Abstract
Marine primary productivity is iron (Fe)-limited in vast regions of the contemporary oceans, most notably the high nutrient low chlorophyll (HNLC) regions. Diatoms often form large blooms upon the relief of Fe limitation in HNLC regions despite their prebloom low cell density. Although Fe plays an important role in controlling diatom distribution, the mechanisms of Fe uptake and adaptation to low iron availability are largely unknown. Through a combination of nontargeted transcriptomic and metabolomic approaches, we have explored the biochemical strategies preferred by Phaeo dactylum tricornutum at growth-limiting levels of dissolved Fe. Processes carried out by components rich in Fe, such as photosynthesis, mitochondrial electron transport, and nitrate assimilation, were down-regulated. Our results show that this retrenchment is compensated by nitrogen (N) and carbon (C) reallocation from protein and carbohydrate degradation, adaptations to chlorophyll biosynthesis and pigment metabolism, removal of excess electrons by mitochondrial alternative oxidase (AOX) and non-photochemical quenching (NPQ), and augmented Fe-independent oxidative stress responses. Iron limitation leads to the elevated expression of at least three gene clusters absent from the Thalassiosira pseudonana genome that encode for components of iron capture and uptake mechanisms.
Keywords: genome, metabalomics, photosynthesis, transcriptomics, nutrients
The oxidizing conditions of contemporary marine ecosystems result in exceedingly low levels of dissolved Fe (1), and the cellular Fe demand of modern phytoplankton is frequently in excess of Fe availability (1, 2). This paradox reflects the likelihood that their biochemical machinery evolved in the Fe-replete, reducing Proterozoic oceans. Fe limitation is a major factor controlling phytoplankton growth in the large, perennially high nutrient low chlorophyll (HNLC) regions in the subarctic Pacific, equatorial Pacific, and Southern Oceans (3–5) and may be important in the North Atlantic (6).
Marine diatoms are the most important eukaryotic phytoplankton for carbon sequestration, contributing ≈40% of global oceanic organic carbon production per year. More than 70% of blooms stimulated by mesoscale in situ Fe fertilization of Fe-limited HNLC waters were dominated by diatoms (7), indicating that diatoms persist in chronically Fe-limited environments and resume rapid growth when the limitation pressure is alleviated. The tolerance of diatoms to Fe limitation varies widely between species (8–11).
Phaeodactylum tricornutum is highly tolerant to Fe limitation and can grow in steady-state laboratory cultures at iron levels 50 times lower than those tolerated by the centric diatom Thalassiosira pseudonana (12). Steady-state growth of P. tricornutum is Fe limited in the range of 10–30 pmol·liter−1 Fe′ (Fe′ is the sum of all unchelated Fe species) (Table 1), similar to T. oceanica and Pseudonitzschia spp., two open ocean diatoms that are commonly found in the most severely Fe limited regions of the world's oceans (13).
Table 1.
General cellular, physiological, and biochemical characteristics of Fe-limited P. tricornutum cells and cultures
| Parameter | Fe-limited | Fe-replete | Fe-limited/Fe-replete |
|---|---|---|---|
| Growth rate | 0.18 ± 0.05 | 0.88 ± 0.01 | 0.2 ± 0.06 |
| Ferric reductase assay, AU per cell | 0.717 ± 0.03 | 0.020 ± 0.004 | 35.9 ± 7.3 |
| Ferric reductase assay, AU/μm3 | 0.01 | 0.0002 | 50 |
| Carbon fixation, pmol C per cell × hour | 0.010 ± 0.006 | 0.141 ± 0.022 | 0.07 ± 0.04 |
| Cell diameter, μm | 2.5 | 3.5 | 0.71 |
| Cell volume, μm3 | 60 | 100 | 0.60 |
| Chl per cell, pg per cell | 0.22 | 0.5 | 0.44 |
| Chl per volume, fg/μm3 | 3.67 | 5.0 | 0.73 |
| Fv/Fm | 0.18 ± 0.1 | 0.5 ± 0.05 | 0.36 ± 0.2 |
| PSII | 1.2 ± 0.14 | 1 | 1.2 ± 0.14 |
| PSI | 0.43 ± 0.1 | 1.0 | 0.43 ± 0.10 |
| Cytochrome b6f | 0.75 ± 0.13 | 1.0 | 0.75 ± 0.13 |
| Cytochrome c6 | 0.7 ± 0.05 | 1.0 | 0.70 ± 0.05 |
| NPQ | 4.8 ± 0.8 | 3.1 ± 0.51 | 1.5 ± 0.4 |
| AOX activity | 0.6 ± 0.15 | 0.25 ± 0.18 | 2.4 ± 1.83 |
Photosystem and electron carrier concentrations are given in relative units normalized to Fe-replete levels. AOX activity is given as a percentage of total respiration (see SI Methods for details). Fe levels in Fe-limited and Fe-replete cultures corresponded to 13.4 pmol·liter−1 Fe′ and 2.6 nmol·liter−1 Fe′, respectively.
The recently completed genome sequence of P. tricornutum (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html) sheds light on some important differences between P. tricornutum and T. pseudonana, which may account for their respective thresholds for Fe limitation. We evaluated the whole-cell response of P. tricornutum to Fe limitation with multiple approaches combining gene expression profiling and comparative genomics with gas chromatography-mass spectroscopy (GC-MS)-aided nontargeted metabolomic analysis and a range of physiological measurements. Genes responsive to Fe limitation were identified through a statistically-verified quantitative comparison (14) of 8,669 expressed sequenced tags (ESTs) derived from Fe-limited P. tricornutum cells with 104,783 ESTs derived from cells grown in 11 different (all iron-replete) culture conditions (www.biologie.ens.fr/diatomics/EST). A partial genome microarray and qRT-PCR provided further assessment and verification of differential regulation, leading to the identification of 212 up-regulated and 26 down-regulated genes. The represented acclimation strategies were grouped into three categories: down-regulation, compensation, and acquisition.
Results and Discussion
Down-Regulation of Photosynthesis.
Low Fe supply leads to cellular energy limitation in P. tricornutum (15) and causes significant changes in carbon metabolism. Carbon fixation rates per cell were 14-fold lower in Fe-limited P. tricornutum cells compared with Fe-replete cells (Table 1). This difference remained significant (8-fold) despite normalization to the smaller cell volume observed in Fe-limited conditions [Table 1 and supporting information (SI) Fig. S1]. Reductions in cell volume, chlorophyll (Chl) per cell, photosynthetic efficiency of PSII (Fv/Fm), and content of Fe-rich complexes and/or electron carriers of the electron transfer chain are all common responses to Fe limitation (Table 1) (15, 16), reflecting compromised photosystem reaction centers, reduced photosynthetic electron transport rates, decreased reductant production, and an inability to process absorbed photons (4, 17, 18).
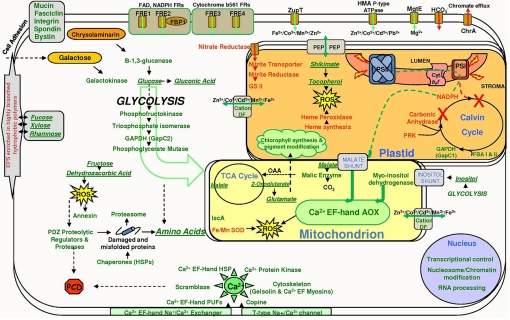
Severe down-regulation of genes encoding plastid targeted copies of β-carbonic anhydrase (CA) and phosphoribulokinase (PRK), two enzymes supplying substrate for RuBisCO, and a decrease in expression of a HCO3− transporter (Fig. 1 and Table S1) suggest that carbon fluxes into the cell and toward RuBisCO are adjusted to match reductant supply. In accordance, Calvin/Benson cycle-related genes downstream of RuBisCO, such as plastid localized glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and plastid fructose bisphosphate aldolases (FBA) I and II were up-regulated under Fe stress (Fig. 1 and Table S1), likely in response to reductions in substrate abundance.
Fig. 1.
Hypothetical cellular pathways and processes in iron-limited pennate diatom cells. All roman green or red type depicts gene transcripts found to be up- or down-regulated, respectively. Italicized and underlined green type indicates metabolites found to be enriched relative to total protein in iron-limited cells. DF, diffusion factor; EPS, extracellular polymeric substances; FR, ferric reductase; HMA, heavy metal-associated; PCD, programmed cell death; ROS, reactive oxygen species.
Higher levels of expression of genes encoding galactokinase and endo-1,3-beta-glucanase under Fe limitation (Fig. 1 and Table S1) point to increased degradation of carbohydrates relative to Fe-replete conditions. Significant increases in cellular glucose and in the intermediate sugars maltose and trehalose (Fig. S2 and Table S2), coupled with the elevated levels of genes encoding the cytosolic enzymes phosphofructokinase, GAPDH, and phosphoglycerate mutase (Fig. 1 and Table S1) are strong indicators that the conversion of polysaccharides to glucose fuels increased glycolytic activity under Fe limitation (Fig. 1). The altered cell size (Table 1 and Fig. S1) and the observed increase in expression of genes putatively encoding polysaccharide-associated cell surface proteins (e.g., integrins, fasciclins and mucins) (Fig. 1 and Table S1) are also indicative of the reallocation of carbon from structural molecules.
Chlorophyll Biosynthesis and Pigment Metabolism.
Fe limitation induces chlorosis in photoautotrophs (19), including diatoms (20). Although cellular Chl content decreased by more than half in Fe-limited P. tricornutum cells, this difference was much less striking when the parallel reduction in cell volume was taken into account (Table 1). However, as observed by the dramatic alterations in plastid ultrastructure (Fig. S1), the cell size reduction alone is not sufficient to account for the changes in physiology observed under Fe limitation. Although cellular Chl pools decreased under Fe limitation, the expression of 16 genes involved in Chl/heme biosynthesis pathways were enhanced (Table S1). Conversely, genes encoding proteins that contain heme cofactors, such as those involved in mitochondrial cytochrome c biogenesis and b5 electron transport, nitrate reductase, and heme peroxidase, were strongly down-regulated, indicating that Chl- rather than heme-dependent processes were up-regulated.
Glutamate, the precursor for the tetrapyrrole moiety of the Chl and heme biosynthesis pathways, was one of the most elevated metabolites under Fe limitation (≈16-fold) (Table S2). Down-regulation of the gene encoding succinyl-CoA-ligase may account for the accumulation of TCA intermediates such as the glutamate precursor 2-oxoglutarate and thus for glutamate production.
In line with the possibility that high cellular glutamate levels were fueling Chl biosynthesis and the subsequent changes in the chromophore composition of pigment-protein complexes, differences in expression were detected in a variety of other genes involved in pigment metabolism and light harvesting. Genes encoding the enzymes geranylgeranyl reductase, geranylgeranyl pyrophosphate synthase, phytoene dehydrogenase, and phytoene synthase, which are involved in the production of carotenoids, were significantly up-regulated. Geranylgeranyl reductase is also a key regulator of the phytol moiety of Chl and is involved in tocopherol production (see below).
Diatoms contain light-harvesting pigment-binding proteins (LHC) that are phylogenetically related to the green/red lineage chlorophyll a/b-binding protein (CAB) family and diatom-specific fucoxanthin-chlorophyll a/c-binding proteins (Fcp). Two genes that encode the former, one of the latter (FcpD), and a member of the phylogenetically and functionally distinct LI818 family of LHC-like proteins (21) were significantly up-regulated in Fe-limited conditions. Structural modifications to the light-harvesting system and biochemical adjustment of the Fe stoichiometry of photosystem constituents under Fe limitation are further supported by the severe down-regulation of genes encoding high light inducible protein (Hlip 2), zeaxanthin epoxidase, photosystem II subunit M (psbM), a ferredoxin subunit, FcpB, and a non-Fcp-type LHC protein. As expected, the gene for flavodoxin, a small electron-transfer protein capable of replacing ferredoxin during Fe deficiency, was significantly up-regulated (Table S1). The ratio of PSII to PSI complexes and non-photochemical quenching (NPQ) capacity are both elevated under Fe deprivation (Table 1).
The onset of Fe deficiency has been shown to induce remodeling of the photosynthetic apparatus in cyanobacteria and green algae; PSI antennas are modified with specialized accessory Chl-protein complexes that are thought to play a photoprotective role. Fe stress responsive LHC gene products, such as CP43′ and Tidi, which are found in cyanobacteria and green algae (22), respectively, function as peripheral light-harvesting antennas coupled to PSI reaction centers. Large-scale patterns in open ocean variable fluorescence suggest that such specialized pigment-protein complexes are typical in phytoplankton populations found in iron-limited HNLC waters (17). Thus, although levels of Chl a per cell decreased, we propose that de novo pigment synthesis and changes in the antenna polypeptide composition may be required to compensate for the relative decrease of PSI units under periods of Fe limitation (10) and to enhance the NPQ response (Table 1). Although we have not found a direct ortholog of either CP43′ or Tidi in diatoms, one of the four up-regulated Chl binding proteins identified here may fulfill this role in P. tricornutum.
Reprogramming of Nitrogen Metabolism.
Under optimal growth conditions, nitrate is efficiently used by diatoms. However, under Fe limitation genes for nitrate assimilation, such as nitrate reductase (NR), two forms of nitrite reductase, and a plastid-targeted nitrite transporter, were down-regulated, suggesting a reduced capacity for NO3− assimilation (Fig. 1 and Table S1). This may reflect both the high Fe costs associated with NO3− assimilation and the reduction in the supply of organic carbon substrates for amino acid biosynthesis associated with the down-regulation of photosynthesis. To compensate for the reduced capacity to assimilate nitrate, our data suggest that amino acids are obtained by reallocation from protein recycling and from glycolysis. Supporting this view, the amino acids isoleucine, leucine, valine, and alanine, which are derived from the glycolytic end product pyruvate, were all significantly elevated under low Fe conditions, as was the proteolytically-derived hydroxyproline (Fig. S2 and Table S2). The apparent increased flow of carbon through glycolysis may therefore be indicative of a demand for amino acids required for the synthesis of a cellular proteome adapted to iron-limited conditions.
In agreement with proteome remodeling, genes encoding proteases or proteins linked to proteolytic processes (a proteasome subunit, a metalloprotease, a cathepsin/cysteine protease, a ubiquitin ligase, and two PDZ-domain proteases) were strongly induced under Fe limitation. In addition, nine heat shock protein (HSP) genes were preferentially accumulated in response to reduced Fe levels, including an HSP containing a Ca2+-binding EF-hand domain (Table S1) (see below).
Multiple Responses to Oxidative Stress.
Accumulation of reactive oxygen species (ROS) under Fe limitation has been suggested to stem from inefficient activity of an Fe-impoverished photosynthetic electron transport chain, which results in an increased quantity of electrons accepted by O2 (23). Typical ROS defense proteins, such as heme peroxidase, superoxide dismutase (SOD), and aconitase, which is also a key component of the TCA cycle, require iron as a cofactor and are of limited value in iron-limited conditions (24), as revealed by the down-regulation of the genes encoding these proteins under Fe limitation (Fig. 1 and Table S1). Our results suggest alternate biochemical pathways that may compensate for the decrease in conventional, Fe-rich ROS scavenging machinery. Notably, tocopherol, a powerful antioxidant, increased 5-fold under Fe limitation (Table S2), whereas tyrosine, a precursor of tocopherol, was reduced twofold. Tryptophan synthase transcript levels, and shikimate, a metabolite involved in the synthesis of aromatic amino acids, were increased. Fe limitation provoked a 12-fold increase in the glycolytic derivative dehydroascorbate (Table S2), which is also a strong antioxidant, and transcripts encoding a 2-phosphoglycolate phosphatase (GPH), involved in repairing a class of DNA lesions induced by oxidative stress (25), were also elevated (Table S1).
Photorespiration is thought to play a role in excess energy dissipation under stress conditions, such as high light and low temperature (26), and may have a similar function in Fe-limited diatoms. Combined with the strong down-regulation of β-plastid carbonic anhydrase, the large increases observed in the photo respiratory metabolic intermediates serine, glycine, and glycerate (Fig. S2 and Table S2) may suggest an increased role for photo respiration under Fe limitation.
Respiratory electron transfer chain activity, one of the most Fe-demanding cellular processes, is restricted during Fe limitation (27). In P. tricornutum, genes encoding mitochondria-targeted proteins involved in cytochrome-based respiration were significantly down-regulated. A cytochrome pathway restriction would promote ROS accumulation in mitochondria, and our results indicate that Fe-limited diatoms may employ at least three mechanisms effective in reducing mitochondrial ROS production and facilitating respiratory processes despite impairment of cytochrome-based respiration (Fig. 1).
Mitochondrial alternative oxidase (AOX) activity and mRNA levels were significantly higher under Fe starvation (Table 1 and Table S1). AOX can lower mitochondrial reactive oxygen production in plant cells and may have a regulatory role in balancing carbon metabolism, including glycolysis and TCA cycle activity, when the cytochrome pathway is compromised (28).
Second, the exchange of reducing equivalents between the mitochondria and cytosol may be achieved via an alternative NADH dehydrogenase (NDH)-based shuttle system. NDH, a critical site for superoxide production, is thought to be particularly important during times of intensified glycine and serine metabolism. Under Fe limitation, the glycolytic derivative myo-inositol and mRNA levels of a gene encoding mitochondrial myo-inositol dehydrogenase were both elevated 6-fold (Table S1 and Table S2). The distribution of myo-inositol dehydrogenase among algal classes and the fact that red algal mitochondria are able to use myo-inositol for respiration suggest that red algae and chromalveolates have the ability to use an inositol/inosase shuttle system for reducing equivalents (29) in addition to the NDH system common in the green lineage. The importance of mitigating ROS production and the apparent intensified flux of the photorespiratory metabolites glycine and serine under Fe stress could lead to the utilization of AOX and myo-inositol dehydrogenase as respiratory alternatives.
Finally, photosynthetic electrons can be directed into the mitochondrial electron transport chain through the malate:aspartate shuttle, where AOX functions as a sink for excess reducing equivalents (30). Increased levels of malate and AOX activity may indicate that this shunt is used to transfer excess electrons from the chloroplast toward AOX, further reducing ROS-mediated insult and at the same time generating ATP (Fig. 1).
Intracellular Signaling.
The importance of calcium (Ca2+)-based second messenger signaling pathways in P. tricornutum has been shown for nitric oxide (NO)-mediated death and defense responses and for cellular perception of bioavailable Fe concentrations (31, 32). Diatom AOX proteins contain calcium-binding EF-hand domains. This suggests that AOX activation in diatoms is partially controlled by calcium-based signaling mechanisms. One protein possibly involved in Ca2+-activated ROS defense is the oxidative stress responsive protein annexin (33). mRNA levels for annexin were significantly up-regulated in P. tricornutum under Fe limitation (Fig. 1 and Table S1). The importance of Ca2+ signaling under Fe limitation was further reinforced by the up-regulation of Ca2+ responsive genes encoding an HSP and proteins, such as myosin, gelsolin, copine, and scramblase (Fig. 1 and Table S1), involved in cytoskeleton and cell membrane regulation and modification.
Metal Assimilation and Homeostasis.
Fe-limited P. tricornutum cells showed a 20-fold increase in levels of the strong iron chelating glucose oxidation derivative gluconate (Table S2). Internal chelators are important for the intracellular control of Fe, permitting efficient Fe utilization particularly under Fe limitation, and for alleviating oxidative DNA lesions, which may occur through leaching of Fe from storage proteins and enzymatic clusters ([4Fe–4S]) (34). In marine Synechococcus sp., IscA has recently emerged as a major component in regulating cellular Fe homeostasis and delivery of iron from organic ligands and chelators to iron–sulfur assembly cluster proteins under conditions of depleted cellular iron pools (35). The observed increase in IscA mRNA in response to Fe limitation (Table S1) suggests a similar role in P. tricornutum. Although very little is known about the intracellular handling of iron in eukaryotes, IscA and gluconate may be important components for controlling the intracellular availability of Fe during periods of limitation. Ferritin is present in the P. tricornutum genome sequence but absent from the T. pseudonana genome. This prompts speculation that fundamental differences in Fe storage capacity and turnover exist between these two diatom species and may partly explain the ability of P. tricornutum to grow at lower Fe concentrations than T. pseudonana.
Physiological and molecular evidence from T. pseudonana and P. tricornutum support an important role for high affinity cell surface ferric-chelate reductases in diatom iron uptake systems (8). P. tricornutum and T. pseudonana each contain two such transmembrane oxidoreductases (FRE1 and FRE2) with FAD and NADPH-ribose binding signatures and requisite histidine residues for heme coordination (12). In P. tricornutum, three additional Fe-responsive genes encoding cell surface transmembrane proteins may be involved in iron uptake. Two DcytB-type ferric reductases belonging to the cytochrome b561 family, FRE3 and FRE4, contain five to six transmembrane domains each. Furthermore, an opisthokont-type decycling heme oxygenase (Table S3) containing two TM domains, if localized to the cell surface, may help destabilize the Fe bound in porphyrins, thus corroborating the report in ref. 36 of heme-based Fe acquisition systems in diatoms. Four gene transcripts encoding proteins for various metal transporters were enriched under Fe limitation (Table S1). These included one member of the ZRT, IRT-like protein family (ZIP), one heavy metal ATPase (HMA), and two, putatively organelle targeted, cation diffusion facilitators (CDF). We also observed down-regulation of a gene that encodes the bacterial chromate efflux protein ChrA (Fig. 1 and Table S1).
Iron-Regulated Gene Clusters.
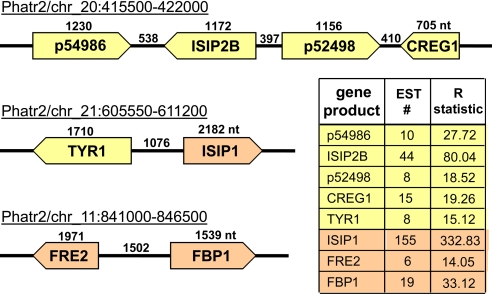
The P. tricornutum genome contains at least three clusters of Fe-responsive genes (Fig. 2). Genes within these clusters were among the most highly expressed under Fe limitation and, with the exception of ferric reductase (FRE2), are absent in T. pseudonana. FRE2 is immediately adjacent to a highly Fe-responsive gene encoding a putative ferrichrome-binding protein (FBP1), similar to FhuD found in prokaryotic ferrichrome ABC transport systems. In bacteria, FhuD enables the direct transport of Fe(III)-hydroxymate chelates across the plasma membrane. The identification of a diatom-type FBP1 is consistent with kinetic measurements of siderophore uptake in P. tricornutum (37) and supports the presence of a coupled FBP1/FRE-dependent siderophore-based system for the destabilization and subsequent reduction of organically complexed Fe(III)-hydoxymates at the cell surface. Although standard assays (38) failed to detect the production of siderophores in P. tricornutum (data not shown), we discovered the genetic basis for Fe-uptake mechanisms that are a combination of classical eukaryotic components and those commonly found in bacteria but not usually present in eukaryotes.
Fig. 2.
Iron-regulated gene clusters in P. tricornutum. With the exception of FRE2, the iron-regulated cluster genes are absent from the T. pseudonana genome. Putative components of the Fe uptake system are shown in brown. The number of ESTs retrieved from the Fe limited EST library and the R statistic are provided. R > 12 indicates a statistically significant enrichment of ESTs in the Fe limited library relative to 11 other Fe-replete EST libraries (14). qRT-PCR data are given in Table S1.
A second Fe-regulated cluster contained genes encoding a copper(Cu)-tyrosinase (TYR1) and a protein of unknown function, ISIP1 (Fig. 2 and Table S1). The role of Cu-nutrition in Fe-limited phytoplankton has become increasingly apparent (39, 40). Cu-tyrosinase represents an undescribed Fe-sensitive Cu-protein. The ISIP1 encoding gene was the most highly expressed gene on the microarray of Fe-limited cRNA target and was the most commonly sequenced transcript in the Fe-limited EST library. An orthologous Fe-responsive gene in Dunaliella salina encodes a protein thought to associate with plasma membrane transferrins where it functions, along with a multicopper ferroxidase, as part of a complex that enhances binding and uptake of ferric ions (41). However, there is no evidence for the occurrence of genes that encode transferrin in the genome of P. tricornutum.
A third cluster of Fe-regulated genes contains five genes of unknown function with the exception of a gene encoding an ortholog of the cellular repressor of E1A-stimulated genes (CREG). CREG is a secreted glycoprotein that acts as a regulatory ligand for signaling kinases and reduces cell proliferation. In Fe-deficient P. tricornutum cells, CREG could be central in confining cell growth to match resource availability. Each of the nine genes found within the Fe-regulated clusters are predicted to encode signal peptides (Fig. 2 and Table S1), suggesting a role for the gene products in cell surface processes.
Comparative Genomics of the Fe-Responsive P. tricornutum Gene Repertoire.
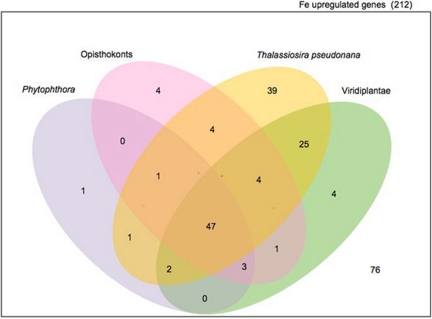
Out of 120 P. tricornutum Fe-responsive up-regulated genes that have an ortholog in T. pseudonana, 39 (nearly 20% of genes identified) appear to be diatom specific (Fig. 3). Whole-genome expression profiling in T. pseudonana in response to various nutrient stresses identified a nearly identical percentage of differentially expressed diatom-specific genes (42). Fifteen of the diatom-specific Fe-sensitive genes reported here have identifiable domain signatures (Table S3). The diatom-specific gene products containing Fe–S binding sites and ferredoxin-dependent bilin reductase domains (Table S3) are likely involved directly in Fe metabolism.
Fig. 3.
Venn diagram of the distribution of proteins upregulated by iron limitation in P. tricornutum. Distribution of homologs (E ≤ 10−5, ≥50% coverage, ≥30% percent identity) to proteins in other lineages and the centric diatom T. pseudonana is depicted.
Although diatom genes have at least as many orthologs in opisthokonts as in Viridiplantae and red algae (43), the Fe-responsive gene repertoire in P. tricornutum does not fit this trend. The higher number of orthologs in Viridiplantae for the Fe-responsive subset reflects the high proportion of genes involved in light harvesting and pigment metabolism (Fig. 3 and Table S3). Many of these, such as flavodoxin, are only found in other algae and appear to have been lost in vascular plants (Table S3).
The complement of annotated Fe-sensitive genes (up- or down-regulated) in P. tricornutum contains 38 genes that are absent in the genome of T. pseudonana (Table S1). Five of the 38 genes encode proteins of unknown function, whereas the remaining genes have orthologs in fungi, bacteria, and chlorophytes. Among these, plastid targeted β-carbonic anhydrase (CA) and fructose bisphosphate aldolase I (FBA I) are likely important for controlling down-regulation of photosynthetic activity and mediating overall partitioning of electron flow. Many genes involved in signaling, transcription, and cytoskeleton modification do not have orthologs in T. pseudonana (Table S1 and Table S3). The putative ferrichrome binding protein (FBP1) and ISIP1 are also absent from T. pseudonana, suggesting important differences in the Fe-acquisition systems of these two diatoms.
Conclusions
P. tricornutum has evolved novel strategies for acquiring different forms of iron, as demonstrated by the presence of Fe-regulated gene clusters containing genes such as FBP1. However, the sole presence of specialized uptake systems is not sufficient to ensure survival, as demonstrated by the need for reducing cellular Fe requirements through metal or protein substitution and for overall physiological adjustment centered on down-regulation of photosynthetic activity. Compensation is evidenced by the down-regulation of Fe-requiring proteins and pathways, such as respiration and ROS defense, and the simultaneous up-regulation of Fe-free ROS defense alternatives and substrate shuttles for channeling excess electrons from the chloroplast and cytosol to the AOX in the mitochondria.
Perhaps the most important physiological adjustment of P. tricornutum to Fe limitation is the ability to cope with sustained illumination despite the dramatic down-regulation of photosynthetic activity. In contrast to T. pseudonana, P. tricornutum is able to survive Fe starvation at levels of Fe availability similar to those tolerated by oceanic centric and common HNLC pennate diatoms (12, 13). A range of Fe-sensitive gene products and metabolites involved in light harvesting complex modification and various alternative electron cycling pathways appear to arbitrate an Fe limitation induced equilibrium between metabolic retrenchment and electron and ROS dissipation. In contrast to the oceanic diatom T. oceanica, which has constitutively low levels of Fe-rich photosystem I complexes and substitutes plastocyanin for cytochrome C6 in Fe limiting conditions (10, 39), P. tricornutum appears to use metabolic reconfigurations to acclimate to low Fe levels. In conclusion, the cellular strategies identified in P. tricornutum provide a foundation for understanding the constraints on diatom metabolism in iron-limited regions of today's oceans and provide a basis for understanding how diatom growth will be affected by nutrient fertilization strategies to enhance CO2 sequestration from the atmosphere.
Methods
Growth Conditions and Physiological Parameters.
Semicontinuous batch cultures of Phaeodactylum tricornutum (CCMP2561) were grown in f/2 ASW medium modified to contain either 13.4 pmol·liters−1 Fe′ (Fe-limited) or 2.6 nmol·liters−1 Fe′. Fe′ is the sum of all unchelated Fe species. The degree of iron limitation in steady-state cultures was assessed from various physiological parameters and cellular indicators (Table 1 and Fig. S1) and by following the recovery of high growth rates upon Fe addition to a small aliquot of the culture at the time of harvest.
Metabolite Extraction and GC-MS Analysis.
Approximately 1 × 109 Fe-limited and Fe-replete cells were pelleted and used for metabolite analysis. Metabolites were extracted and derivatization and GC-MS analysis were carried out as described in ref. 44. Metabolites were identified compared with database entries of authentic MSRI libraries (45).
Gene Expression.
Genes responsive to Fe limitation were evaluated through a combination of statistical analysis of EST libraries, a partial genome microarray, and qRT-PCR. In the case of EST library analysis, a log-likelihood ratio test statistic, designed specifically for the purpose of statistically evaluating gene expression level across multiple cDNA libraries, was computed (14). The array platform with all of the probe sequences and corresponding genome browser protein IDs and all of the normalized hybridization data are available in Minimum Information About a Microarray Experiment (MIAME) compliant format at the National Center for Biotechnology Information Gene Expression Omnibus (GEO).
Annotation and Comparative Analysis.
Two hundred twenty-eight Fe-responsive EST contigs were mapped to 212 predicted proteins on the P. tricornutum genome browser. The 212 predicted proteins corresponding to the up-regulated transcripts were used for the further comparative analysis against different taxonomical lineages (see SI Methods).
Supplementary Material
Acknowledgments.
We thank Mathangi Thiagarajan for assistance with statistical analysis of microarray data and Guangzuo Luo for microarray development. This work was supported by National Science Foundation postdoctoral fellowship in microbial biology DBI-0301626 and Biological Oceanography grant OCE-0727997 (to A.E.A.), Deutches Forschungs Gemeinschaft Grant RO-2138/7-1 (awarded to J.L.R. and supporting M.L.), European Union-funded FP6 Diatomics Project Grant LSHG-CT-2004-512035, EU-FP6 Marine Genomics Network of Excellence Grant GOCE-CT-2004-505403, an ATIP “Blanche” grant from Centre National de la Recherche Scientifique, and the Agence Nationale de la Recherche (France). Diatom genome sequencing at the Joint Genome Institute (Walnut Creek, CA) was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under Contract DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344, and Los Alamos National Laboratory under Contract DE-AC02-06NA25396. P. tricornutum ESTs were generated at Genoscope (Evry, Paris).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8675).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711370105/DCSupplemental.
References
- 1.Anbar AD, Knoll AH. Proterozoic ocean chemistry and evolution: A bioinorganic bridge? Science. 2002;297:1137–1142. doi: 10.1126/science.1069651. [DOI] [PubMed] [Google Scholar]
- 2.Quigg A, et al. The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature. 2003;425:291–294. doi: 10.1038/nature01953. [DOI] [PubMed] [Google Scholar]
- 3.Archer DE, Johnson K. A model of the iron cycle in the ocean. Global Biogeochem Cy. 2000;14:269–279. [Google Scholar]
- 4.Behrenfeld MJ, Bale AJ, Kolber ZS, Aiken J, Falkowski PG. Confirmation of iron limitation of phytoplankton photosynthesis in the equatorial Pacific Ocean. Nature. 1996;383:508–511. [Google Scholar]
- 5.de Baar HJW, et al. Synthesis of iron fertilization experiments: From the iron age in the age of enlightenment. J Geophys Res Oceans. 2005;110:C09S16. [Google Scholar]
- 6.Moore C, Hickman A, Poulton A, Seeyave S, Lucas M. Iron-light interactions during the CROZet natural iron bloom and EXport experiment (CROZEX): II - Taxonimc responses and elemental stoichiometry. Deep-Sea Res Pt II. 2007;54:2066–2084. [Google Scholar]
- 7.Boyd PW, et al. Mesoscale iron enrichment experiments 1993–2005: Synthesis and future directions. Science. 2007;315:612–617. doi: 10.1126/science.1131669. [DOI] [PubMed] [Google Scholar]
- 8.Brand LE, Sunda WG, Guillard RL. Limitation of marine primary production rates by zinc, manganese, and iron. Limnol Oceanogr. 1983;28:1182–1198. [Google Scholar]
- 9.McKay RML, Geider RJ, LaRoche J. Physiological and biochemical response of the photosynthetic apparatus of two marine diatoms to Fe stress. Plant Physiol. 1997;114:615–622. doi: 10.1104/pp.114.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strzepek RF, Harrison PJ. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature. 2004;431:689–692. doi: 10.1038/nature02954. [DOI] [PubMed] [Google Scholar]
- 11.Sunda WG, Huntsman SA. Iron uptake and growth limitation in oceanic and coastal phytoplankton. Mar Chem. 1995;50:189–206. [Google Scholar]
- 12.Kustka A, Allen AE, Morel FMM. Sequence analysis and transcriptional regulation of Fe acquisition genes in two marine diatoms. J Phycol. 2007;43:715–729. [Google Scholar]
- 13.Marchetti A, Maldonado MT, Lane ES, Harrison PJ. Iron requirements of the pennate diatom Pseudo-nitzschia: Comparison of oceanic (high-nitrate, low-chlorophyll waters) and coastal species. Limnol Oceanogr. 2006;51:2092–2101. [Google Scholar]
- 14.Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–2061. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene RM, Geider RJ, Kolber Z, Falkowski PG. Iron-Induced changes in light harvesting and photochemical energy conversion processes in eukaryotic marine algae. Plant Physiol. 1992;100:565–575. doi: 10.1104/pp.100.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geider RJ, LaRoche J, Greene RM, Olaizola M. Response of the photosynthetic apparatus of. Phaeodactylum tricornutum (BACILLARIOPHYCEAE) to nitrate, phosphate, or iron starvation. J Phycol. 1993;29:755–766. [Google Scholar]
- 17.Behrenfeld MJ, et al. Controls on tropical Pacific Ocean productivity revealed through nutrient stress diagnostics. Nature. 2006;442:1025–1028. doi: 10.1038/nature05083. [DOI] [PubMed] [Google Scholar]
- 18.Milligan AJ, Harrison PJ. Effects of non-steady-state iron limitation on nitrogen assimilatory enzymes in the marine diatom Thalassiosira weissflogii (Bacillariophyceae) J Phycol. 2000;36:78–86. [Google Scholar]
- 19.Moseley JL, et al. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 2002;21:6709–6720. doi: 10.1093/emboj/cdf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene RM, Geider RJ, Falkowski PG. Effect of iron limitation on photosynthesis in a marine diatom. Limnol Oceanogr. 1991;36:1772–1782. [Google Scholar]
- 21.Koziol AG, et al. Tracing the evolution of the light harvesting antennae in chorophyll. a/b-containing organisms. Plant Physiol. 2007;143:1802–1816. doi: 10.1104/pp.106.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varsano T, Wolf S, Pick U. A chlorophyll. a/b-binding protein homolog that is induced by iron deficiency is associated with enlarged photosystem I units in the eukaryotic alga Dunaliella salina. J Biol Chem. 2006;281:10305–10315. doi: 10.1074/jbc.M511057200. [DOI] [PubMed] [Google Scholar]
- 23.Niyogi KK. Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 24.Baxter CJ, et al. The metabolic eesponse of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol. 2007;143:312–325. doi: 10.1104/pp.106.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellicer MT, Nunez MF, Aguilar J, Badia J, Baldoma L. Role of 2-phosphoglycolate phosphatase of Escherichia coli in metabolism of the 2-phosphoglycolate formed in DNA repair. J Bacteriol. 2003;185:5815–5821. doi: 10.1128/JB.185.19.5815-5821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker MS, Armbrust EV. Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae) J Phycol. 2005;41:1142–1153. [Google Scholar]
- 27.Kudo I, Miyamoto M, Noiri Y, Maita Y. Combined effects of temperature and iron on the growth and physiology of the marine diatom, Phaeodactylum tricornutum (Bacillariophyceae) J Phycol. 2000;36:1096–1102. [Google Scholar]
- 28.Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Gross W, Meyer A. Distribution of myo-inositol dehydrogenase in algae. Eur J Phycol. 2003;38:191–194. [Google Scholar]
- 30.Yoshida K, Terashima I, Noguchi K. Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 2007;48:606–614. doi: 10.1093/pcp/pcm033. [DOI] [PubMed] [Google Scholar]
- 31.Falciatore A, d'Alcala M. R., Croot P, Bowler C. Perception of environmental signal by a marine diatom. Science. 2000;288:2363–2366. doi: 10.1126/science.288.5475.2363. [DOI] [PubMed] [Google Scholar]
- 32.Vardi A, et al. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 2006;4:411–419. doi: 10.1371/journal.pbio.0040060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee NJ, Kim GY, Huh JW, Kim SW, Na DS. Annexin I is a stress protein induced by heat, oxidative stress and a sulfhydryl-reactive agent. Eur J Biochem. 2000;267:3220–3225. doi: 10.1046/j.1432-1327.2000.01345.x. [DOI] [PubMed] [Google Scholar]
- 34.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balasubramanian R, Shen G, Bryant DA, Golbeck JH. Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 2006;188:3182–3191. doi: 10.1128/JB.188.9.3182-3191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchins DA, Witter AE, Butler A, Luther GW. Competition among marine phytoplankton for different chelated iron species. Nature. 1999;400:858–861. [Google Scholar]
- 37.Soria-Dengg S, Horstmann U. Ferrioxamines B and E as iron sources for the marine diatom Phaeodactylum tricornutum. Mar Ecol Prog Ser. 1995;127:269–277. [Google Scholar]
- 38.Schwyn B, Neilands J. UNiversal chemical assay for the detection and determination of siderophores. Analyt Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 39.Peers G, Price NM. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature. 2006;441:341–344. doi: 10.1038/nature04630. [DOI] [PubMed] [Google Scholar]
- 40.Maldonado MT, et al. Copper-dependent iron transport in coastal and oceanic diatoms. Limnol Oceanogr. 2006;51:1729–1745. [Google Scholar]
- 41.Paz Y, Katz A, Pick U. A multicopper ferroxidase involved in iron binding to transferrins in Dunaliella salina plasma membranes. J Biol Chem. 2007;282:8658–8666. doi: 10.1074/jbc.M609756200. [DOI] [PubMed] [Google Scholar]
- 42.Mock T, et al. Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc Natl Acad Sci USA. 2008;105:1579–1584. doi: 10.1073/pnas.0707946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 44.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 45.Schauer N, et al. GC-MS librraies for the rapid indentification of metabolites in complex biological samples. FEBS Lett. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 46.Maheswari U, et al. The diatom EST database. Nucleic Acids Res. 2005;33:344–347. doi: 10.1093/nar/gki121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.