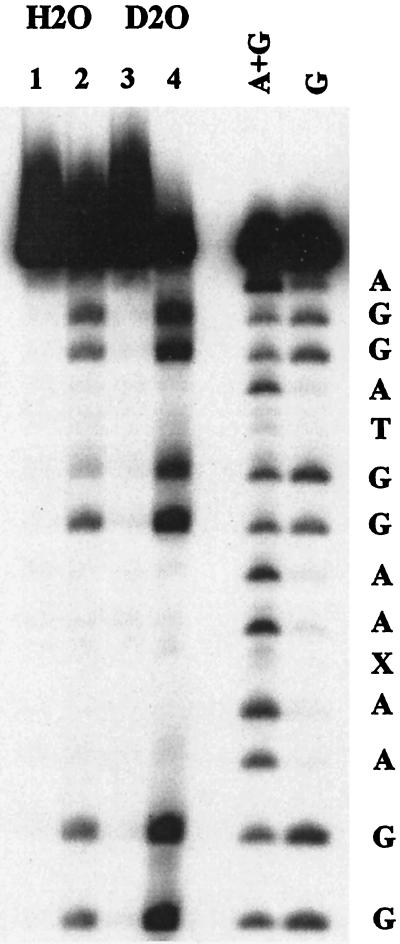
Selective cleavage of DNA at guanine has been observed in reactions initiated by 1-electron oxidation and by reaction of singlet oxygen (1O2) (29). The G-selective cleavage of the PNA–DNA hybrid cannot be caused by freely diffusing 1O2, but a recent report raises the possibility of one-dimensional intramolecular diffusion of 1O2 in a groove of duplex DNA (30), and this could account for the long range cleavage that is observed. However, this path is unlikely in the present case because the lifetime of the requisite AQ triplet is shortened by electron transfer quenching. Additional control experiments were performed that compare the cleavage of PNA-1(AQ1)–DNA hybrids caused by irradiation of methylene blue, a known 1O2 generator (31), with direct irradiation of the quinone in the hybrid duplex. Inspection of Fig. 5 shows cleavage of DNA upon irradiation of methylene blue that is enhanced significantly when D2O is substituted for H2O (compare Fig. 5, lanes 4 and 2), proving that the cleavage in this case is caused by reaction with 1O2 generated by excited methylene blue (32). No effect of D2O is seen for the quinone-initiated cleavage (data not shown). It is important to note that the cleavage pattern due to the reaction of 1O2 is significantly different from that seen for irradiation of the quinone. In particular, densitometric analysis indicates that the ratio of cleavage of the 5′-G to the 3′-G at site A is 1.6 for 1O2 and 0.7 for the AQ-initiated reaction. Clearly, the long range G-selective cleavage reaction of the PNA–DNA hybrid duplex cannot involve 1O2 but must occur by electron transfer to generate a base radical cation and long distance migration of the hole as has been previously proposed for anthraquinones in duplex DNA.
Figure 5.
Methylene blue-sensitized photocleavage of PNA(AQ1)/DNA-1X hybrid. Two samples were prepared, each containing 5000 cpm 5′-32P-end-labeled DNA-1X and 5.0 μM each of unlabeled DNA-1X and PNA in 10 mM sodium phosphate buffer (pH 7.0) with 10 μM methylene blue. The water was evaporated and replaced with D2O in one case, H2O in the other. The solvent was evaporated and replaced twice more to minimize the amount of H2O present in the D2O sample. Samples were then divided in two; one set of samples was kept in the dark, and the other set was irradiated for 15 min in microcentrifuge tubes with the filtered (λ > 600 nm) output of a 150-W Hg arc lamp. All samples were treated with piperidine after precipitation. Samples for lanes 1 and 3 were kept in the dark, and those for lanes 2 and 4 were irradiated. Control samples lacking methylene blue showed no cleavage of the DNA after irradiation under these conditions (data not shown).
Mechanistic interpretation of reactions with extraordinarily low quantum yields is risky because minor structural isomers or impurities can confound the analysis. The quantum yields for cleavage at the GG step (ΦGG) by photonucleases range from 1.4% for an intercalated AQ derivative (with a 10-fold preference for cleavage at the 5′-G) (16) to 0.000005% reported for a Rh(phi)2DMB+3 complex linked covalently to a 5′ terminus of duplex DNA and presumed to be intercalated (14). We determined ΦGG in a PNA–DNA hybrid duplex by an HPLC technique.
The PNA-2(AQ1)/DNA-3 hybrid duplex (Fig. 3) was designed for measurement of ΦGG. Cleavage at the 5′-G gives, after 5′-dephosphorylation, 5′-GAAT-3′, and cleavage at the 3′-G gives 5′-AAT-3′. These oligonucleotides are separated easily and quantified by reversed phase HPLC. In air-saturated solution, the ΦGG for PNA-2(AQ1)/DNA-3 is 0.17%. The ratio of 5′ to 3′ G cleavage is 1:3, a preference opposite to that seen in duplex DNA. Clearly, the efficiency of radical cation generation, migration, and conversion to a piperidine-cleavable lesion is relatively high in the quinone-containing hybrid PNA–DNA duplex.
The 3′-G at site A of the PNA-1(AQ1)/DNA-1X hybrid is more than 25 Å from the AQ group, and the short linkage between the quinone and the PNA backbone prohibits direct contact between the donor (G) and acceptor (AQ), yet cleavage is observed at this and other remote sites within the duplex. One model for long range oxidative damage involves instantaneous delocalization of the radical cation, i.e., the electron is transferred directly from the GG site to the excited state photosensitizer. Selective GG cleavage in such a process would reflect a higher radical cation density at these most easily oxidized sites. An alternative mechanism postulates oxidation of a base at a distinct position (e.g., adjacent to the photosensitizer) followed by migration of the radical cation by sequential electron transfers, i.e., hole-hopping. In this model, the hole will be distributed among the various low oxidation potential sites only if the rate of hopping is faster than the rate of irreversible chemical reaction (e.g., addition of water or O2 to the radical cation) at a particular site. The data presented thus far cannot distinguish between these two mechanistic models.
A Deep Trap Reveals the Mechanism.
Introduction of a low oxidation potential trap in the hybrid duplex allows a clear test of the mechanism for migration of oxidative damage. In particular, the central position of the AQ acceptor between two GG reaction sites (as opposed to the linkage of the photosensitizer to the duplex terminus) allows the placement of a trap so that instantaneous delocalization of the hole into the trap can be distinguished from its arrival at the trap by a series of hops. The delocalization model predicts inhibition of cleavage at all GG sites by the trap because the duplex is considered to be one continuous orbital system. Instantaneous connection of the hole and the trap decreases the likelihood of reaction at all other sites. On the other hand, migration by the hopping mechanism will exhibit a distinct directional preference for cleavage inhibition because the hole cannot “know” of the trap’s existence until it hops into it.
The ideal trap is a modified base that does not distort the structure of the PNA–DNA duplex and that has an oxidation potential significantly below that of the GG sequence. We selected the guanine derivative 7,8-dihydro-8-oxoguanine (8-OxoG; Fig. 3) as the trap on this basis. This modified base is often detected as a byproduct of oxidative damage (14, 20, 23, 31, 33–36). Although structural information is not available for PNA-containing duplexes, Williams and coworkers (37) recently determined the structure of a DNA duplex having an 8-OxoG substitution by x-ray crystallography and found that the oxidized base caused little perturbation. We found that substitution of an 8-OxoG for G on DNA in a PNA/DNA hybrid lowers the melting temperature of the duplex only 1°C. Foote and Sheu (36) report that the oxidation potential of 8-OxoG is 0.4–0.5 V below that of guanosine (as their t-butyldimethylsilyl-protected nucleoside derivatives). Considering that the difference in oxidation potential between G and A is only ≈0.1 V (38), 8-OxoG should provide a deep trap for holes in the PNA/DNA hybrid.
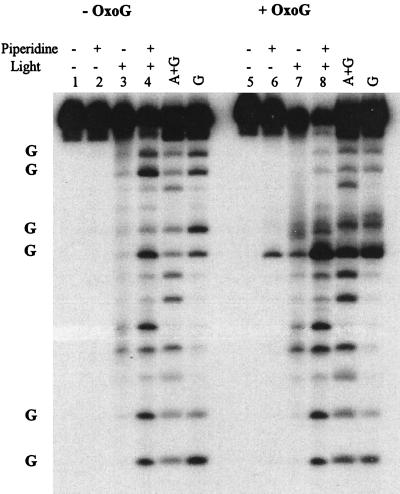
Photoinduced cleavage of the PNA-1(AQ1)/DNA-1T hybrid yields a cleavage pattern nearly identical to that observed for DNA-1X (Fig. 6, lane 4; compare with Fig. 4, lane 8). DNA-2 is analogous to DNA-1T except that 8-OxoG is substituted for G at site B. Irradiation of the PNA-1(AQ1)/DNA-2 hybrid shows significantly enhanced cleavage at site B, but the more dramatic effect is observed at site A, where cleavage is almost completely inhibited (lane 10). The 8-OxoG acts as a barrier to cleavage at this distal GG site. It is important to note that 8-OxoG substitution has no effect on cleavage at site C, which is located in the opposite direction from the trap. Clearly, the hole does not sense the presence of the 8-OxoG trap when the selection of migration direction occurs. Consequently, the hole cannot be in electronic contact with the trap as is required by the instantaneous delocalization model. These findings provide evidence that radical cations in a PNA–DNA duplex migrate by a discrete hopping mechanism.
Figure 6.
Effect of a hole trap on migration of oxidative damage in PNA/DNA. Samples containing 5.0 μM each of PNA(AQ1) and DNA-1T or DNA-3 (8-OxoG) were prepared and treated as described in Fig. 4. Conditions of irradiation and piperidine treatment are indicated above individual lanes. Lanes: 1–4, DNA-1T; 5–8, DNA-3. Note the lone cleavage band observed in lane 6; this band corresponds to the 8-OxoG site, which is cleaved slowly by piperidine treatment (23). The three GG sites are indicated at the left of the figure, with site A at the top and site C at the bottom.
CONCLUSIONS
The results described above illustrate the exciting potential of PNA. In this case, it provides a tool for probing long range electron transfer through stacked π-electron systems as well as for the photomodification of complementary DNA strands. The ability to cleave DNA in PNA/DNA hybrids by the same mechanisms that operate in DNA/DNA duplexes is a further demonstration of the remarkable ability of PNA to mimic DNA. Although there are reports of the use of PNA–nuclease conjugates for the site-specific cleavage of double-stranded DNA (26, 39), these experiments provide the first example in which PNA cleaves single-stranded DNA targets. Extension of these studies to electron transport within PNA/PNA duplexes and PNA2/DNA triplexes is planned as are experiments that will assess the ability of the PNA strand to trap oxidative damage. The ease of synthesizing internally modified PNA, the stability of the resulting hybrid duplexes, and their ability to mimic the reactivity of duplex DNA should contribute to the growing application of PNA in chemistry, biochemistry, and medicine.
Acknowledgments
We thank Professor Loren Williams of Georgia Tech for many helpful discussions and Professor W. David Wilson and Dr. Daoyuan Ding of Georgia State University for preparation of the PNA(AQ)/DNA hybrid computer model. Densitometry was performed in the Wilson lab with the assistance of Katherine Hopkins. This work was supported by funding from the National Institutes of Health and the National Science Foundation, for which we are grateful.
ABBREVIATIONS
- PNA
peptide nucleic acid
- AQ
anthraquinone derivative
- 8-OxoG
7,8-dihydro-8-oxoguanine
References
- 1.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 2.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Nordén B, Nielsen P E. Nature (London) 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 3.Hyrup B, Egholm M, Nielsen P E, Wittung P, Nordén B, Buchardt O. J Am Chem Soc. 1994;116:7964–7970. [Google Scholar]
- 4.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen P E, Nordén B, Gräslund A. J Am Chem Soc. 1996;117:5544–5552. [Google Scholar]
- 5.Dandliker P J, Holmlin R E, Barton J K. Science. 1997;275:1465–1468. doi: 10.1126/science.275.5305.1465. [DOI] [PubMed] [Google Scholar]
- 6.Beratan D N, Priyadarshy S, Risser S M. Chem Biol. 1997;4:3–8. doi: 10.1016/s1074-5521(97)90230-1. [DOI] [PubMed] [Google Scholar]
- 7.Sinden R R. DNA Structure and Function. San Diego: Academic; 1994. [Google Scholar]
- 8.Melvin T, Plumb M A, Botchway S W, O’Neill P, Parker A W. Photochem Photobiol. 1995;61:584–591. doi: 10.1111/j.1751-1097.1995.tb09873.x. [DOI] [PubMed] [Google Scholar]
- 9.Boon P J, Cullis P M, Symons M C R, Wren B W. J Chem Soc Perkin Trans. 1984;2:1393–1399. [Google Scholar]
- 10.Wolf P G, Jones D D, Candeias L P, O’Neill P. Int J Rad Biol. 1993;64:7–18. doi: 10.1080/09553009314551061. [DOI] [PubMed] [Google Scholar]
- 11.Becker D, Sevilla M D. Adv Radiat Biol. 1993;17:121–180. [Google Scholar]
- 12.Friedberg E C, Walker G C. Repairs and Mutagenesis. Washington, D.C.: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 13.Wilson E K. Chem Eng News. 1997;75:33–40. [Google Scholar]
- 14.Hall D B, Holmlin R E, Barton J K. Nature (London) 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 15.Hall D B, Barton J K. J Am Chem Soc. 1997;119:5045–5046. [Google Scholar]
- 16.Ly D, Kan Y, Armitage B, Schuster G B. J Am Chem Soc. 1996;118:8747–8748. [Google Scholar]
- 17.Breslin D T, Schuster G B. J Am Chem Soc. 1996;118:2311–2319. [Google Scholar]
- 18.Matsugo S, Kawanishi S, Yamamoto K, Sugiyama H, Matsuura T, Saito I. Angew Chem Int Ed Engl. 1991;30:1351–1353. [Google Scholar]
- 19.Saito I, Takayama M, Kawanishi S. J Am Chem Soc. 1995;117:5590–5591. [Google Scholar]
- 20.Ito K, Inoue S, Yamamoto K, Kawanishi S. J Biol Chem. 1993;268:13221–13227. [PubMed] [Google Scholar]
- 21.Armitage B A, Yu C, Devadoss C, Schuster G B. J Am Chem Soc. 1994;116:9847–9859. [Google Scholar]
- 22.Kasai H, Yamaizumi Z, Berger M, Cadet J. J Am Chem Soc. 1992;114:9692–9694. [Google Scholar]
- 23.Cullis P M, Malone M E, Merson-Davies L A. J Am Chem Soc. 1996;118:2775–2781. [Google Scholar]
- 24.Wittung P, Kim S K, Buchardt O, Nielsen P E, Nordén B. Nucleic Acids Res. 1994;22:5371–5377. doi: 10.1093/nar/22.24.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen L, Fitzpatrick R, Gildea B, Petersen K H, Hansen H F, Koch T, Egholm M, Buchardt O, Nielsen P E, Coull J, Berg R H. J Peptide Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 26.Lohse J, Hui C, Sönnichsen S H, Nielsen P E. In: DNA and RNA Cleavers and Chemotherapy of Cancer and Viral Diseases. Meunier B, editor. Dordrecht: Kluwer; 1996. pp. 133–141. [Google Scholar]
- 27.Erikkson M, Nielsen P E. Nat Struct Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Paillous N, Vicendo P. J Photochem Photobiol B. 1993;20:203–209. [Google Scholar]
- 30.Boutorine A S, Brault D, Takasugi M, Delgado O, Hélène C. J Am Chem Soc. 1996;118:9469–9476. [Google Scholar]
- 31.Devasagayam T P A, Steenken S, Obendorf M S W, Schulz W A, Sies H. Biochemistry. 1991;30:6283–6289. doi: 10.1021/bi00239a029. [DOI] [PubMed] [Google Scholar]
- 32.Merkel P B, Kearns D R. J Am Chem Soc. 1972;94:1029–1031. [Google Scholar]
- 33.Adam W, Saha-Möller C R, Schönberger A, Berger M, Cadet J. Photochem Photobiol. 1995;62:231–238. doi: 10.1111/j.1751-1097.1995.tb05263.x. [DOI] [PubMed] [Google Scholar]
- 34.Floyd R A, West M S, Eneff K L, Schneider J E. Arch Biochem Biophys. 1989;273:106–111. doi: 10.1016/0003-9861(89)90167-7. [DOI] [PubMed] [Google Scholar]
- 35.Schneider J E, Price S, Maidt L, Gutteridge J M C, Floyd R A. Nucleic Acids Res. 1990;18:631–635. doi: 10.1093/nar/18.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheu C, Foote C S. J Am Chem Soc. 1995;117:6439–6442. [Google Scholar]
- 37.Lipscomb L A, Peek M E, Morningstar M L, Verghis S M, Miller E M, Rich A, Essigmann J M, Williams L D. Proc Natl Acad Sci USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jovanovich S V, Simic M G. J Phys Chem. 1986;90:974–979. [Google Scholar]
- 39.Footer M, Egholm M, Kron S, Coull J M, Matsudaira P. Biochemistry. 1996;35:10673–10679. doi: 10.1021/bi960486p. [DOI] [PubMed] [Google Scholar]