Abstract
Stem cells hold great promise for regenerative medicine, but remain elusive in many tissues in part because universal markers of “stemness” have not been identified. The ribonucleoprotein complex telomerase catalyzes the extension of chromosome ends, and its expression is associated with failure of cells to undergo cellular senescence. Because such resistance to senescence is a common characteristic of many stem cells, we hypothesized that telomerase expression may provide a selective biomarker for stem cells in multiple tissues. In fact, telomerase expression has been demonstrated within hematopoietic stem cells. We therefore generated mouse telomerase reverse transcriptase (mTert)-GFP-transgenic mice and assayed the ability of mTert-driven GFP to mark tissue stem cells in testis, bone marrow (BM), and intestine. mTert-GFP mice were generated by using a two-step embryonic stem cell-based strategy, which enabled primary and secondary screening of stably transfected clones before blastocyst injection, greatly increasing the probability of obtaining mTert reporter mice with physiologically appropriate regulation of GFP expression. Analysis of adult mice showed that GFP is expressed in differentiating male germ cells, is enriched among BM-derived hematopoietic stem cells, and specifically marks long-term BrdU-retaining intestinal crypt cells. In addition, telomerase-expressing GFP+ BM cells showed long-term, serial, multilineage BM reconstitution, fulfilling the functional definition of hematopoietic stem cells. Together, these data provide direct evidence that mTert-GFP expression marks progenitor cells in blood and small intestine, validating these mice as a useful tool for the prospective identification, isolation, and functional characterization of progenitor/stem cells from multiple tissues.
Keywords: intestinal stem cell, telomerase, tissue stem cells
Stem cells hold great promise for regenerative medicine and tissue repair, but have been difficult to identify in many tissues. Traditional methods used to isolate stem cells have largely relied on FACS by using complex combinations of antibodies to cell surface antigens. This approach has enabled the characterization of hematopoietic stem cells (HSCs), along with a detailed understanding of hematopoietic lineage development (1). More recently, the development of methods based on particular biochemical properties of stem cells (e.g., the side-population phenotype) (2) has provided additional tools to enrich for stem cell populations. A model system that enables the direct isolation and/or enrichment of stem cells from multiple tissues would greatly facilitate the identification, purification, and functional characterization of novel stem cell populations.
Relative resistance to cellular senescence despite multiple rounds of cell division is a common characteristic of stem cells (3). Telomerase is a ribonucleoprotein complex that helps maintain the telomeric ends of chromosomes, normally shortened with each cell division. Because loss of telomeric DNA beyond a critical threshold induces senescence in most somatic cells, maintenance or induction of telomerase activity provides a means of preventing cellular senescence (4) that may be relevant for the self-renewal of tissue stem cells. Consistent with this hypothesis, telomerase activity has been demonstrated in self-renewing ES cells (5), HSCs (6), hematopoietic progenitor and lymphoid cells (6, 7), germ cells (8), regenerating tissues such as intestine and skin (9–11), and self-renewing tissues from urochordates, suggesting an evolutionarily conserved mechanism (12).
Mouse telomerase reverse transcriptase (mTert) is tightly regulated and correlates with telomerase activity (11). mTert expression is down-regulated upon differentiation in most somatic cells (11), and telomerase-deficient mice exhibit a defect in stem cell maintenance, such that tissues highly dependent on stem cell function throughout life [e.g., bone marrow (BM), testis, and intestine] undergo organ failure (10). Several reports have suggested a direct role for mTert in the regulation of adult stem cell proliferation and mobilization (13, 14). Because mTert gene expression is the regulatory step in the assembly of a functional telomerase complex (11), we hypothesized that a reporter gene system using the mTert promoter would allow for the identification of telomerase-expressing cells and thus provide a useful biomarker for stem cells in adult tissues.
In this article, we describe the generation of mTert-GFP-transgenic mice using a two-step ES cell-based screening strategy and validate GFP expression as a marker of two classical stem cells: male germ cells and HSCs. In addition, these mice have been used to mark the intestinal stem cell and may allow for the subsequent isolation and characterization of this highly elusive cell. Thus, mTert-GFP-transgenic mice represent a model system to facilitate the identification, purification, and functional analysis of novel stem cell populations.
Results
Generation of mTert-GFP-Transgenic Mice.
Given that previous attempts to generate mTert-GFP mice (using the same 4.4-kb promoter fragment used in this article) failed to show reporter gene expression in adult tissues (15), we used an alternative method to generate transgenic mice with the goal to screen for optimal levels of GFP expression before the production of live animals. Because ES cells normally express telomerase at high levels (5), the use of ES cell transgenesis (rather than pronuclear oocyte injection) allowed for the selection of maximal transgene expression (1° screen) and exclusion of clones that underwent transgene silencing upon genomic integration. G418-resistent ES cell clones expressing high levels of GFP fluorescence were visualized by using standard epifluorescence microscopy (Fig. 1a). Of 100 G418-resistent colonies examined, only half demonstrated GFP expression, ranging from low to high. Fourteen clones demonstrating the highest level of GFP fluorescence were expanded for further analysis.
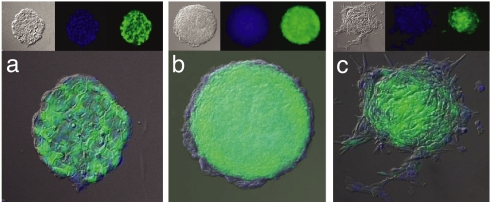
Fig. 1.
mTert-GFP expression during EB formation. (a) Representative undifferentiated ES cell colony stably transfected with mTert-GFP. (b) Representative EB 7 days after LIF withdrawal. Note the outer layer of differentiated cells no longer expressing GFP. (c) Representative EB after 48 h of DMSO exposure and 7 days of LIF withdrawal. Note lack of GFP in the differentiated cell outgrowth. Each panel is a merge of Nomarski imaging (Upper Left Insets), DAPI stained nuclei (Upper Center Insets), and GFP expression (Upper Right Insets). (Magnification: ×20.)
Because ES cells down-regulate telomerase expression upon in vitro differentiation (5), each clone was secondarily screened by using embryoid body (EB) formation to ensure proper down-regulation of GFP expression and to prevent selection of clones demonstrating constitutive or dysregulated mTert-GFP expression (2° screen). Upon differentiation, a decrease in GFP expression could be visualized in cells forming the outer layer of primitive endoderm (Fig. 1 b and c).
Of the 14 clones studied, 9 showed appropriate down-regulation of the GFP transgene upon differentiation, and the 2 brightest (clones 14 and 22) were selected for blastocyst injection. Chimeric mice were generated and bred to germ-line heterozygosity. Male and female mice from both lines are healthy, are fertile, demonstrate normal longevity, and can be bred to homozygosity. In addition, Southern blot analyses revealed 1–3 transgene copies for both lines (data not shown). Extensive analysis of both lines confirmed an overlapping GFP expression profile.
mTert-GFP Expression in Testis.
To validate transgene expression in a tissue known to express telomerase at high levels (16), GFP expression was analyzed in testis. GFP was detected in both lines 14 and 22 by Western blot analysis of whole testis protein extract by using an anti-GFP antibody. Protein extracts from wild-type and constitutively expressing Actin-GFP-transgenic mice served as negative and positive controls, respectively (Fig. 2a). The amount of total protein loaded from the Actin-GFP control was empirically reduced to allow for a more direct comparison with the less abundant GFP signal present in the mTert-GFP lanes (Fig. 2a).
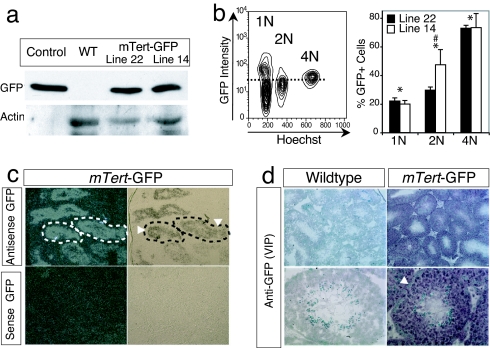
Fig. 2.
mTert-GFP expression in testis. (a) Western blotting was performed by using whole testis protein extract from WT, mTert-GFP, or Actin-GFP mice and rabbit anti-GFP antibody. The blot was stripped and reblotted by using anti-Actin antibody as a loading control. (b) Representative FACS analysis for GFP expression performed on single-cell isolates from seminiferous tubules of adult mTert-GFP and WT testes. Cells were gated into 1N, 2N, and 4N populations. The dashed line illustrates the GFP threshold as defined by WT control cells. Histogram represents the mean ± SEM from two independent experiments with 3–5 replicates; ANOVA, P < 0.001, post hoc Fisher's (PLSD) analysis revealed significant differences among (*) groups (1N, 2N, and 4N) for both lines and between (#) the 2N populations. (c) ISH for GFP expression in mTert-GFP adult testis. Dotted lines outline the seminiferous tubules. Arrowheads demarcate the basal layer of spermatogonial stem cells. (Left) Dark field images. (Right) Bright field images. (Magnification: ×10.) (d) Immunohistochemistry for GFP expression in mTert-GFP testis. Arrowhead indicates the basal layer of spermatogonial stem cells. (Magnification: Upper, ×10; Lower, ×40.)
To establish transgene expression within the germ cell population, we used flow cytometry. Single-cell suspensions were purified from transgenic and wild-type seminiferous tubules, labeled with Hoechst dye 33342, and analyzed by flow cytometry based on DNA content [i.e., haploid (1N), diploid (2N), or tetraploid (4N)] (Fig. 2b). Analysis of both lines demonstrated GFP+ cells in each cell fraction, with the largest percentage of GFP+ cells (as compared with GFP− cells) found in the 4N population (line 22, 72.9 ± 2.0%; line 14, 73.3 ± 9.8%), indicating that a substantial fraction of meiotically active primary spermatocytes express GFP (Fig. 2b). A smaller fraction of 2N cells (line 22, 29.6 ± 2.1%; line 14, 47.4 ± 10.5%), representing spermatogonia, secondary spermatocytes, and/or somatic cells, and 1N cells (line 22, 21.1 ± 2.2%; line 14, 19.9 ± 2.5%), representing spermatids, was observed to be GFP positive (Fig. 2b). Given that 4N cells represented the smallest fraction of the isolated cells (9.4 ± 2.3%) when compared with 2N (14.6 ± 1.3%) or 1N (67.2 ± 2.2%) cells, the fraction of GFP+ cells as a function of all isolated germ cells was 4N (line 22, 8.3 ± 0.7%; line 14, 4.9 ± 1.6%), 2N (line 22, 4.0 ± 0.7%; line 14, 6.6 ± 0.6%), and 1N (line 22, 14.2 ± 1.6%; line 14, 13.8 ± 1.5%). These data are consistent with previous reports demonstrating telomerase activity in each germ cell population (16) and validate GFP expression as a marker of male germ cells.
To further localize GFP expression, in situ hybridization (ISH) (Fig. 2c) and immunohistochemistry (Fig. 2d) were performed. GFP mRNA and protein were localized within the seminiferous tubules and corresponded to primary and secondary spermatocytes, consistent with the flow-cytometry analysis. Surprisingly, the majority of spermatogonial stem cells lining the basement membrane (arrowhead) did not express GFP (Fig. 2 c and d), suggesting that most slowly cycling spermatogonial stem cells do not express mTert-GFP at baseline. In addition, GFP was not detected within the interstitial (non-germ cell) compartment (Fig. 2 c and d). No GFP expression was detected using WT control testis (Fig. 2d). These data indicate that GFP is differentially expressed in male germ cells during meiosis.
mTert-GFP Expression in BM and Peripheral Blood.
To establish mTert-GFP expression in a second tissue where stem and progenitor cells have been shown to express telomerase (6, 7), whole BM was analyzed for GFP expression. Flow-cytometric analysis revealed ≈2% of BM cells to be GFP+ (see Figs. 3 and 4 for phenotypic and functional analysis). In addition, both lines of mice display stable expression of GFP in a small subset (≈1%) of peripheral blood cells (PBCs), which include B cells (≈45%), T cells (≈25%), and myeloid cells (≈25%) (data not shown). These circulating GFP+ PBCs express CD45 (data not shown), allowing them to be readily distinguished from GFP-expressing putative tissue stem cells.
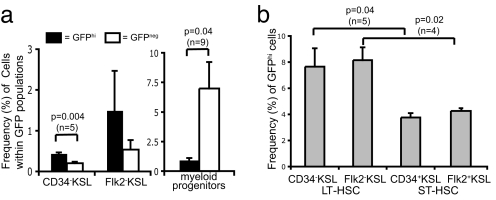
Fig. 3.
Phenotypic Analysis of mTert-GFP expression in HSCs. (a) Increased frequency of LT HSCs and decreased frequency of myeloid progenitors among GFPhi populations by using multicolor FACS analysis of LT HSCs. Pooled results (mean ± SEM) from two independent experiments; Student's t test indicated. (b) Increased frequency of GFPhi cells within the LT HSC population compared with the ST HSC population. Pooled results from two independent experiments (mean ± SEM), Student's t test indicated.
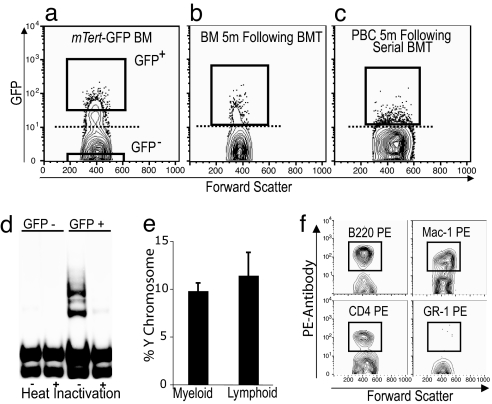
Fig. 4.
Functional analysis of mTert-GFP expression in HSCs. (a) FACS scatter plot of BM from a male mTert-GFP mouse demonstrating GFP+ cells above the dotted line (set using WT BM). GFP+ or GFP− cells were sorted according to the gates shown and transplanted into sublethally irradiated female recipient mice. (b) FACS analysis of BM from female recipient mice 5 months after transplant. GFP+ cells were sorted and transplanted into secondary female recipients. (c) FACS analysis of PBCs 5 months after serial BMT confirm serial LT engraftment of GFP+ cells. (d) TRAP assay was performed by using isolated GFP− and GFP+ BM cells (Fig. 4a) to determine telomerase activity. Heat inactivation samples were used as a negative control. (e) FISH for Y chromosome demonstrated engraftment into both myeloid (Gr-1+ and Mac-1+) and lymphoid (B220+ and CD4+) lineages 2 months after serial BMT. (f) FACS analysis of GFP+ PBCs 5 months after transplantation confirm GFP in both myeloid (Mac-1+) and lymphoid (B220+, CD4+) lineages at levels comparable to donor animals.
Phenotypic Analysis of GFP Expression in HSCs.
To establish that GFP is expressed specifically within the HSC, previously shown to express telomerase (6), whole BM from mTert-GFP mice was analyzed by using multicolor flow cytometry. Although only long-term (LT) HSCs have unlimited self-renewal potential (17), additional hematopoietic cells [e.g., transiently self-renewing short-term (ST) HSCs, hematopoietic progenitor cells, and lymphoid cells] also express telomerase (6, 7). To determine at the single-cell level the expression of mTert in these hematopoietic subsets, BM cells were analyzed based on the presence or absence of two distinct cell surface markers (CD34 or FLK2) within the cKit+Sca1+Lineage− (KSL) subset of hematopoietic progenitor cells. CD34−KSL and Flk2−KSL cells correspond to LT HSCs, whereas CD34+KSL and Flk2+KSL cells correspond to ST HSCs (18–20). Extensive analysis of lines 14 and 22 demonstrated similar GFP expression profiles, and so, for simplicity, results from only line 22 are presented here.
To determine whether GFP fluorescence intensity correlated with self-renewal capacity, LT HSCs were gated into GFPhi, GFPlo, and GFP− populations [see supporting information (SI) Fig. S1a for representative FACS plots], and the frequency of cells within each population was determined. A 2-fold enrichment of LT HSCs was detected within the GFPhi population compared with the GFP− population: CD34−KSL (0.44 ± 0.02% vs. 0.20 ± 0.04%; P = 0.004) (Fig. 3a). No enrichment of LT HSCs was observed in the GFPlo population compared with the GFP− population (data not shown). We also assessed the frequency of myeloid progenitor cells (MPCs) (cKit+Sca1−Lineage−), a population of lineage-committed progenitors without self-renewal potential (21) within the GFPhi and GFP− populations. The results showed a significant enrichment of MPC in the GFP− subset of BM cells compared with GFPhi cells (0.93 ± 0.18% vs. 6.99 ± 2.23%; P = 0.04) (Fig. 3a). Together, these results indicate that the GFPhi subset of BM cells in mTert-GFP mice is enriched for self-renewing LT HSCs and contains few non-self-renewing MPCs. Thus, in mTert-GFP BM, fluorescence intensity correlates with self-renewal capacity, consistent with a previous report that telomerase activity is highest in LT HSCs and decreases with hematopoietic cell differentiation (6).
To analyze the fraction of HSCs expressing GFP and the relative intensity of GFP expression, BM cells were gated into LT or ST HSC populations and then plotted to display GFP fluorescence. A 2-fold higher frequency of GFPhi cells was detected within LT HSCs, compared with ST HSCs: (CD34−KSL vs. CD34+KSL, 7.7 ± 1.4% vs. 3.8 ± 0.3%; P = 0.04; Flk2−KSL vs. Flk2+KSL, 8.2 ± 1.0% vs. 4.3 ± 0.1%; P = 0.02) (Fig. 3b). No differences were observed between GFPlo and GFP− populations (data not shown). Representative FACS plots for LT and ST HSCs illustrate the gates corresponding to the three GFP populations: GFPhi, GFPlo, and GFP− (Fig. S1a). These results suggest that GFP marks a subset of highly self-renewing HSCs.
Failure to detect GFP in all LT HSCs may be due to additional regulation of telomerase activity within these cells. One possible explanation is that telomerase activity is regulated by cell cycle progression (22). To assess whether GFP expression is induced in HSCs during the proliferative phase of the cell cycle, we performed cell cycle analysis on BM cells using Hoechst dye labeling, followed by multicolor FACS analysis. Cells were gated into LT HSCs, ST HSCs, or MPCs and fractionated into G0/G1 (enriched for noncycling cells) or S/G2-M (proliferating cells) according to their DNA content (2N vs. 4N, respectively). Each group was then analyzed according to GFP fluorescence intensity (Fig. S1b). This analysis confirmed our previous finding that the frequency of GFPhi cells is greatest among LT HSCs when compared with either ST HSCs or MPCs. Despite a trend toward an increased frequency of GFPhi cells in the proliferating fractions, no statistical differences were observed between G0/G1 and S/G2-M within a given population. No differences were observed between GFPlo and GFP− populations (data not shown). To further define whether GFP is preferentially expressed during the proliferative phase of the cell cycle, we used Ki67 staining (expressed during late G1-S/G2-M, but not in G0) in conjunction with flow cytometry; ≈30% of the GFPhi population stained positive for Ki67, which was no different from the control population (Fig. S2). Therefore, taken together, these results demonstrate that GFP expression in LT HSCs is not a function of cell cycle regulation.
Functional Analysis of GFP Expression in HSCs.
To validate that mTert-GFP expression marks a functionally significant subset of HSCs, we assessed the capacity of GFP+ BM cells to exhibit long-term, serial, multilineage reconstitution, the functional definition of the HSC. BM was harvested from adult mTert-GFP male mice and sorted by FACS into GFP+ and GFP− fractions (Fig. 4a). The cell-sorting gate for GFP+ cells corresponds to the GFPhi population defined to be enriched for HSCs (Fig. S1a). An equal number of GFP+ or GFP− cells were injected into the tail vein of prewarmed, sublethally irradiated isogenic (129/SvImJ) female mice (GFP+ recipients, n = 4; GFP− recipients, n = 5). Long-term engraftment, defined as the persistence of donor cells >5 months after BM transplantation (BMT), was demonstrated in 50% of recipient mice receiving GFP+ cells by FACS analysis of BM and PBCs (data not shown). In contrast, recipient mice receiving GFP− BM cells showed no evidence for engraftment as late as 3.5 months after BMT (data not shown). Together, these data support the conclusion that (i) mTert-GFP+ HSCs maintain their capacity to compete for the HSC niche, (ii) give rise to long-term engraftment, and (iii) are enriched in the GFPhi population. These data do not, however, rule out the presence of HSCs within the GFP− population, as demonstrated by the phenotypic analysis (see Fig. 3).
To further demonstrate that GFP marks HSCs in mTert-GFP mice, we next performed BMT into secondary female recipients using GFP+ BM cells obtained 5 months after the initial BMT (Fig. 4b). FACS-purified GFP+ cells (10,000 or 40,000) were injected into lethally irradiated isogenic female recipients (to establish the capacity of GFP+ cells for radioprotection). The secondary recipient receiving 10,000 GFP+ cells survived for 28 days, compared with control mice, which died at 13–15 days (data not shown). In contrast, the female recipient receiving 40,000 GFP+ cells demonstrated long-term HSC engraftment, surviving 5 months after 2° BMT, when she was killed for analysis of blood chimerism. Multilineage reconstitution was assessed at 2 and 5 months after 2° BMT (Fig. 4c). At 2 months, PBCs were sorted into myeloid (Mac-1+ and Gr-1+) or lymphoid (CD4+ and B220+) populations, followed by FISH analysis for the Y chromosome, to confirm that cells were derived from the original male donor. The results demonstrated chimerism for both myeloid (9.8% ± 0.8) as well as lymphoid cells (11.4% ± 2.4) (Fig. 4e). The presence of Y chromosome-containing myeloid cells 2 months after serial BMT (7 months after the initial BMT) demonstrates GFP is expressed within the HSC because any transplanted myeloid progenitors would be exhausted by this time (21). Chimerism also was confirmed at 5 months (10 months after the initial BMT) by demonstrating GFP+ myeloid (Mac-1+) and lymphoid (CD4+ and B220+) cells in the peripheral blood (Fig. 4f) at levels comparable to the frequency of GFP expression in these subsets in unmanipulated donor mice (data not shown). These results demonstrate the capacity of GFP+ BM cells to support long-term, multilineage reconstitution in 2° transplants, and thus functionally establish mTert-GFP expression as a marker for the HSC.
Analysis of GFP Expression in Intestinal Stem Cells.
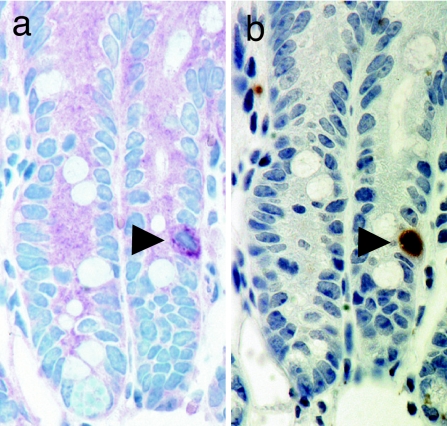
For >30 years, it has been accepted that the continuously renewing intestinal epithelium is maintained by pluripotent stem cells located in the intestinal crypts (23). Despite this well established model, intestinal stem cells (ISCs) have remained highly elusive (24). To determine whether GFP expression could serve as a marker for ISCs, we used the only established method to label these cells, long-term (LT) BrdU retention (25). mTert-GFP mice were administered a BrdU pulse during intestinal regeneration, followed by a period of chase. Immunohistochemical analysis of sequential sections from small intestine revealed single GFP+BrdU+ cells in the lower portion of the intestinal crypts (Fig. 5, arrowhead), consistent with previous reports demonstrating single, LT BrdU-retaining cells in the lower crypt (25). Analysis of ≈15,700 crypts demonstrated 1 in 157 to contain single GFP+ cells and 1 in 25 to contain single LT BrdU+ cells, similar to prior estimates of intestinal stem cell frequency (25). Of the identified GFP-expressing crypt cells, 17% coexpressed LT BrdU, suggesting that GFP is marking a subpopulation of ISCs. Expression of GFP within single cells provides us with a useful model system in which to isolate viable cells and functionally characterize this elusive stem cell population.
Fig. 5.
GFP expression in intestinal stem cells. Immunohistochemical analysis for GFP and BrdU using serial sections from mTert-GFP mice having undergone BrdU pulse–chase. (a) Staining with anti-GFP antibody, VIP (dark purple) chromagen substrate. (b) Staining with anti-BrdU antibody, DAB (dark brown) chromagen substrate. (Magnification: ×40.)
Telomerase Activity Colocalizes with GFP-Expressing Cells.
Finally, to ensure that GFP expression in vivo correlates with telomerase activity, isolated cells from BM, testis, and intestine were fractionated into GFP+ and GFP− populations by FACS and assayed for telomerase activity by using the Telomeric Repeat Amplification Protocol (TRAP) (Fig. 4d and Fig. S3). Telomerase activity was detected within each GFP+ cell fraction, whereas low or undetectable levels of activity were observed in each GFP− cell fraction, indicating that mTert-GFP is a valid biomarker for mTert activity.
Discussion
We have generated mTert-GFP-transgenic mice as a model system to facilitate the identification, isolation, and functional analysis of adult stem cells. To ensure adequate levels of transgene expression in adult mice, we used a two-step ES cell-based visual screening method, which resulted in the efficient generation of two informative transgenic founder lines. GFP expression was then shown to mark three well accepted stem cell populations (male germ cells, HSCs, and ISCs). These results illustrate the power of this model system to identify and enrich for putative stem cell populations and to subsequently validate their functional significance.
Using multiple techniques, mTert-GFP-transgene expression was demonstrated in male germ cells at various stages of spermatogenesis, with the greatest percentage of GFP+ cells demonstrated within meiotically active primary spermatocytes, followed by secondary spermatocytes. Haploid spermatids showed the lowest percentage of GFP+ cells. These results are consistent with previous reports describing telomerase expression in meitotically active male germ cells (16). The expression of GFP within spermatids, previously suggested to be mTert-negative (26), may be explained by the long half-life of GFP (≈24 h) or by aberrant transgene expression within these cells. The slight increase in the percentage of GFP+ cells in the 2N population from line 14, compared with line 22, may be explained by the site of transgene integration and/or by differential expression in the somatic cell compartment.
GFP expression also was shown to mark the LT HSC using both phenotypic and functional definitions. The functional analysis confirmed the presence of LT HSCs within the GFP+ population based on the capacity of these cells to give rise to long-term, serial, multilineage BM reconstitution. Analysis of mTert-GFP-expressing BM cells, however, is complicated by the fact that other hematopoietic cells, in addition to the HSC, express telomerase (e.g., ST HSCs and lymphocytes) (6, 7). Therefore, the finding that ≈2% of whole BM cells express GFP was not surprising.
Phenotypic analysis of BM cells using multicolor flow cytometry demonstrated an enrichment of LT HSCs within the GFPhi population, as well as the reciprocal, an enrichment of GFPhi cells within the LT HSC population compared with the ST HSC population. In contrast, no enrichment of LT HSCs was observed in the GFPlo population when compared with the GFP− population, suggesting a correlation between GFP fluorescence intensity and self-renewal potential. This observation is consistent with a previous report demonstrating a correlation between the frequency of telomerase-expressing cells within a given population and their degree of self-renewal potential (6). Furthermore, the decreasing frequency of GFP+ cells in populations of progressively differentiated cells is consistent with a decreasing role for telomerase as cells mature.
The low overall percentage of GFP+ LT HSCs was unanticipated and led us to speculate that mTert-GFP might be differentially regulated throughout the cell cycle. Previous reports have indicated that quiescent stem cells might be telomerase-negative (6, 9) whereas proliferative stem cells may be telomerase-positive (22, 27). To address this hypothesis, we performed cell cycle analysis using Hoechst dye on HSCs harvested from mTert-GFP mice, but we were unable to demonstrate a statistically higher percentage of GFP+ cells in the proliferative (S/G2-M) phase of the cycle compared with the (largely) noncycling (G0/G1) phase. These results may be complicated by coanalysis (via Hoechst staining) of G0- and G1-phase HSCs particularly if telomerase expression is induced during late G1 phase. To further assess whether GFP was preferentially expressed during the proliferative phase of the cycle, we performed Ki67 staining on LT HSCs, which showed no difference between groups. These results demonstrate that the regulation of mTert-GFP expression in LT HSCs is not a function of cell cycle. Alternatively, it remains possible that (i) the promoter fragment used to generate these mice may not contain all of the necessary regulatory elements required for mTert expression in HSCs, and/or (ii) stem cell heterogeneity may explain differential telomerase expression. For example, it has recently been shown that ES cells display marked heterogeneity in their expression of key pluripotency factors, demonstrating a level of regulation not previously appreciated (28). Consistent with this notion, our recent analysis of mTert-GFP expression after the generation of induced pluripotent stem (iPS) cells revealed significant heterogeneity in GFP expression along with other markers (29). These results suggest that heterogeneity of mTert-GFP expression within stem cell compartments may be explained by, as yet undefined, self-renewal mechanisms.
To identify the highly elusive ISC, we used LT BrdU retention and demonstrated GFP coexpression within this population. The ability to mark single intestinal crypt cells in this highly regenerative tissue, in contrast to the multiple cell types marked in BM and testis, where the telomerase expression profile is complicated, demonstrates the utility of this model system. The capacity to express GFP within this intestinal population provides us with a unique opportunity, not previously possible, to prospectively isolate and functionally validate ISCs.
To summarize, we have generated mTert-GFP-transgenic mice, a reporter system to provide a straightforward method for the identification, prospective isolation, and functional validation of adult stem cells from multiple tissues. In addition, this model can be used to seek out novel progenitor/stem cells in tissues where their presence remains to be established and to potentially investigate mechanisms underlying stem cell heterogeneity. These mice also may be useful to investigate other phenomena related to telomerase activation, such as oncogenesis, where GFP might serve as a sensitive tool to study the initial stages of tumor formation, identify cancer stem cells, or establish minimal residual disease status after specific treatment regimens.
Methods
Generation of Mice.
Linearized mTert-GFP transgene (5) was electroporated into mouse (J1) ES cells, selected with G418, and screened for GFP expression (1° screen). EBs were generated in suspension culture in the absence of LIF for 7 days ± 1% DMSO as described previously (5). Nuclei were counterstained with DAPI, and images were obtained by using a Nikon Eclipse E800.
Mice were maintained on a pure 129S1/SvIMJ background. All animal procedures were approved by the Children's Hospital Institutional Animal Care and Use Committee.
Western Blot Analysis.
Western blotting was performed by using 50 μg (WT and mTert-GFP) or 2.5 μg (Actin-GFP) of whole testis protein extract. Immunoblotting was performed by using a rabbit anti-GFP antibody (Abcam 6556) and the ECL reagent (Bio-Rad). The blot was stripped and reblotted by using anti-Actin antibody (Cell Signaling) as a loading control.
FACS Analysis of Germ Cells.
Seminiferous tubules were digested with collagenase (100 IU/ml, Worthington), washed to remove interstitial cells, dissociated to obtain germ cells, incubated with verapamil (Sigma) and Hoechst 33342 (Invitrogen) as described previously (30), and sorted by using a FACS VantageSE flow cytometer.
Histochemistry.
In situ hybridization was performed as described previously (31). Briefly, 10-μm frozen sections of adult testes were hybridized with 35S-labeled anti-sense or sense cRNA GFP riboprobes (≈350 bp). For testis, immunohistochemistry was performed on 10-μm frozen sections by using rabbit anti-GFP antibody (MBL), Vectastain ABC kit (Vector Laboratories), VIP substrate reagent (Vector Laboratories), and 1% methyl green. For intestine, immunohistochemistry was performed on 4-μm sequential paraffin sections by using rabbit anti-GFP (Abcam 6556), Vectastain ABC kit, VIP substrate reagent, and 1% methyl green. BrdU staining used the BrdU in situ detection kit (BD PharMingen) as recommended, with DAB and hematoxylin. Light and dark field images were obtained by using a Nikon Eclipse E800.
FACS Analysis of BM and PBCs.
Whole BM and PBCs were isolated from adult mice as described previously (32). Briefly, mature cells were depleted from BM by using lineage markers CD3 (clone KT31.1), CD4 (GK1.5), CD5 (53–7.8), CD8 (53–6.7), B220 (6B2), Mac-1 (M1/70), Gr-1 (8C5), and Ter119, followed by magnet bead (Dynal) separation. Depleted cells were stained with goat anti-rat PECy5 to visualize lineage marker staining. HSCs and progenitors were stained with PECy7-conjugated c-Kit (2B8) and APC-conjugated Sca1 (D7) with either biotinylated CD34 or biotinylated Flk2, followed by PE-conjugated streptavidin. Cells were analyzed by using a Dako-Cytomation MoFlo and/or a BD FACSAria flow cytometer. Data were analyzed by using FlowJo (Treestar), with frequencies of CD34− or Flk2− KSL (ckit+sca1+lineage−) HSCs and CD34+ or Flk2+ KSL progenitors determined after gating on live (PI−) cells. For cell cycle analysis, lineage-depleted BM was stained additionally with Hoechst 33342 and verapamil (32).
BM Transplantation.
Primary BMT recipients were x-irradiated (12 Gy, delivered as a split dose 3 h apart) before BMT of ≈1.8 × 105 GFP+ or GFP− cells by tail vein injection. 2° BMT recipients received γ-irradiation (11 Gy, delivered as a split dose 3 h apart) before BMT of 1 × 104 or 4 × 104 GFP+ cells by retro-orbital injection. Mice were maintained in autoclaved cages with acidified (pH 2.5), Bactrim-treated water.
Fluorescence In Situ Hybridization.
For FISH analysis of PBC nuclei, FACS-isolated cells were treated with hypotonic solution and fixed in methanol/acetic acid before slide preparation as described previously (33). The Y chromosome FISH probe (kindly provided by E. Gussoni, Children's Hospital, Boston) was labeled with digoxigenin-11-dUTP as described previously (33) and standardized on male and female control cells with a hybridization efficiency of >90%. Slides were examined by using a Zeiss Axiophot microscope.
BrdU Labeling of ISCs.
LT (pulse–chase) BrdU labeling was performed by using a modification of Potten et al. (25). Adult mice received a single dose of γ-irradiation (10 Gy) in the morning, followed by administration of BrdU for 48 h. During the day, mice received BrdU IP (100 mg/kg every 6 h) beginning immediately after the dose of radiation. During the overnight (active) period, BrdU was provided in the drinking water (1 mg/ml in water). Intestines were harvested for analysis 10 days after irradiation.
Telomerase Activity in BM.
Whole BM was sorted into GFP− and GFP+ populations, and 2,500 cells from each group were assayed for telomerase activity by using the TRAPeze Telomerase Detection Kit (Chemicon) per the manufacturer's protocol. Heat-inactivated samples were used as controls.
TRAP Assay, RT-PCR, and Ki67 FACS Analysis.
These procedures are described in SI Methods.
Supplementary Material
Acknowledgments.
We thank J. A. Majzoub, E. Gussoni, S. Bonner-Weir, M. Thompson, and R.A. DePinho for key insights; H. Ye, Y. Zhou, H. El Tomi, A. Payne-Tobin, and A. Flint for expert assistance; and S. Mayack, F. Kim, J. LaVecchio, and G. Buruzula at the Joslin-HSCI Flow Core. This work was supported by National Institutes of Health Grants 5T32 DK007699, K08 DK066305, JDRF 5–2005-216, and 1–2007-116 (to D.T.B.), R37 DK32658 (to R.K.M.), and 2T32 DK07260 (to I.M.M.); a Harvard Stem Cell Institute Seed Grant (to D.T.B.); National Institute of Child Health and Human Development Grant P30 HD18655 (to D.T.B.); the Timothy Murphy Fund; a Burroughs Wellcome Fund Career Award (to A.J.W.); a Smith Family New Investigator Award (to A.J.W.); and the Leukemia Research Foundation (to N.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804800105/DCSupplemental.
References
- 1.Kondo M, et al. Biology of hematopoietic stem cells and progenitors: Implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 2.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovell-Badge R. The future for stem cell research. Nature. 2001;414:88–91. doi: 10.1038/35102150. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong L, Lako M, Lincoln J, Cairns PM, Hole N. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech Dev. 2000;97:109–116. doi: 10.1016/s0925-4773(00)00423-8. [DOI] [PubMed] [Google Scholar]
- 6.Morrison SJ, Prowse KR, Ho P, Weissman IL. Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity. 1996;5:207–216. doi: 10.1016/s1074-7613(00)80316-7. [DOI] [PubMed] [Google Scholar]
- 7.Hiyama K, et al. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J Immunol. 1995;155:3711–3715. [PubMed] [Google Scholar]
- 8.Eisenhauer KM, Gerstein RM, Chiu CP, Conti M, Hsueh AJ. Telomerase activity in female and male rat germ cells undergoing meiosis and in early embryos. Biol Reprod. 1997;56:1120–1125. doi: 10.1095/biolreprod56.5.1120. [DOI] [PubMed] [Google Scholar]
- 9.Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation in multicellular organisms: Turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee HW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 12.Laird DJ, Weissman IL. Telomerase maintained in self-renewing tissues during serial regeneration of the urochordate Botryllus schlosseri. Dev Biol. 2004;273:185–194. doi: 10.1016/j.ydbio.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 13.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 14.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pericuesta E, et al. The proximal promoter region of mTert is sufficient to regulate telomerase activity in ES cells and transgenic animals. Reprod Biol Endocrinol. 2006;4:5. doi: 10.1186/1477-7827-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekaert S, Derradji H, Baatout S. Telomere biology in mammalian germ cells and during development. Dev Biol. 2004;274:15–30. doi: 10.1016/j.ydbio.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 18.Rossi DJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, et al. Identification of Lin(−)ScaI(+)kit(+)CD34(+)Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 21.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 22.Belair CD, Yeager TR, Lopez PM, Reznikoff CA. Telomerase activity: A biomarker of cell proliferation, not malignant transformation. Proc Natl Acad Sci USA. 1997;94:13677–13682. doi: 10.1073/pnas.94.25.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 24.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 25.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 26.Riou L, et al. The telomerase activity of adult mouse testis resides in the spermatogonial alpha6-integrin-positive side population enriched in germinal stem cells. Endocrinology. 2005;146:3926–3932. doi: 10.1210/en.2005-0502. [DOI] [PubMed] [Google Scholar]
- 27.Yui J, Chiu CP, Lansdorp PM. Telomerase activity in candidate stem cells from fetal liver and adult bone marrow. Blood. 1998;91:3255–3262. [PubMed] [Google Scholar]
- 28.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:1–11. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastos H, et al. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- 31.Venihaki M, Carrigan A, Dikkes P, Majzoub JA. Circadian rise in maternal glucocorticoid prevents pulmonary dysplasia in fetal mice with adrenal insufficiency. Proc Natl Acad Sci USA. 2000;97:7336–7341. doi: 10.1073/pnas.97.13.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gussoni E, et al. A method to codetect introduced genes and their products in gene therapy protocols. Nat Biotechnol. 1996;14:1012–1016. doi: 10.1038/nbt0896-1012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.