Abstract
The ESCRT machinery functions in topologically equivalent membrane fission events, namely multivesicular body formation, the terminal stages of cytokinesis and HIV-1 release. Here, we show that the ESCRT-III-binding protein Alix is recruited to the midbody of dividing cells through binding Cep55 via an evolutionarily conserved peptide. Disruption of Cep55/Alix/ESCRT-III interactions causes formation of aberrant midbodies and cytokinetic failure, demonstrating an essential role for these proteins in midbody morphology and cell division. We also show that the C terminus of Alix encodes a multimerization activity that is essential for its function in Alix-dependent HIV-1 release and for interaction with Tsg101. Last, we demonstrate that overexpression of Chmp4b and Chmp4c differentially inhibits HIV-1 release and cytokinesis, suggesting possible reasons for gene expansion within the mammalian Class E VPS pathway.
Keywords: late domain, viral budding, Class E VPS, L-Domain, cell division
Completion of cell division involves the postmitotic separation of daughter cells through the process of cytokinesis and requires abscission of the midbody, a microtubule-rich stalk of membrane that connects daughter cells and forms at a late stage of cell division (1–3). Importantly, abscission must be coordinated by an activity inside the midbody, suggesting that a topologically unique mechanism of membrane fission is used. Recently, Centrosome Associated Protein, 55 kDa (Cep55), a mitotic phosphoprotein that relocalizes from the centrosome to the midbody during late mitosis, has been shown to play an essential role in abscission (4–6). It has also been demonstrated that tumor-susceptibility gene 101 (Tsg101) and Alg2-interacting protein X (Alix), both components of the endosomal sorting complex required for transport (ESCRT) machinery, interact with Cep55, localize to the midbody and are themselves required for abscission (7, 8). These data suggest that the cytokinetic activity of Cep55 may be elicited through recruitment of the Class E VPS pathway.
The ESCRT machinery regulates other membrane scission steps that bear a topological equivalence to midbody abscission, namely biogenesis of intraendosomal vesicles within multivesicular bodies (MVBs) (9) and enveloped virus release (10). During MVB formation, the ESCRT-machinery is recruited to endosomes through the interaction of Tsg101 with ubiquitinated cargo and hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and is thought to generate and sort cargo onto these intraendosomal vesicles. MVB formation is important for the termination of receptor signaling and is a prerequisite for receptor degradation (9, 11). The ESCRT machinery is also recruited by late domains (L domains) within retroviral Gag proteins and provides an activity that allows viral-particle release from the cell surface; HIV-1 Gag encodes two L domains that recruit the ESCRT machinery, a Tsg101-binding PTAP motif (12–15) and an Alix-binding LYPX(n)L motif (16, 17). HIV-1 release largely depends on its PTAP L domain (18, 19), although a virus rendered unable to interact with Tsg101 through mutation of this L domain is released entirely through its auxiliary Alix-dependent L domain (20, 21).
ESCRT-III appears to be the membrane fission machinery recruited by ESCRT-I and Alix (11). Overexpression of ESCRT-III subunits arrests viral budding, recapitulating the phenotype of an L domain-defective virus (17, 22–24), and mutations in the Bro1 domain of Alix that abrogate the Alix/ESCRT-III interaction also abolish the ability of Alix to mediate budding (20, 21) and cell division (8). A requirement for ESCRT-III in cell division has been suggested by the arrest of cytokinesis in cells overexpressing YFP-Chmp4 fusion proteins or a catalytically inactive Vps4 (7, 8). Overall, the anatomy of the Class E VPS pathway is conserved from yeasts to humans, and ESCRT-III is one of the most conserved components in the pathway (9). Intriguingly, the mammalian ESCRT machinery is significantly expanded compared with yeast, with three mammalian isoforms of Vps32 and four of Vps37, suggesting that gene expansion in the mammalian pathway may allow for functional diversification of this machinery. Alix binds numerous trafficking proteins via its proline-rich region (PRR), including Cin85/SETA (25), CD2AP [an adaptor protein involved in MVB biogenesis (26), and cytokinesis (27)], the family of endophilins (28) and the Cep55-binding and ESCRT-I component Tsg101 (7, 8, 29). Additionally, recent studies have identified an activity in Alix's PRR that is essential for Alix-dependent HIV-1 release (20, 21). The nature of this activity remains unclear, but current speculation is that it represents an unknown interaction partner of Alix.
Here, we demonstrate an essential role for Cep55/Alix/ESCRT-III interactions in midbody formation and completion of cytokinesis, define the nature of the Alix-dependent HIV-1 release activity within Alix's PRR, and provide evidence suggesting that gene duplication of ESCRT-III in mammals reflects an adaptation to specialized functions such as midbody abscission.
Results
Cep55 Binds to an Evolutionarily Conserved Peptide in Alix.
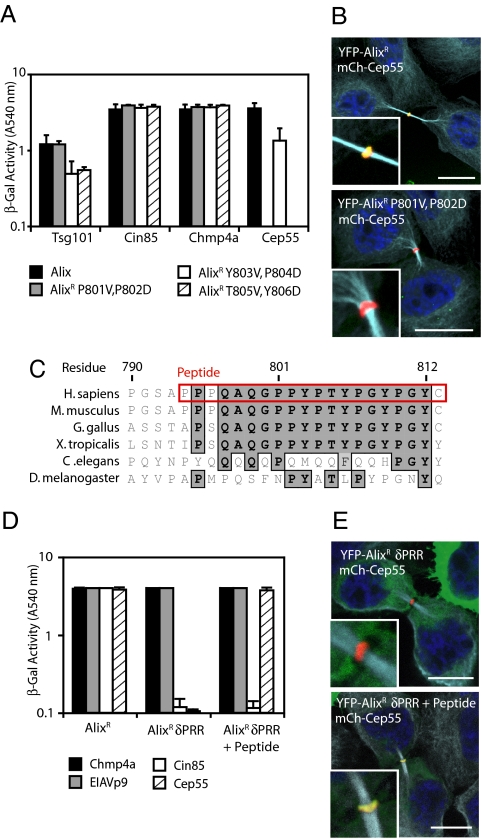
A conserved role for the ESCRT machinery in cell division is suggested by the accumulation of multinucleate trichomes in Arabidopsis thaliana elch/tsg101 mutants (30), indicating that the ESCRT machinery may function in cell division in other organisms. To determine whether the interaction of the ESCRT machinery with Cep55 is conserved in other species, we mapped Cep55's binding site on Alix by using a combination of yeast two-hybrid and pull-down assays. In agreement with Morita et al. (8) Cep55 binds to a repeating PxY motif from residues 801–806 within Alix's PRR [supporting information (SI) Fig. S1]. Mutational analysis of this motif demonstrated that two pairs of amino acids—residues P801, P802 and T805, Y806–were essential for Cep55 binding, suggesting that Alix makes multiple contacts with Cep55 (Fig. 1A). Importantly, mutation of P801V and P802D abrogated Alix's ability to bind Cep55 yet preserved its interactions with known binding partners (Fig. 1A and Fig. S2). This mutation prevented recruitment of Alix to the midbody (Fig. 1B). Sequence analysis of the PRR identified a cluster of evolutionarily conserved amino acids around the Cep55-binding site (Fig. 1C). Indeed, deletion of the PRR abolished the ability of Alix to bind Cep55, and the grafting of a peptide encoding this conserved region onto Alix-δPRR (Alix-δPRR + Peptide) was sufficient to restore both Cep55-binding and midbody localization (Fig. 1 D and E). Cytokinesis was defective in both Alix-depleted cells and in Alix-depleted cells reliant on stably expressed mCh-AlixR δPRR (Fig. 2A and Fig. S3). However, restoration of the ability of Alix δPRR to bind Cep55 (mCh-AlixR δPRR + Peptide) allowed substantial, although incomplete, rescue of these defects (Fig. 2A and Fig. S3). These results suggest that a major role of the Alix PRR in cytokinesis is to bind Cep55 and that, although this interaction is conserved in other vertebrates, it is not present in organisms such as Saccharomyces cerevisiae, Caenorhabditis elegans, or Drosophila melanogaster that lack the Cep55-binding site within Alix's PRR.
Fig. 1.
Cep55 recruits Alix to the midbody through binding a conserved peptide within Alix's PRR. (A) Alix mutants were fused to the VP16-activation domain and tested for interaction with the indicated proteins fused to Gal4 DNA-binding domain by yeast two-hybrid assay (n = 3 ± SD). (B) HeLa cells were transfected with plasmids encoding mCh-Cep55 and either YFP-AlixR or YFP-AlixR P801V, P802D, fixed and stained with α-tubulin. (Scale bars: 10 μm.) (C) Sequence alignment of the Cep55-binding region of the Alix-PRR with the Cep55-binding peptide highlighted in red. (D) Alix, AlixR δPRR, and AlixR δPRR plus peptide were fused to the VP16 DNA-activation domain and tested for interactions with the indicated Gal4 DNA-binding domain fusions by yeast two-hybrid assay (n = 3 ± SD). (E) HeLa cells were transfected with plasmids encoding mCh-Cep55 and either YFP-AlixR or YFP-AlixR δPRR and AlixR δPRR plus peptide, fixed, and stained with α-tubulin. (Scale bars: 10 μm.)
Fig. 2.
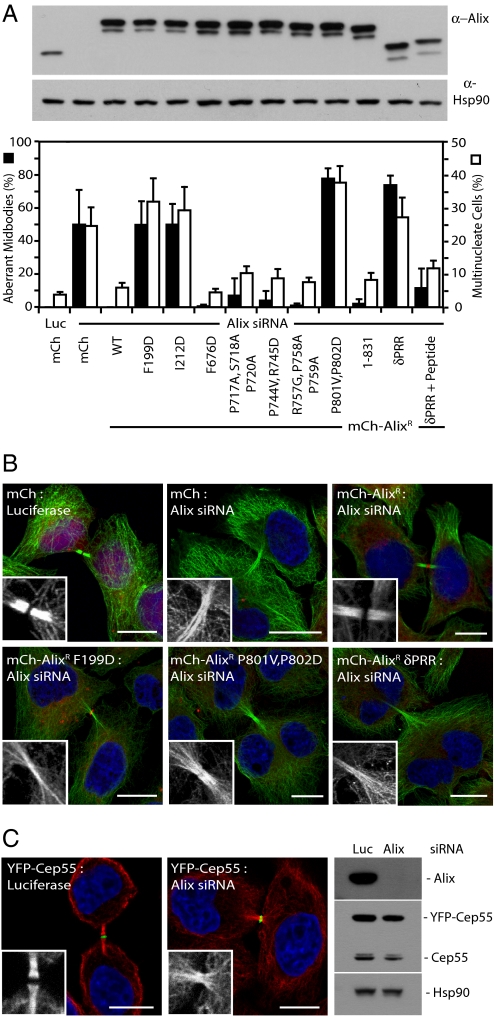
Recruitment of Alix and ESCRT-III to the midbody is essential for normal midbody morphology. (A) HeLa cells stably transduced with retroviral vectors encoding mCh or mCh-AlixR fusions were treated with siRNA targeting Luciferase or Alix, and resolved cell lysates were analyzed with α-Alix or α-Hsp90 antibodies. Alternatively, cells were fixed, stained with α-tubulin, and scored for aberrant midbodies (n = 3 ±SD) and multinucleation (n > 4 ±SD). (B) Representative micrographs of aberrant midbodies from A, enlargement depicting α-tubulin channel. (Scale bars: 10 μm.) (C) Clonal HeLa cells stably expressing YFP-Cep55 were treated with siRNA targeting Luciferase or Alix. Cells were fixed and stained with α-tubulin. Enlargements depict the α-tubulin channel. (Scale bars: 10 μm.) Alternatively, cells were lysed and analyzed with α-Alix, α-Cep55, and α-Hsp90 antisera.
We also tested other functional requirements for Alix in cytokinesis using cell lines stably expressing mCh-tagged versions of AlixR rendered unable to bind certain partners (Fig. S2). Interestingly, mCh-AlixR 1–831 was functional in cytokinesis, showing that, in contrast to a role in Alix-dependent HIV-1 release (20, 21), the extreme C terminus of Alix is not required for cell division (Fig. 2A and Fig. S3). In agreement with Morita et al. (8), we found that Alix constructs that could not be recruited by Cep55 (P801V, P802D) or could not bind the downstream ESCRT-III components Chmp4a, Chmp4b, or Chmp4c (F199D or I212D) were unable to rescue the cytokinesis defects induced by Alix depletion (Fig. S3). Importantly, mCh-AlixR F199D and I212D were still recruited to the midbody (Fig. 2B and Fig. S3C), demonstrating that the cytokinesis defects occur as a direct consequence of a failure to recruit ESCRT-III to the site of abscission.
Recruitment of ESCRT-III by Alix Determines Midbody Morphology.
Tsg101 and Alix are recruited to the Flemming body through the interaction with Cep55; however, it remains unclear whether ESCRT-III is a structural component of the midbody. The Flemming body is a central region of the midbody characterized by a gap in α-tubulin staining, but it is clear from electron micrography studies that interdigitating microtubules persist through this gap (31, 32), indicating that protein density at the Flemming body masks tubulin epitopes known to be present. In Cep55-depleted cells, microtubules at the Flemming body are visible (6, 7), suggesting that Cep55 regulates recruitment of this density. We noted in Alix-depleted cells an accumulation of aberrant midbodies, with α-tubulin staining persisting through the Flemming body (Fig. 2 A and B). Importantly, structural elements of the Flemming body were still present in Alix-depleted cells because YFP-Cep55 was still recruited to the Flemming body (Fig. 2C). Defects in Flemming body formation observed in Alix-depleted cells were reversed through stable expression of mCh-AlixR (Fig. 2 A and B). However, stable expression of mCh-AlixR F199D or mCh-AlixR I212D in Alix-depleted cells could not rescue formation of normal midbodies. Alix mutants that could not be recruited by Cep55 (mCh-AlixR P801V, P802D) or that lack the C-terminal PRR (mCh-AlixR δPRR) also failed to rescue midbody morphology (Fig. 2 A and B). Restoration of the ability of AlixR δPRR to bind Cep55 (mCh-AlixR δPRR + Peptide) rescued midbody formation, and expression of AlixR mutants defective for binding to CD2AP, Cin85, Tsg101, and endophilins or lacking the terminal 37 residues also allowed formation of normal midbodies (Fig. 2 A and B). These data recapitulate exactly the effects of siRNA against Cep55 (6, 7), show that Cep55-dependent recruitment of the ESCRT-III subunits Chmp4a, Chmp4b and Chmp4c via Alix is required for the formation of morphologically normal midbodies, and suggest that ESCRT-III is a major component of the Flemming body.
Differential Requirement for ESCRT-III Subunits in Cytokinesis and HIV-1 Budding.
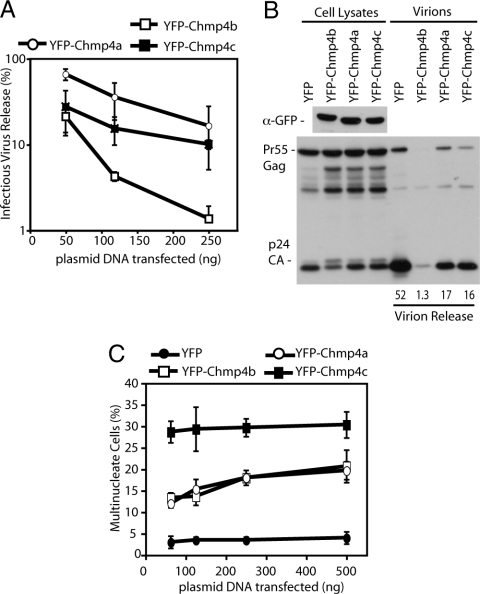
Given the lack of evidence supporting a role for the ESCRT machinery in yeast cell division, we wished to examine whether gene duplication within the ESCRT machinery reflected an adaptation to mammalian-specific functionalities. We examined differential requirements for mammalian Chmp4 isoforms in cytokinesis and HIV-1 release by overexpressing YFP-Chmp4 fusions that have been shown to function as dominant-negative proteins (22). These experiments revealed that, although all Chmp4 isoforms inhibited both HIV-1 release and cytokinesis to some degree, YFP-Chmp4b was the most potent inhibitor of viral budding, producing a large decrease in released infectious virions and a characteristic p24/p25 processing defect in cellular Gag (Fig. 3 A and B), whereas Chmp4c was a much more potent inhibitor of cytokinesis than Chmp4a or Chmp4b (Fig. 3C). The differential inhibition of HIV-1 budding and cytokinesis by heterologous Chmp4 fusions suggests that the Chmp4 isoforms might be differentially adapted to the multiple functions of the ESCRT machinery in mammals.
Fig. 3.
Differential requirements for Chmp4 isoforms in retroviral budding and cytokinesis. (A and B) Cells (293T) were cotransfected with plasmids encoding HIV-1 pNL/HXB and the indicated amount of YFP-Chmp4 constructs. Resolved cell lysates and released virions were examined by Western blot analysis with α-Gag antisera; lysates were also examined by Western blot analysis with α-GFP antisera. Quantification of virion release via infrared imaging is given. β-Galactosidase assay was performed on HeLa-TZM-bl cells infected with 293T supernatant (n = 3 ±SD). (C) HeLa cells were transfected with the indicated amounts of YFP-Chmp4 constructs. Cells were fixed, stained with α-tubulin, and scored for multinucleation (n = 3 ±SD).
Alix Multimerization Is Required for Interaction with Tsg101: Implications for Cytokinesis.
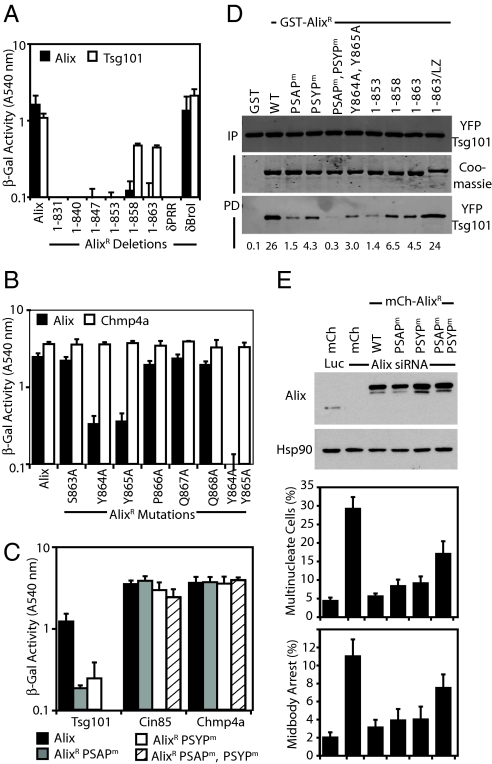
When analyzing our panel of Alix-interaction partners, we noticed that AlixR 1-831 could no longer interact with Alix (Fig. S2), indicating that this region is required for multimerization. We mapped this activity to residues Y864 and Y865 in Alix's extreme C terminus (Fig. 4 A and B). Additionally, we noted that AlixR 1-831 could not interact with Tsg101 (Fig. 4A), although the Tsg101-binding 717PSAP720 motif was present, suggesting that the terminal 37 residues of Alix encode an activity that is required for Tsg101 binding. Mapping this activity identified a requirement for the sequence between residues 853 and 858 (Fig. 4A) overlapping with the residues 852PSYP855. We initially speculated that this sequence represented a pseudoP(S/T)AP motif for coordination by Tsg101's Ubiquitin E2 Variant (UEV) domain. However, scanning mutagenesis of this sequence revealed that these residues made only a limited contribution to direct-Tsg101 binding (Fig. S4 A and B). Intriguingly however, mutation of this entire motif (P852A, Y854G, P855A; AlixR PSYPm) greatly reduced Alix's interaction with Tsg101, and mutation of both PSAP and PSYP sequences (AlixR PSAPm, PSYPm) abolished the ability of Alix to bind Tsg101 (Fig. 4 C and D). We found that AlixR PSYPm could bind all partners tested but was unable to multimerize (Figs. S2 and S4C). The ability of these Alix mutations to interact with Tsg101 was confirmed by coprecipitation assays. Interestingly, these assays demonstrated that Alix-multimerization is required for interaction with Tsg101 (Fig. 4D), because multimerization-defective C-terminal deletions of AlixR, AlixR PSYPm, AlixR Y864A, and Y865A all failed to bind Tsg101. Crucially, restoring the ability of AlixR 1-863 to multimerize through fusion of the heterologous multimerization domain from the GCN4 leucine zipper (33) to AlixR 1-863 (AlixR 1-863/LZ) rescued binding to Tsg101 to control levels (Fig. 4D). These data demonstrate that Alix multimerization is required for efficient Tsg101 binding.
Fig. 4.
Activities in the C terminus of Alix. (A and B) The indicated Alix constructs fused to the VP16-activation domain were tested for interactions with Alix or Tsg101 fused to the Gal4 DNA-binding domain by yeast two-hybrid assay (n = 3 ±SD). (C) The indicated Alix mutants were fused to the VP16-activation domain and tested for interactions with Tsg101, Cin85, and Chmp4a fused to the Gal4 DNA-binding domain by yeast two-hybrid assay (n = 3 ±SD). (D) Coprecipitation assays were performed on lysates of 293T cells cotransfected with plasmids encoding YFP-Tsg101, Myc-Vps28, and the indicated GST-AlixR fusion proteins. Resolved cells lysates (input, IP) and normalized bead-bound fractions (pull down, PD) were analyzed with α-GFP antisera. PD fractions were analyzed by Coomassie staining to visualize GST-AlixR expression, and relative binding was quantified by infrared imaging. (E) HeLa cells stably transduced with retroviral vectors encoding mCh or the mCh-AlixR fusions were treated with siRNA targeting Luciferase or Alix. Cells were fixed, stained with α-tubulin, and scored for midbody arrest and multinucleation (n = 3 ±SD). Resolved cell lysates were analyzed with α-Alix and α-Hsp90 antibodies.
Cytokinetic defects in cells expressing Tsg101 containing a UEV-domain mutation (M95A) abolishing interaction with Alix have been observed (7). However, these effects could be due to loss of binding to other PTAP-containing proteins (34). To establish the requirement for Alix/Tsg101 interaction in cytokinesis, we used the PSAP and PSYP mutations that were rendered unable to interact with Tsg101. We created stable HeLa cells expressing mCh-tagged versions of AlixR PSAPm, AlixR PSYPm, or AlixR PSAPm PSYPm (Fig. 4E) and examined the ability of these proteins to rescue cytokinetic defects induced by Alix depletion. Both mCh-AlixR PSAPm and mCh-AlixR PSYPm could rescue cytokinesis in Alix-depleted cells. However, the joint mutant that was most severely compromised for Tsg101-binding (mCh-AlixR PSAPm, PSYPm) could support only a partial rescue (Fig. 4 D and E), indicating that Alix-Tsg101 interactions are required for cytokinesis.
Alix Multimerization Is Required for HIV-1 Release.
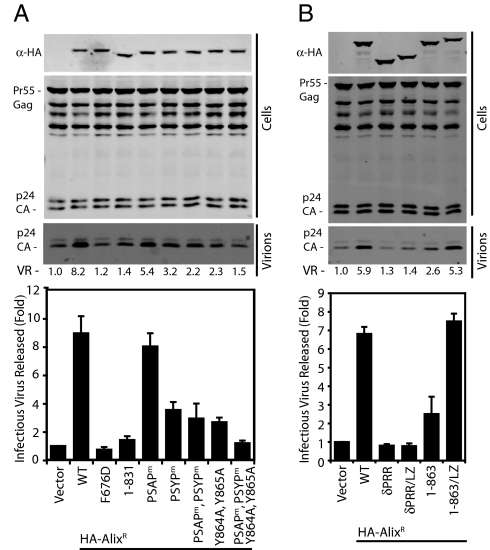
The use of a version of HIV-1 that is released in an Alix-dependent manner has exposed a requirement for the terminal 37 residues of Alix in HIV-1 release (20, 21). We confirmed this and, in agreement with previous reports, demonstrated that Alix's interaction with HIV-1 Gag and ESCRT-III were essential for this activity, but interaction with endophilins CD2AP, Cin85, and Cep55 were dispensable (Fig. S5). We mapped this activity and demonstrated that it correlated well with the site of Alix multimerization (Fig. S6A) (21). However, Alix Y864A and Y865A did not fully recapitulate the phenotype of AlixR 1-831 and additional deletions (for example, AlixR 1-853) resulted in a more pronounced defect in HIV-1 release (Fig. S6A). Mutation of the PSAP motif had little effect on the ability of Alix to support HIV-1 release, suggesting that interaction with Tsg101 is not required for this activity (Fig. 5A). However, as with its effect on multimerization, mutation of the entire PSYP sequence disrupted Alix-dependent HIV-1 release, indicating that multimerization contributes to Alix-dependent HIV-1 release. Indeed, AlixR PSAPm, PSYPm, Y864A, and Y865A recapitulated fully the phenotype of AlixR 1-831 in this assay (Fig. 5A), suggesting that Alix multimerization is required for Alix-dependent HIV-1 release. To extend these studies, a gain-of-function approach was followed whereby the GCN4 LZ was appended to AlixR δPRR or AlixR 1-863, restoring the ability of these proteins to multimerize (Fig. S6B). Similarly to wild-type AlixR, AlixR 1-863/LZ fully rescued Alix-dependent HIV-1 release (Fig. 5B). However, AlixR δPRR/LZ could not support this activity, suggesting that other regions of the PRR are required or that forced oligomerization of AlixR δPRR results in its adoption of a conformation that cannot support HIV-1 release. These data demonstrate that Alix multimerization represents a C-terminal activity that is essential for Alix-dependent HIV-1 release.
Fig. 5.
Alix multimerization is required for Alix-dependent HIV-1 release. Cells (293T) were cotransfected with plasmids encoding the indicated HA-tagged AlixR constructs and an HIV-1 pNL/HXB p6(P7L, P10L) plasmid. Resolved cell lysates and released virions were examined by Western blot analysis with α-Gag antisera. Quantification of virion release (VR) via infrared imaging is given. (n = 3). Lysates were also examined by Western blot analysis with α-HA antisera. β-Galactosidase assay was performed on HeLa-TZM-bl cells infected with 293T supernatants (n = 3 ±SD).
Because neither the C-terminal 37 residues of Alix nor Alix multimerization activity were required for cytokinesis (Fig. 2 and Fig. S3), and, because Alix interacts only weakly with HIV-1p6 (20), we questioned whether multimerization was required for release of viruses employing high-affinity interactions with Alix. Using Alix-siRNA, we suppressed release of a L domain-defective HIV-1 that was complemented in trans with the high-affinity Alix-binding L domain of EIAVp9 (22). We examined the ability of cotransfected HA-AlixR constructs to rescue virus release. When expressed at near-endogenous levels, HA-AlixR and HA-AlixR Y864A, Y865A restored release of EIAVp9-complemented HIV-1, whereas HA-AlixR F676D could not (Fig. S7), suggesting that differential requirements for Alix multimerization exist among retroviruses that engage Alix with differing affinities.
Discussion
In this article, we have explored the role of Alix in two ESCRT-dependent cellular processes, namely cytokinesis and viral budding. We show that Cep55 binds a conserved peptide within Alix's PRR through multiple contacts, demonstrating that this region is an evolutionarily important constituent of the Alix-PRR. Using depletion and rescue experiments, we show that Alix acts as an adaptor protein, linking Cep55 with ESCRT-III to effect completion of cytokinesis. In many ways, this function of Cep55 parallels that of retroviral Gag proteins that elicit particle fission through recruitment of ESCRT-III via various adaptor proteins including Alix itself (16, 17), Tsg101 (12–15), or the HECT ubiquitin ligases (35). In making these observations, we add a degree of processivity to the mechanism of Cep55- and ESCRT-dependent cytokinesis.
We have also shown that Cep55/Alix/ESCRT-III interactions are required for formation of morphologically normal midbodies. Because both Alix depletion and rescue with Alix-constructs that cannot recruit ESCRT-III render midbody microtubules visible to α-tubulin, we reason that the ESCRT machinery is a constituent of the protein density of the Flemming body. Indeed, because YFP-Cep55 and mCh-AlixR F199D, but not mCh-AlixR P801V, P802D, are recruited by the midbody, we can be sure that the Flemming body is present in these Alix-depleted cells but that it is aberrant. Morphologically, these findings have parallels with phenomena observed on the cytosolic face of endosomes, where a proteinaceous Hrs-containing coat is observed adjacent to sites of MVB formation and concentrates ubiquitinated cargo before its inclusion in MVBs (9). Whether this coat contains the ESCRT machinery remains to be established, but because ESCRT-III is proposed to form a proteinaceous lattice on endosomal membranes (23), it is possible that, as in the case of the Flemming body, this visible density represents the ESCRT-machinery. Collectively, these data suggest that the cytokinetic defects that arise as a consequence of Cep55 depletion represent a failure to recruit Alix/ESCRT-III to the midbody.
An important point that remains to be addressed is the mechanism used by the ESCRT machinery to achieve abscission. Electron-microscopic analysis of the terminal stages of cytokinesis reveals that the midbody thins before abscission, reducing its diameter to distances of 200 nm (31). Because the ESCRT machinery is proposed to act in the scission of viral particles tethered to the plasma membrane via 50- to 100-nm diameter stalks, it is possible that the “thinning” process reduces the midbody diameter sufficiently to allow the ESCRT machinery to act directly in abscission. Indeed, Vps4, known to be required for the final stage of virus budding (10) and abscission (7, 8). was recently localized to rings on either side of very thin midbodies (8), suggesting that it acts in late-stages of cytokinesis. Alternatively, the ESCRT machinery may participate in the thinning process itself; close examination of midbodies reveals the presence of budding structures emanating from their surface (31), and it has recently been demonstrated that polarized neuroendocrine cells shed membranous particles from their midbodies and primary cilia in a process topologically equivalent to viral budding (36). Although these budding structures may arise simply as a consequence of the presence of the recruited ESCRT machinery, it is possible that this machinery plays an active role in midbody thinning before abscission.
Additionally, we have defined the nature of the HIV-1 budding activity present within Alix's C terminus. We show that this region represents a multimerization module and that Alix's ability to multimerize represents the essential activity of this region in HIV-1 release. Although the major determinant of multimerization is a dityrosine motif at residues 864 and 865, the loss of multimerization upon mutation of residues 852PSYP855 suggests that multiple Alix–Alix contacts are made throughout Alix's extreme C terminus. Intriguingly, we found that Alix-multimerization is also required for Tsg101 binding, suggesting an important mechanistic requirement for this region in assembly of Alix/ESCRT complexes. However, although full Alix/Tsg101 binding is required for cytokinesis, Alix multimerization appears dispensable; it is possible that Cep55's multimerization activity (4) may override this requirement in this case. Additionally, multimerization appears dispensable for release of HIV rendered dependent on the high-affinity Alix-binding EIAVp9 L domain, suggesting that it may act to enhance the weak interaction between Alix and HIV-1p6 through avidity effects.
Last, using a dominant-negative approach, we also demonstrate that Chmp4c is a more potent inhibitor of cytokinesis than Chmp4a or Chmp4b, yet Chmp4b is the most potent inhibitor of HIV-1 release. The reasons for this differential activity are currently unclear, but it is of note that Chmp4c contains sequence insertions not present within other isoforms that may allow for its differential regulation. These experiments provide evidence suggesting that gene expansion might reflect the functional diversification of the mammalian ESCRT machinery, which will be an important area of future study.
Materials and Methods
siRNA Transfections.
HeLa cells were seeded at a density of 1E5 cells per ml. The 293T cells were seeded at 2.6E5 cells per milliliter and were transfected 2 h later with siRNA targeting Luciferase (CUGCCUGCGUGAGAUUCUCdTdT), Alix (GAAGGAUGCUUUCGAUAAAUU) or Cep55 (GGAGAAGAAUGCUUAUCAAUU) at 100 nM concentration by using Dharmafect-1 (Dharmacon). After 48 h, cells were reseeded and transfected again with siRNA by using Dharmafect-1 or with siRNA and DNA by using Lipofectamine-2000. Cells were assayed 48 hours later.
Virus Budding and Infectivity Assays.
For HIV-1 release assays, 293T cells were transfected with 300 ng of pNL/HXB and mixtures of pCR3.1 and pCR3.1 YFP-Chmp4 constructs totaling 250 ng for 48 h. For Alix-dependent HIV-1 release assays, 293T cells were transfected with 300 ng of pNL/HXB p6 (P7L, P10L), 375 ng of pCR3.1, and 125 ng of pCR3.1 HA-AlixR constructs for 24 h. For p9-dependent HIV-1 release assays, 293T cells were transfected twice with siRNA for 48 h. Included in the second transfection were 40 ng of pCR3.1 or pCR3.1 HA-AlixR constructs, 300 ng of pNL/HXB δp6, and 200 ng of a plasmid encoding a truncated HIV-1 pNL/HXB Gag protein fused to the p9 L domain of EIAV. Virions were harvested from 293T supernatants by filtration (0.2 μm) and centrifugation through 20% sucrose (25,000 × g, 120 min), lysed, resolved by SDS/PAGE, and examined by Western blot analysis. Additionally, HeLa-TZM-bl cells (CD4+, CXCR4+, CCR5+, HIV-1 LTR-LacZ) were infected with 293T supernatants for 48 h. LacZ activities in cell lysates were measured by chemiluminescent detection of β-galactosidase by using Galacto-Star (Applied Biosystems).
Multinucleation Assays.
For overexpression assays, HeLa cells were transfected with mixtures of pCR3.1 and pCR3.1-YFP-Chmp4 plasmids totaling 250 ng, for 48 h (cells were reseeded onto coverslips after 24 h), and 100 well adhered, nonapoptotic cells per coverslip were scored for presence of more than one nucleus. Alternatively, siRNA treated cells were fixed and stained with α-tubulin. For siRNA assays, 300 cells per coverslip were scored for presence of more than one nucleus or arrest at midbody. In all cases, cells unambiguously connected by midbodies were considered multinucleated. Fifty individual midbodies were scored for morphology per experiment.
Additional methods are described in SI Text.
Supplementary Material
Acknowledgments.
We thank W. Sundquist (University of Utah, Salt Lake City), M. Malim (King's College London, London), and H. McMahon (Medical Research Council-Laboratory of Molecular Biology, Cambridge, UK) for reagents. This work was supported by Career Establishment Grant G0400207 from the Medical Research Council U.K. J.G.C. is a Beit Memorial Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802008105/DCSupplemental.
References
- 1.Glotzer M. Animal cell cytokinesis. Annu Rev Cell Dev Biol. 2001;17:351–386. doi: 10.1146/annurev.cellbio.17.1.351. [DOI] [PubMed] [Google Scholar]
- 2.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: From parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 3.Barr FA, Gruneberg U. Cytokinesis: Placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Fabbro M, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Garay I, Rustom A, Gerdes HH, Kutsche K. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics. 2006;87:243–253. doi: 10.1016/j.ygeno.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhao WM, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell. 2006;17:3881–3896. doi: 10.1091/mbc.E06-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 8.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- 11.Hurley JH, Emr SD. The ESCRT complexes: Structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 13.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 14.VerPlank L, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Serrano J, Bieniasz PD. A bipartite late-budding domain in human immunodeficiency virus type 1. J Virol. 2003;77:12373–12377. doi: 10.1128/JVI.77.22.12373-12377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 18.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 21.Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muziol T, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Zamborlini A, et al. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, Borinstein SC, Gillis J, Sykes VW, Bogler O. The glioma-associated protein SETA interacts with AIP1/Alix and ALG-2 and modulates apoptosis in astrocytes. J Biol Chem. 2000;275:19275–19281. doi: 10.1074/jbc.M908994199. [DOI] [PubMed] [Google Scholar]
- 26.Kim JM, et al. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science. 2003;300:1298–1300. doi: 10.1126/science.1081068. [DOI] [PubMed] [Google Scholar]
- 27.Monzo P, et al. Clues to CD2-associated protein involvement in cytokinesis. Mol Biol Cell. 2005;16:2891–2902. doi: 10.1091/mbc.E04-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatellard-Causse C, et al. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- 29.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 30.Spitzer C, et al. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- 31.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matuliene J, Kuriyama R. Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol Biol Cell. 2002;13:1832–1845. doi: 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 34.McDonald B, Martin-Serrano J. Regulation of Tsg101 expression by the steadiness box: A role of Tsg101-associated ligase. Mol Biol Cell. 2008;19:754–763. doi: 10.1091/mbc.E07-09-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Brauninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.