Abstract
The pathways that allow quiescent disseminated cancer cells to survive during prolonged dormancy periods are unknown. Here, we identify the transcription factor ATF6α as a pivotal survival factor for quiescent but not proliferative squamous carcinoma cells. ATF6α is essential for the adaptation of dormant cells to chemotherapy, nutritional stress, and, most importantly, the in vivo microenvironment. Mechanism analysis showed that MKK6 and p38α/β contribute to regulating nuclear translocation and transcriptional activation of ATF6α in dormant cancer cells. Downstream, ATF6α induces survival through the up-regulation of Rheb and activation of mTOR signaling independent of Akt. Down-regulation of ATF6α or Rheb reverted dormant tumor cell resistance to rapamycin and induced pronounced killing only of dormant cancer cells in vivo. Knocking down ATF6α also prolonged the survival of nude mice bearing dormant tumor cells. Targeting survival signaling by the ATF6α-Rheb-mTOR pathway in dormant tumor cells may favor the eradication of residual disease during dormancy periods.
Keywords: dormancy, endoplasmic reticulum stress, quiescence, p38, MKK6
Minimal residual disease caused by solitary disseminated tumor cells (DTC) is a well recognized event associated with unfavorable patient prognosis (1, 2). These cells, which usually stain negative for proliferation markers (e.g., Ki67) (3, 4), may be the source of tumor recurrence that can develop up to decades after treatment of the primary tumor (3, 4). These findings suggest that tumor cells may be able to reprogram into a quiescent behavior upon specific cues from the microenvironment (5). This may result from growth arrest and survival programs that allow dormant tumor cells to resist therapy and survive for long periods of time. Understanding how dormant DTCs survive dissemination and therapy and persist in a viable state for prolonged periods is of fundamental clinical interest.
Despite clinical evidence that dormancy may arise from tumor cell quiescence, models that recapitulate such a behavior are scarce (5, 6). We hypothesized that tumor cells able to reversibly acquire a nontumorigenic phenotype may recapitulate the programs of stress resistance that result in tumor cell quiescence. We previously identified a highly tumorigenic and metastatic human squamous carcinoma cell line (T-HEp3) that, when passaged in culture for >40 generations, reprograms into a reversible dormant phenotype (D-HEp3). This behavior is revealed upon inoculation of the cells in vivo and is durable for months (7–9). Mechanistic studies showed that dormancy is due to a G0–G1 arrest triggered in part by a low ERK/p38 signaling ratio and activation of the endoplasmic reticulum (ER) stress kinase PERK (10). However, the signals responsible for the prolonged survival of these quiescent tumor cells in vivo had remained unknown.
The transcription factor ATF6 is an important survival mediator upon ER stress. Although D-HEp3 cells display an ER stress response characterized by PERK activation and chaperone up-regulation (10), the role of ATF6 for these cells was unknown. Upon ER stress, ATF6, which is anchored to the ER membrane, translocates from the ER to the Golgi apparatus, where it is cleaved by S1P and S2P proteases (11). The cytosolic domain of ATF6 then translocates into the nucleus and activates transcription of unfolded protein response genes. Unlike ATF6β, ATF6α is required for cellular adaptation to acute and chronic ER stress in mouse embryonic fibroblasts (MEFs) (12). These data suggest that ATF6α is an important mediator of survival upon ER stress required for cell and tissue homeostasis.
Little is known about the functional role of ATF6 isoforms in cancer. Query of the Oncomine (13) database showed that head and neck cancer patients whose primary tumors expressed higher ATF6 mRNA levels were more likely to be lymph node positive, which could be interpreted as a positive association with enhanced survival of DTC (14). Similarly, a multicancer study showed higher ATF6 expression in metastases vs. primary lesions (15), and colon cancer patients with increased expression of ATF6 in their primary tumors had higher chances of relapse (16). This led us to hypothesize that ATF6α might mediate adaptation and survival either during quiescence or later during growth phases in secondary sites. We further hypothesized that our model of HEp3 carcinoma dormancy is amenable for testing such mechanisms. Here, we report that ATF6α serves as a basal and stress survival factor for dormant but not tumorigenic squamous carcinoma cells. We show further that ATF6α contributes to Rheb-mTOR pathway activation and rapamycin resistance and mediates survival signaling independently of Akt. Finally, we show that genetic ablation of ATF6α results in dormant cancer cell apoptosis and an extension of the dormancy period with delayed onset of recurrent growth.
Results
ATF6α Signaling Is Constitutive in Dormant but Not Malignant Variants of HEp3 Cells.
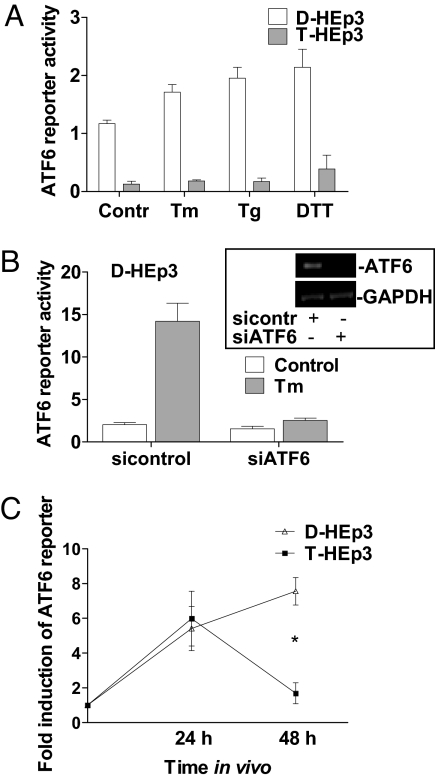
We first analyzed ATF6 signaling in dormant D-HEp3 and tumorigenic T-HEp3 cells by monitoring the activity of a 5xATF6-GL3 Luciferase construct containing the ATF6 consensus binding site that had been previously used to measure ATF6 and XBP-1 signaling (see Materials and Methods and refs. 11 and 17). We found ≈10-fold-higher ATF6 reporter activation in D-HEp3 vs. T-HEp3 cells under basal conditions (Fig. 1A). Furthermore, treatments with the ER stressors tunicamycin (Tm), thapsigargin (Tg), or DTT in serum-free media could further induce the ATF6 reporter activity in D-HEp3 cells, but only by ≈2-fold. These stressors had no effect on ATF6 reporter activity in T-HEp3 cells, suggesting that the low basal activity is not inducible. Using a siRNA that targets ATF6α in D-HEp3 cells, we could inhibit basal ATF6 reporter activation up to 50% and its Tm-inducibility almost completely (Fig. 1B). A siRNA to XBP-1 only had a marginal effect on Tm-induced reporter activation [supporting information (SI) Fig. S1A]. These results suggest that in D-HEp3 cells, the ATF6 reporter reflects mostly ATF6α and not XBP-1 activation.
Fig. 1.
In vitro and in vivo ATF6α transcriptional activity in HEp3 cells. (A) Basal 5xATF6-GL3 activity (serum-free) or treated with 5 μg/ml Tm, 1 μM Tg, or 1 mM DTT for 6 h. (B) 5xATF6-GL3 activity in D-HEp3 cells (full serum) with a siRNA to ATF6α with or without 5 μg/ml Tm for 6 h. The Inset shows ATF6α mRNA knockdown (RT-PCR). (C) Induction of 5xATF6-GL3 activity in D-HEp3 and T-HEp3 in vitro (time point 0) or 24 and 48 h after inoculation on CAMs in vivo. *, P < 0.0001.
To determine whether ATF6 is activated during adaptation to the tissue microenvironment, we monitored the ATF6 reporter in D-HEp3 and T-HEp3 cells inoculated in vivo on the chorioallantoic membrane (CAM) of live E9-E10 chicken embryos. This extraembryonic tissue provides a highly vascularized stromal microenvironment supportive of tumor cell survival and/or growth (7). Inoculation of HEp3 cells on CAMs resulted in a 6-fold induction of ATF6 reporter activity after 24 h in vivo in both cell lines. At 48 h, this response persisted only in D-HEp3 cells (Fig. 1C). This induction was much stronger than that obtained with ER stressors (6-fold versus 2-fold) suggesting that the in vivo microenvironment imposes high levels of stress that result in ATF6 signaling in both HEp3 cell lines. The persistent activation of ATF6 signaling in dormant tumor cells paralleled that observed for p38 in vivo (8, 9). Activation of p38 is functionally linked to the quiescence and survival of dormant cells (8, 10) opening the question as to how ATF6 is regulated and whether it depends on p38 signaling.
Subcellular Localization and Activation of ATF6α in Quiescent HEp3 Cells Depends on p38 Signaling.
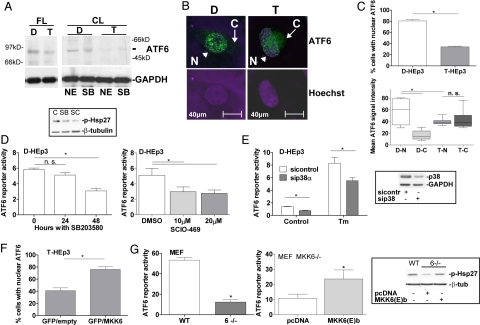
We previously showed that D-HEp3 cell quiescence is largely due to high p38 and ER-stress signaling (10). Thus, we measured expression levels, activation, localization, and p38 dependence of ATF6α in T-HEp3 and D-HEp3 cells. Western blots showed that basal full-length ATF6α levels were higher in D-HEp3 cells than in T-HEp3 cells and that processing into the cleaved nuclear form of ATF6α was more abundant in D-HEp3 than in T-HEp3 cells (Fig. 2A). Confocal microscopy and immunofluorescence (IF) revealed, that in D-HEp3 cells, ATF6α is predominantly localized in the nucleus (Fig. 2B, arrowhead). In contrast, whereas some nuclear translocation is observed in T-HEp3 cells, ATF6α can also be prominently detected in their cytoplasm, a signal almost absent in D-HEp3 cells (Fig. 2B, arrow and 2C). These findings suggest that lower ATF6α nuclear translocation may reflect the low basal ATF6 reporter activity in T-HEp3 cells and that ATF6α might play a role in the D-HEp3 cell dormant phenotype.
Fig. 2.
ATF6α localization in HEp3 is partly MKK6-p38-dependent. (A) Detection of full-length (FL) ATF6α by Western blot in D-HEp3 (D) and T-HEp3 (T) cell RIPA lysates (Left). Cleaved (CL) ATF6α in extracts enriched in nuclear components (commercial kit) (NE) or Laemmli buffer (SB) (Right). (B) IF for ATF6α and nuclear detection with Hoechst-33342 in D- vs. T-HEp3 [N (nuclear), arrowhead; C (cytoplasmic), arrow]. (C) Percentage of nuclear ATF6α-staining. A total of ≈300 cells in three independent experiments was scored. *, P = 0.0015 (Upper). Nuclear (N) vs. cytoplasmic (C) densitometric fluorescence quantification of ATF6α signal for D-HEp3 (D) and T-HEp3 (T) by using ImageJ. *, P < 0.05 (Lower). (D) Basal ATF6 reporter activity in D-HEp3 treated with 10 μM SB203580 (SB) for 24/48 h (Left) and 10–20 μM SCIO-469 (SC) for 48 h (Right). *, P < 0.05. The Inset shows decreased p-Hsp27 with both drugs by Western blot. (E) Basal and Tm-induced ATF6 reporter activity in D-HEp3 with and without siRNAs to p38α. *, P < 0.05. The Inset shows the level of p38 knockdown by Western blot. (F) Percentage of T-HEp3 cells with nuclear ATF6α after cotransfection of GFP with constitutively active MKK6(E)b. A total of ≈300 GFP-positive cells were scored in three experiments. *, P < 0.001. (G) Basal ATF6 reporter activity in WT or MKK6−/− MEFs (Left). *, P < 0.001. ATF6 reporter activity in MKK6−/− MEFs transfected with pcDNA3.1 or constitutively active MKK6(E)b (Right). *, P < 0.05. The Inset shows a Western blot for p-Hsp27 in these cells.
Next, we determined whether ATF6α activation depends on p38 in D-HEp3 cells. Pharmacologic or genetic inhibition of p38 with 10 μM SB203580, 10–20 μM SCIO-469 (a specific p38α inhibitor) (18), or siRNA to p38α resulted in a 50% reduction of ATF6 reporter activity, suggesting that ATF6α activity is at least in part p38α/β-dependent (Fig. 2 D and E). SB203580 treatment for 24 or 48 h also reduced ATF6α nuclear translocation in D-HEp3 cells (Fig. S1B). Conversely, cotransfection of T-HEp3 cells that have low p38 activity (8) with a constitutively active MKK6b(E), an upstream activator of p38 (19), significantly increased the percentage of cells with ATF6α nuclear localization (Fig. 2F). To further support the involvement of MKK6 in ATF6 activation, we tested ATF6 reporter activity in WT and MKK6−/− MEFs (20). Compared with WT MEFs from control littermates, normalized ATF6 reporter activity was significantly reduced in MKK6−/− MEFs (Fig. 2G Left). Transfection of MKK6b(E) in MKK6−/− cells resulted in rescue of p38 activity to WT levels as shown by Western blot analysis for phosphorylated Hsp27 and, more importantly, ATF6 reporter activity (Fig. 2G Right and Inset). These data show that in D-HEp3 cells or MEFs, ATF6α transcriptional activity shows dependence on MKK6-p38α/β signaling. We conclude that in contrast to T-HEp3, D-HEp3 cells express more ATF6α, which is nuclear at a higher frequency and activated by a pathway that depends on MKK6 and p38α/β.
ATF6α Is Required for the Prolonged Survival of Dormant Tumor Cells in Vivo.
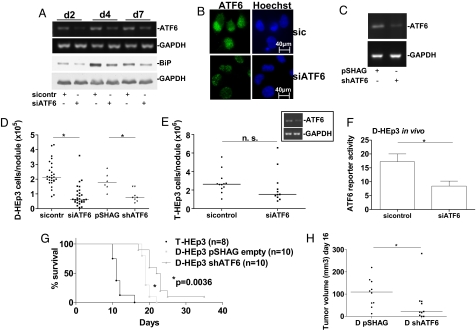
In vitro studies showed that ATF6α is required for D-HEp3 cells to withstand various types of stress, including the TOPO II inhibitor doxorubicin, Tm, and glucose restriction (SI Text and Fig. S1C). These data and our findings that ATF6α signaling is activated in vivo led us to ask whether ATF6α plays a role in prolonged D-HEp3 cell survival or growth arrest. We tested the survival/growth of D-HEp3 or T-HEp3 cells with knockdown of ATF6α after 1 week on the CAM. At this time, D-HEp3 cells are fully arrested, and T-HEp3 cells have undergone at least six population doublings (8, 21). The siRNA to ATF6α persistently down-regulated the ATF6α gene transcript, its target gene BiP (Fig. 3A and Fig. S1D), and ATF6α protein levels (Fig. 3B). D-HEp3 cells stably expressing a shRNA to ATF6α after retroviral delivery also showed significant ATF6α mRNA knockdown (Fig. 3C). We found that after 1 week in vivo, transient or stable RNAi targeting of ATF6α caused a statistically significant decrease in the number of viable cells per tumor nodule formed by D-HEp3 cells (Fig. 3D). This suggests that whereas arrested control cells remain viable (8, 9), D-HEp3 cells with ATF6α down-regulation are unable to survive in the new tissue microenvironment. ATF6α knockdown resulted in ≈3-fold higher cleaved caspase-3 levels in D-HEp3 cells as early as 24 h after in vivo inoculation (Fig. S1E). ATF6α knockdown in the tumorigenic T-HEp3 cells did not affect apoptosis (Fig. S1E and data not shown) and growth on CAMs (Fig. 3E). This indicates that ATF6α is dispensable for T-HEp3 survival. The increased in vivo apoptosis of D-HEp3 cells with ATF6α down-regulation correlated with reduced ATF6 reporter activity 48 h after in vivo inoculation (Fig. 3F). These results strongly suggest that survival of D-HEp3 cells in vivo critically depends on ATF6α signaling.
Fig. 3.
ATF6α is required for survival of D-HEp3 cells in vivo. (A) ATF6α mRNA and BiP protein expression after ATF6α knockdown [RT-PCR for ATF6α (Upper) and Western blot for BiP (Lower)]. (B) IF for ATF6α in D-HEp3 expressing a control siRNA or a siRNA to ATF6α, nuclear staining with Hoechst-33342. Mean nuclear pixel intensity was 40.7 ± 3.6 for sicontrol vs. 16.4 ± 1.5 for siATF6α. *, P = 0.0002. (C) shRNA-mediated down-regulation of ATF6α mRNA (RT-PCR). (D and E) Number of D-HEp3 cells treated with and without ATF6α siRNA (left two columns) and shRNA (right two columns) (D) and T-HEp3 cells treated with and without ATF6α siRNA (E) or controls after 7 days on CAMs. *, P < 0.001 for siRNA and P = 0.008 for shRNA in D-HEp3. The Inset in E shows ATF6α knockdown by RT-PCR in T-HEp3. (F) In vivo ATF6 activity in D-HEp3 cells expressing siATF6α 48 h after inoculation on CAMs, duplicate experiments and SEM. *, P = 0.008. (G) Disease-free survival of BALB/c nude mice injected with T-HEp3 (positive control) and D-HEp3 expressing shATF6α or a pSHAG empty vector. P < 0.001 for T- vs. D-pSHAG and P = 0.0036 for D-pSHAG vs. D-shATF6α. (H) Median tumor volume on day 16 in D-HEp3 cells expressing the pSHAG empty vector or shATF6α. *, P < 0.05.
To determine whether ATF6α down-regulation enhances the survival of mice carrying dormant tumor cells, animals were injected s.c. with T-HEp3 cells or D-HEp3 cells expressing shATF6α or the pSHAG empty vector. Mice carrying D-HEp3 cells expressing shATF6α showed a statistically significant delayed tumorigenic take (16 vs. 20 days, data not shown) and a prolonged disease-free survival (19 vs. 22.5 days, Fig. 3G) when compared with control cells. One mouse in the ATF6α shRNA group never developed a tumor up to 35 days after injection. Although the median T-HEp3 tumor volume at 16 days, when all T-HEp3 bearing mice had been killed, was 590 mm3 (data not shown), median D-HEp3 tumor volume was 109.5 mm3 for control cells and 20.7 mm3 for cells expressing shATF6α (Fig. 3H). Tumors originating from D-HEp3 cells expressing shATF6α had no detectable ATF6α knockdown as determined by RT-PCR (data not shown), suggesting a selection against low levels of this transcript. Taken together, our in vivo data support that ATF6α is a critical survival factor in dormant HEp3 cells. Its down-regulation causes in vivo apoptosis of dormant tumor cells, therefore significantly prolonging overall survival of mice carrying dormant disease.
ATF6α Mediates Survival Through Rheb and mTOR Independently of Akt-Signaling.
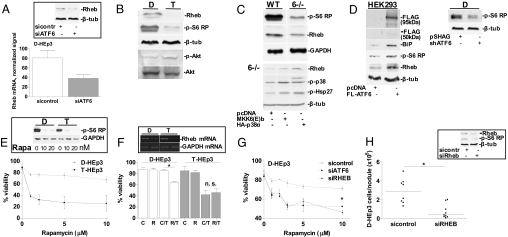
D-HEp3 cells display a chronic unfolded protein response (UPR) (10), suggesting that ATF6α activation is regulating a survival program under basal conditions. Gene expression profiling of D-HEp3 cells after ATF6α knockdown by siRNA, revealed 193 and 20 genes to be significantly up- and down-regulated, respectively. As reported recently (12), ATF6α targets included genes in the general categories of protein folding, protein trafficking/secretion, protein degradation, and nutrient transport (Table S1). Knockdown of ATF6α-regulated genes in these categories, including SCG II, SLC2A3 (Table S1), and BiP/Grp78 did not significantly affect in vivo survival of D-HEp3 cells (data not shown). This suggested that other less obvious ATF6α target genes mediated the observed effect. The expression profiles revealed that ATF6α induces the expression of Ras homolog enriched in brain (Rheb), a critical activator of the mammalian target of rapamycin (mTOR), a pathway that propagates strong cell survival signals (22). Rheb mRNA levels in the arrays were down-regulated 2- to 2.5-fold by ATF6α knockdown (Fig. 4A), a result that was confirmed by Western blot analysis (Fig. 4A Inset). This suggested that in D-HEp3 cells, ATF6α may signal for survival through a Rheb-mTOR pathway. We also found that Rheb expression levels in T-HEp3 cells are lower than in D-HEp3 cells and that levels of phosphorylated S6 ribosomal protein were reduced in T-HEp3 than D-HEp3 cells (Fig. 4B). However, T-HEp3 cells showed higher p-Akt levels than D-HEp3 cells (Fig. 4B). This suggested that D-HEp3 cells are able to achieve strong mTOR-S6 signaling through up-regulation of Rheb and by creating perhaps less dependence on active Akt. To explore the generality of Rheb-S6 regulation by ATF6α, we compared WT vs. MKK6−/− MEFs that show reduced ATF6α signaling (Fig. 2G). We found MKK6−/− MEFs to have lower levels of Rheb and markedly decreased phosphorylation of S6 ribosomal protein (Fig. 4C Upper), suggesting that the p38-ATF6α-mTOR pathway can be active in murine cells. Expression of a constitutively active MKK6(E)b or overexpression of p38α (23) in MKK6−/− cells resulted in higher Hsp27 phosphorylation and increased Rheb expression levels (Fig. 4C Lower). Accordingly, overexpression of ATF6 in HEK293 cells caused up-regulation of Rheb and increased phosphorylation of S6 ribosomal protein (Fig. 4D Left). Conversely, stable or transient down-regulation of ATF6 (shRNA vs. siRNA) caused decreased phosphorylation of S6 ribosomal protein in D-HEp3 cells (Fig. 4D Right and Fig. S1F). Taken together, these data suggest that Rheb signaling to S6 ribosomal protein can be regulated by ATF6α and this is not only limited to the HEp3 model but may be operating to a certain extent in immortalized MEFs. The higher mTOR signaling in D-HEp3 cells is consistent with the markedly higher resistance of these cells to rapamycin when compared with T-HEp3, even at high concentrations of the drug (Fig. 4E).
Fig. 4.
Identification of an ATF6α-Rheb-mTOR survival pathway in D-HEp3. (A) Normalized Rheb signal in the gene arrays of D-HEp3 with or without ATF6α knockdown. The Inset shows Rheb protein levels. (B) Western blots for Rheb, p-S6 ribosomal protein (p-S6 RP), p-Akt, and Akt in D-HEp3 vs. T-HEp3. (C) Western blots for Rheb and p-S6 RP in WT and MKK6−/− MEFs (Upper). Western blots for Rheb, p-p38 and p-Hsp27 in MKK6−/− MEFs transfected with pcDNA3.1 Neo, constitutively active MKK6(E)b or HA-p38α (Lower). (D) Western blots for FLAG, BiP, p-S6 RP, and Rheb in HEK293 cells transfected with pcDNA3.1 or 3xFLAG-ATF6 (Left). p-S6-RP in D-HEp3 cells stably expressing shATF6 (Right). (E) Viability curve for rapamycin in D- vs. T-HEp3. (Inset) mTOR inhibition by 10–20 nM rapamycin measured by Western blot for p-S6 RP. (F) Tm sensitivity in D-HEp3 vs. T-HEp3 after Rheb knockdown by siRNA (R) or in control siRNA cells (C), viability after Tm (T) 5 μg/ml for 24 h. *, P < 0.0001. (Insets) RT-PCR in both cell lines showing Rheb mRNA knockdown. (G) Rapamycin sensitivity in D-HEp3 cells expressing a control siRNA, a siRNA to ATF6α, or a siRNA to Rheb. Viability was assessed by using Trypan blue exclusion test. *, P < 0.05. (H) Number of D-HEp3 cells with or without a Rheb siRNA recovered after 7 d on CAMs. *, P = 0.0025. The Inset shows a Western blot for Rheb and p-S6 RP.
We next tested whether Rheb signaling is functionally linked to D-HEp3 cell survival. Although siRNAs to Rheb did not affect basal viability in T- or D-HEp3 cells, Rheb knockdown resulted in a sensitization to Tm in D-HEp3 cells only (Fig. 4F). Also, both ATF6α and Rheb knockdown sensitized D-HEp3 cells to rapamycin, showing the importance of mTOR activation by an ATF6α-Rheb signal (Fig. 4G). In agreement with the hypothesis that ATF6α up-regulation of Rheb confers D-HEp3 cells with a selective survival advantage, knockdown of Rheb that also caused a reduction in p-S6 ribosomal protein phosphorylation (Fig. 4H Inset) resulted in a marked decrease only in D-HEp3 survival in vivo (Fig. 4H). T-HEp3 cells remained unaffected by the siRNA to Rheb (Fig. S1G). All in all, this indicates that ATF6α is not only an important transducer of ER stress signals but also regulates Rheb signaling to promote survival. In D-HEp3 cells, this leads to stress resistance in vitro and prolonged survival in a dormant state in vivo.
Discussion
The present study demonstrates that in dormant carcinoma cells, ATF6α transduces survival signals through a ATF6α-Rheb-mTOR pathway. We explored under which stimuli ATF6α may serve as a survival factor and found that like BiP (10), ATF6α signaling is important for protection against TOPO II inhibitors, ER, and low glucose stress. However, a very important function of ATF6α that diverged from BiP was that disruption of its signaling prevents dormant, but not proliferative, tumor cells to adapt to the in vivo microenvironment. ATF6 reporter measurements in vivo further substantiate this notion because both T-HEp3 and D-HEp3 cells activated the ATF6 reporter to the same extent, but only D-HEp3 cells maintained this activation. Thus, it is possible that although T-HEp3 cells might use ATF6α for the initial adaptation to the in vivo microenvironment, they ultimately do not rely on it for their expansion. This suggests that redundant survival signals overcome ATF6α deficiency in expanding tumor masses but that high ATF6α-Rheb-mTOR signaling is a survival advantage under situations of adaptation such as tumor cell quiescence.
The higher levels of ATF6α in D-HEp3 than in T-HEp3 cells are consistent with previous studies (10) showing that D-HEp3 cells display a UPR characterized by higher p-PERK, XBP-1 splicing, and chaperone expression that depend in part on p38α/β activation (10). Our data support that p38α/β signaling is an important contributor, although not the only one, to ATF6α activation. Our results using MEFs with homozygous deletion for MKK6, and HEp3 cells, in which p38 activity was stimulated by an active MKK6 mutant or inhibited with SB203580 or SCIO-469, showed that ATF6α nuclear translocation and transcriptional activation depends on MKK6 and p38α/β signaling. The p38α RNAi and SCIO-469 experiments further support that this particular isoform is required for ATF6α activation in D-HEp3 cells. These findings are in agreement with those in normal cells showing that p38 directly phosphorylates ATF6α to regulate chaperone expression (24, 25). Our studies further define a contribution of MKK6 and at least p38α in the activation of ATF6α in dormant carcinoma cells. However, because BiP was not mediating the ATF6α survival signal in vivo, it became important to further identify the mechanism.
Our gene array and validation studies revealed that the GTPase Rheb was induced by ATF6α. Rheb is an important mediator of survival linked to the sensing of nutritional stress and an immediate upstream activator of mTOR, a strong survival regulator in tumors (26, 27). Knockdown of Rheb sensitized only D-HEp3 cells to Tm, reduced their Rapamycin-resistance, and, most importantly, resulted in a marked reduction in D-HEp3 cell survival in vivo. Unlike BiP or other genes involved in secretion or glucose transport (data not shown), which are major pathways regulated by ATF6α (12), Rheb might be an important regulator of ATF6α-induced in vivo survival. D-HEp3 cells express higher levels of Rheb and display stronger S6 ribosomal protein phosphorylation than T-HEp3 cells. Furthermore, in MKK6−/− MEFs with reduced ATF6α activity, Rheb protein levels and p-S6 were reduced, suggesting that transcriptional activation of ATF6α by MKK6-p38 is functional in murine cells for inducing Rheb expression and mTOR signaling. A distinguishing feature was that T-HEp3 cells have higher p-Akt levels than D-HEp3 cells, but S6 phosphorylation is still higher in the latter. This suggests that although in D-HEp3 cells Akt activation is low, probably because of low EGFR and FAK signaling (28), these cells are able to maintain strong mTOR activation and survival through an alternative induction of Rheb by ATF6α.
There is limited evidence of the role of ATF6α in cancer. Our studies show that p38 signaling through ATF6α, Rheb, and mTOR may have a prosurvival function in solid tumor cells, particularly during a quiescent phase. These findings may be of relevance because cancer patient gene expression profiling data in the Oncomine database (13) revealed a positive correlation between high ATF6 expression in head and neck and colorectal cancer primary tumors and the propensity of patients to develop lymph node metastasis (14) and relapse (16) (Fig. S2). A weaker but similar trend was found for Rheb in head and neck cancer studies (29) (Fig. S2). It is possible that ATF6α and Rheb may have a survival function that allows dormant HNSCC cells to resist changes in the tissue microenvironment but also to survive nutritional or chemotherapy-induced stress. The latter is substantiated by the fact that ATF6α knockdown sensitizes D-HEp3 cells to doxorubicin-induced cell death. This might occur through the up-regulation of BiP, as in D-HEp3 cells BiP protected from TOPO II inhibitors, through the inhibition of Bax (10). Our data suggest that ATF6α regulates two arms of a survival pathway; one that will respond to ER-, nutritional- or chemotherapy-induced stress through BiP and another that may protect against these insults and/or microenvironment-derived stress through Rheb and mTOR.
In summary, our findings reveal a pathway where ATF6α up-regulates Rheb to activate mTOR, selectively enhancing dormant cell survival in vivo. In addition, we identified a previously unrecognized cross-talk among p38, ATF6α, and mTOR signaling pathways that may be of general relevance because it is active in both human and mouse cells. The interaction between ATF6α and mTOR signaling appears to confer resistance of D-HEp3 cells to doxorubicin and to the mTOR inhibitor rapamycin, revealing a potential drug resistance mechanism. Taken together, our data identifies the ATF6α-Rheb-mTOR axis as a new target to inhibit the survival of dormant tumor cells.
Materials and Methods
Reagents, Antibodies, and Cell Lines.
For a full list of reagents and antibodies, see SI Text. T- and D-HEp3 human squamous carcinoma cells were cultured as described (7, 8). MKK6−/− and wt MEFs were kindly provided by Roger Davis (University of Massachusetts Medical School, Worcester).
Stable shRNA-Mediated ATF6α Knockdown and siRNA Transfection and RT-PCR.
Transduction of D-HEp3 cells with the shRNA plasmid to ATF6α in the pSHAG-MAGIC cassette was performed as described (30); and see SI Text. All resistant clones were pooled to avoid clonal variability. SiRNA transfections were performed by using siPORT NeoFX (Ambion). For RT-PCR, primer sequences, and siRNAs, see SI Text.
IF and Immunoblotting (IB).
IF for ATF6α and p-S6 RP was performed as described (10, 30); and see SI Text. For Caspase-3 IF, cells from tumor nodules in vivo were attached to poly-l-lysine (Sigma) -coated coverslips. Images were captured by using a Leica Confocal or a Nikon Eclipse TS100 microscope. In some experiments, D-HEp3 cells were treated with 10 μM SB203580 for 24/48 h, and T-HEp3 cells were transfected with eGFP (Clontech) and the MKK6(E)b plasmid or with pCDNA3.1Neo (Clontech). Quantification was performed by using NIH ImageJ 1.38. IB was performed as described (8).
Luciferase (Luc) Assays.
Luc assays were performed as described (31). The 5xATF6-GL3 and 3xFLAG-ATF6 plasmids were kindly provided by Ron Prywes (Columbia University, New York), the pcDNA3–3HA-p38αWT plasmid by David Engelberg (The Hebrew University of Jerusalem, Israel), the Renilla-Luc plasmid was from Clontech, and activity was assayed by using the Dual Luciferase Assay kit (Promega).
Xenograft Studies.
Cells were grown on CAMs as described (8, 9); and see SI Text. For nude mice experiments, 0.5 × 106 cells were inoculated s.c. in the interscapular region of 2- to 3-month-old female BALB/c nude mice (Taconic Farms). Tumor growth was measured daily by using a caliper. All experiments were approved by the State University of New York Institutional Animal Care and Use Committee.
Microarray.
Four Affymetrix Hgu133a chips were run in the State University of New York (Albany, NY) Center for Functional Genomics with the following samples: two siRNA to ATF6α (48-h transfection) and two siRNA control. Raw data were background-corrected, normalized, and summarized by using mas5 and Genespring. Statistically significant differences were expressed as ≥1.5-fold changes in cells with ATF6α knockdown compared with the control.
Statistical Analysis.
Statistical analysis was performed by using GraphPad Prism 5.0 for Windows from GraphPad Software. Two means were compared by using the unpaired t test or ANOVA with Bonferroni multiple comparison, whereas in vivo data were analyzed by using the Mann–Whitney or Kruskall–Wallis test followed by Dunn's multiple comparison. Survival differences were calculated by using the Mantel–Cox log-rank test. P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments.
We thank Dr. Roger Davis for the WT and MKK6−/− MEFs and Dr. Nabeel Bardeesy (Massachusetts General Hospital, Harvard University, Boston, MA) for providing the initial evidence on S6-phosphorylation and for fruitful discussions. This work was supported by grants from the Samuel Waxman Cancer Research Foundation Tumor Dormancy Program and National Institutes of Health/National Cancer Institute Grant CA109182 (to J.A.A.-G.) and a Dr. Mildred-Scheel postdoctoral grant by the Deutsche Krebshilfe (to D.M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800939105/DCSupplemental.
References
- 1.Lindemann F, Schlimok G, Dirschedl P, Witte J, Riethmuller G. Prognostic significance of micrometastatic tumour cells in bone marrow of colorectal cancer patients. Lancet. 1992;340:685–689. doi: 10.1016/0140-6736(92)92230-d. [DOI] [PubMed] [Google Scholar]
- 2.Izbicki JR, et al. Prognostic value of immunohistochemically identifiable tumor cells in lymph nodes of patients with completely resected esophageal cancer. N Engl J Med. 1997;337:1188–1194. doi: 10.1056/NEJM199710233371702. [DOI] [PubMed] [Google Scholar]
- 3.Meng S, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 4.Schardt JA, et al. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell. 2005;8:227–239. doi: 10.1016/j.ccr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Ossowski L, Reich E. Changes in malignant phenotype of a human carcinoma conditioned by growth environment. Cell. 1983;33:323–333. doi: 10.1016/0092-8674(83)90414-2. [DOI] [PubMed] [Google Scholar]
- 8.Aguirre-Ghiso JA, Liu D, Mignatti A, Kovalski K, Ossowski L. Urokinase receptor and fibronectin regulate the ERK(MAPK) to p38(MAPK) activity ratios that determine carcinoma cell proliferation or dormancy in vivo. Mol Biol Cell. 2001;12:863–879. doi: 10.1091/mbc.12.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguirre-Ghiso JA, Ossowski L, Rosenbaum SK. Green fluorescent protein tagging of extracellular signal-regulated kinase and p38 pathways reveals novel dynamics of pathway activation during primary and metastatic growth. Cancer Res. 2004;64:7336–7345. doi: 10.1158/0008-5472.CAN-04-0113. [DOI] [PubMed] [Google Scholar]
- 10.Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006;66:1702–1711. doi: 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen J, Prywes R. ER stress signaling by regulated proteolysis of ATF6. Methods. 2005;35:382–389. doi: 10.1016/j.ymeth.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes DR, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginos MA, et al. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 15.Ramaswamy S, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YH, et al. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin Cancer Res. 2007;13:498–507. doi: 10.1158/1078-0432.CCR-05-2734. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 18.Hideshima T, et al. p38 MAPK inhibition enhances PS-341 (bortezomib)-induced cytotoxicity against multiple myeloma cells. Oncogene. 2004;23:8766–8776. doi: 10.1038/sj.onc.1208118. [DOI] [PubMed] [Google Scholar]
- 19.Alpert D, Schwenger P, Han J, Vilcek J. Cell stress and MKK6b-mediated p38 MAP kinase activation inhibit tumor necrosis factor-induced IkappaB phosphorylation and NF-kappaB activation. J Biol Chem. 1999;274:22176–22183. doi: 10.1074/jbc.274.32.22176. [DOI] [PubMed] [Google Scholar]
- 20.Brancho D, et al. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003;17:1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguirre-Ghiso JA, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol. 1999;147:89–104. doi: 10.1083/jcb.147.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Diskin R, Askari N, Capone R, Engelberg D, Livnah O. Active mutants of the human p38alpha mitogen-activated protein kinase. J Biol Chem. 2004;279:47040–47049. doi: 10.1074/jbc.M404595200. [DOI] [PubMed] [Google Scholar]
- 24.Thuerauf DJ, et al. p38 Mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum response element. A potential role for the transcription factor ATF6. J Biol Chem. 1998;273:20636–20643. doi: 10.1074/jbc.273.32.20636. [DOI] [PubMed] [Google Scholar]
- 25.Luo S, Lee AS. Requirement of the p38 mitogen-activated protein kinase signalling pathway for the induction of the 78-kDa glucose-regulated protein/immunoglobulin heavy-chain binding protein by azetidine stress: Activating transcription factor 6 as a target for stress-induced phosphorylation. Biochem J. 2002;366:787–795. doi: 10.1042/BJ20011802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Hanrahan J, Blenis J. Rheb activation of mTOR and S6K1 signaling. Methods Enzymol. 2005;407:542–555. doi: 10.1016/S0076-6879(05)07044-8. [DOI] [PubMed] [Google Scholar]
- 28.Aguirre-Ghiso JA. Inhibition of FAK signaling activated by urokinase receptor induces dormancy in human carcinoma cells in vivo. Oncogene. 2002;21:2513–2524. doi: 10.1038/sj.onc.1205342. [DOI] [PubMed] [Google Scholar]
- 29.Roepman P, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37:182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 30.Sequeira SJ, et al. Inhibition of proliferation by PERK regulates mammary acinar morphogenesis and tumor formation. PLoS ONE. 2007;2:e615. doi: 10.1371/journal.pone.0000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguirre-Ghiso JA, Estrada Y, Liu D, Ossowski L. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK) Cancer Res. 2003;63:1684–1695. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.