Abstract
The concerted action of four viral glycoproteins and at least one cellular receptor is required to catalyze herpes simplex virus 1 entry into host cells either by fusion at the plasma membrane or intracellularly after internalization by endocytosis. Here, we applied cryo electron tomography to capture 3D intermediates from Herpes simplex virus 1 fusion at the plasma membrane in their native environment by using two model systems: adherent cells and synaptosomes. The fusion process was delineated as a series of structurally different steps. The incoming capsid separated from the tegument and was closely surrounded by the cortical cytoskeleton. After entry, the viral membrane curvature changed concomitantly with a reorganization of the envelope glycoprotein spikes. Individual glycoprotein complexes in transitional conformations during pore formation and dilation revealed the complex viral fusion mechanism in action. Snapshots of the fusion intermediates provide unprecedented details concerning the overall structural changes occurring during herpesvirus entry. Moreover, our data suggest that there are two functional “poles” of the asymmetric herpesvirion: one related to cell entry, and the other formed during virus assembly.
Keywords: cryo electron tomography, herpesvirus, synaptosomes, glycoproteins
Membrane fusion is essential for cell–cell fusion during development, intracellular membrane trafficking, exo- and endocytosis as well as virus entry into and egress from cells. Herpes simplex virus type 1 (HSV-1) enters host cells via membrane fusion. HSV-1 is the prototype of the family Herpesviridae, which includes a number of other human pathogens like varicella zoster virus, cytomegalovirus, Epstein–Barr virus, and Kaposi's sarcoma-associated herpesvirus. It has a complex structure consisting of an icosahedral, DNA-containing capsid, which is asymmetrically located within the virion and surrounded by an amorphous protein layer called the tegument, and a membrane envelope heterogeneously studded with morphologically distinct spikes formed by 12 different glycoprotein species (1). Five of these glycoproteins (gB, gC, gD, and the complex of gH and gL) mediate the entry of HSV-1 into host cells (2). The specific mode of entry is cell-type dependent. Neurons and Vero cells (an African monkey kidney cell line) are infected by direct fusion at the plasma membrane and subsequent release of the capsid into the cytosol, whereas in keratinocytes and HeLa cells, the primary step in entry involves endocytosis (3). After initial attachment by a nonessential interaction of the viral glycoprotein gC with heparan sulfate proteoglycans on the plasma membrane, entry proceeds by binding of gD to one of its three known cellular receptors: herpesvirus entry mediator (HVEM), nectin-1, or modified heparan sulfate (2). The heterodimer gH/gL and the homotrimer gB are required for membrane fusion (2). So far, it is unclear how the interactions between the four essential entry-associated viral glycoproteins are coordinated to mediate membrane fusion (4, 5) and what role the recently discovered gB receptor PILRα (6) has.
Here, we applied cryo electron tomography (cryo-ET) to probe the early events of HSV-1 infection comprehensively and in unprecedented detail. Using several adherent cell lines and synaptosomes as host systems, we addressed the structural reorganization of pathogen and host upon interaction. In particular, a systematic study addressing the intermediate states, i.e., the native dynamics of the fusion process in three dimensions at close to macromolecular resolution was missing. We characterized (i) entry sites at the plasma membrane, including changes in the membrane curvature during entry, and the fate of the glycoprotein spikes; (ii) the orientation of the asymmetric virion in relation to the plasma membrane during fusion; (iii) structural clues as to how the virus traverses the dense cortical actin barrier underneath the plasma membrane; and (iv) the details of the functional “gymnastics” of the herpesvirus glycoprotein spikes during fusion, that have been suggested (4) based on the available crystal structures (7, 8) and similarities to other viral fusion systems (9).
Results
HSV-1 Enters Vero, PtK2, and HFF Cells by Fusion at the Plasma Membrane.
Cells grown on a holey carbon support film of an electron microscopy grid were inoculated with HSV-1 and vitrified in growth medium. These native samples were directly examined in the frozen hydrated state by electron microscopy. We initially chose Vero cells as an established model system for HSV-1 fusion at the plasma membrane (10, 11). These adherent cells are not especially thin. Cellular cryo-ET (12), however, is limited to specimen areas with a maximal thickness of ≈1 μm (13), and therefore we could only analyze the outer rims of Vero cells. Two other cell types susceptible to HSV-1 infection, human foreskin fibroblasts (HFF) and rat kangaroo kidney cells (PtK2) (11, 14), provided an extended, sufficiently flat cell periphery, and thus were also included in our study, although their mode of entry was unknown. The cells were pretreated with nocodazole, a microtubule depolymerizing drug, to prevent dynein driven transport of incoming capsids from the periphery toward the cell nucleus (10, 11).
Entry events were captured in the cell lines Vero, PtK2, and HFF. All three were found to be entered by fusion of the HSV-1 envelope with the plasma membrane (Fig. 1 for PtK2; data for HFF and Vero cells not shown) as demonstrated by the presence of nonenveloped cytosolic capsids 2 minutes after the onset of infection, and recognition of the corresponding fusion sites at the plasma membrane. Taking advantage of the 3D tomographic information, we identified the sites of virus entry at the plasma membrane by virtue of the unique structural signature of the viral glycoproteins and the tegument (Fig. 1 D–G).
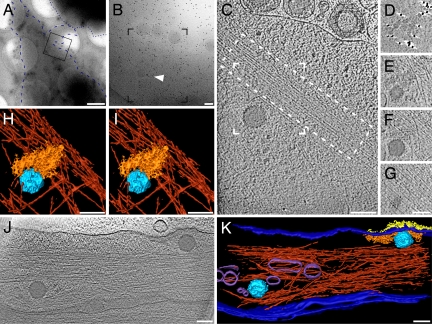
Fig. 1.
HSV-1 enters PtK2 cells by fusion at the plasma membrane. (A) Low-magnification electron micrograph of a PtK2 cell grown on a grid and inoculated with HSV-1 virions for 5 min at 37°C after virus binding on ice for 2 h and pretreatment with nocodazole. Whereas the central cellular areas were too thick for imaging and appear black, the cell periphery was sufficiently thin to allow tomographic imaging. The cell outline is accentuated by a blue, dashed line, a neighboring cell by a dotted blue line. The frame marks the area of a higher magnification projection image shown in B. Some virions are visible along the plasma membrane; the arrowhead highlights a capsid hardly recognizable in the projection image. Black dots are colloidal gold markers used for alignment of the tilt series. (C) A 3.2-nm-thick slice of the tomographic reconstruction of the boxed area in B reveals the cytosolic capsid and intracellular structures like macromolecular assemblies and actin filaments (dashed box). (D–G) Four different 3.2-nm-thick slices of the partial, denoised volume containing the capsid. (D) In a slice just above the plasma membrane, glycoprotein spikes cut in the stalk region appear as dots. (E) Directly underneath the plasma membrane, tegument protein density and the upper end of the capsid are visible. (F) An actin bundle neighbors the viral capsid and occupies the area underneath the capsid (G). (H–I) Stereoview of the surface rendering from the volume boxed in C. A dense actin network (red) underlies the incoming capsid (light blue) and the tegument (orange). The cell membrane, the glycoproteins, and macromolecular complexes are not displayed. (J) An 8.1-nm-thick slice from a tomogram of a PtK2 cell inoculated for 2 min in the absence of nocodazole. Two capsids of recently entered virions are visible. The one on the left entered from the top, and the right one from the side at the area where the glycoprotein spikes emerge from the membrane. (K) Surface rendering of the tomogram presented in J: capsid (light blue), tegument (orange), glycoproteins (yellow), cell membrane/viral membrane (dark blue), actin (dark red; upper part cut away), and cellular vesicles (purple). (Scale bars: A, 1 μm; and B–K, 100 nm.)
The glycoprotein spikes protruded in dense clusters from the plasma membrane at the entry site. The majority of the tegument density—the so-called outer tegument (15)—formed a dense layer just underneath the plasma membrane, clearly corresponding in shape and extent to the patch of glycoprotein spikes on the outer face of the plasma membrane. The volume of this tegument density was consistent with the volume of the tegument in complete virions (1). The capsid was typically separated from most of the tegument density and did not show evidence for any densities that could be ascribed to the tegument (Fig. 1 C–G and J).
Capsids released into the cell were located close to actin filaments [Fig. 1C and supporting information (SI) Movie S1]. To exclude the possibility that the actin organization had been modified by nocodazole, which might affect the cortical cytoskeleton, we performed experiments in the absence of nocodazole. The overall actin network organization in untreated cells near the viral entry sites resembled that of nocodazole treated cells (Fig. 1 J and K), but the number of capsids in the peripheries of untreated cells was lower (14).
HSV-1 Entry into Synaptosomes.
Due to the specimen thickness, tomography of virus entry events was restricted to the cell periphery, resulting in a low number of analyzable events. Viruses were rarely observed entering at the outer rim of adherent cells, hence there was a shortage of side-views of the glycoprotein patches on the cell membrane and the underlying tegument (Fig. 1 J and K). To analyze the structural dynamics of entry by fusion more extensively, we therefore shifted to synaptosomes, a smaller host system that was completely accessible by cryo-ET. Synaptosomes are physiologically active endings of neurons prepared by homogenization and fractionation of brain tissue (16, 17). Neurons are authentic host cells for herpesviruses, and they have been characterized as a model host system for HSV-1 cell entry (C. H. Nagel, A. Binz, S. Borzsutzky, R. Bauerfeind, and B.S., unpublished data). Moreover, presynaptic regions of neurons express nectin-1, an HSV-1 entry receptor, at high levels at the plasma membrane (18).
Viruses entered exclusively into the presynaptic part (Fig. 2 A and B and Movie S2). The overall morphology of glycoprotein spikes and tegument at the entry site was analogous to our observations in intact cells. The tomograms from synaptosomes have higher quality due to the thinner specimens; therefore, we exploited them for a more detailed analysis. To trap different intermediate stages of the fusion process and thereby reveal its structural dynamics, we analyzed tomograms of synaptosomes inoculated with HSV-1 at a variety of infection times and temperatures ranging from 1 to 60 min and from 10 to 37°C, respectively (for details see Materials and Methods and Table S1). The actual fusion at the plasma membrane occurred within seconds. We detected cytosolic capsids inside synaptosomes after only 1 min of incubation at 37°C. Surprisingly, the membrane fluidity at 10°C was sufficient to enable entry, whereas no fusion intermediates were detected at 4°C. Capsids were located close to the entry site, judged by glycoprotein spikes and tegument at the plasma membrane, even after 60-min incubation at 37°C.
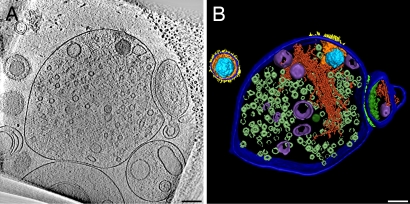
Fig. 2.
HSV-1 enters the presynaptic part of synaptosomes. (A) A 16-nm-thick slice of a tomogram from a synaptosome inoculated with HSV-1 for 60 min at 25°C. A capsid is localized inside the presynaptic element on the left characterized by abundant synaptic vesicles. A smaller postsynaptic fraction (right) is still attached via adhesion molecules to the synaptic cleft. Glycoprotein spikes are visible on the outer phase; tegument proteins correspond to the local densities near the cytoplasmic phase of the plasma membrane. Note structural changes between the entire virion (upper left corner) and the one that had entered the synaptosome. (B) Surface rendering of one virion and the synaptosome from the tomogram in A: capsid (light blue), tegument (orange), glycoproteins (yellow), cell membrane/viral membrane (dark blue), actin (dark red), vesicles (purple), synaptic vesicles (only partially segmented—metallic green), synaptic cleft (light green), and postsynaptic density (green). (Scale bars, 100 nm.)
Structural Dynamics of HSV-1 During Entry.
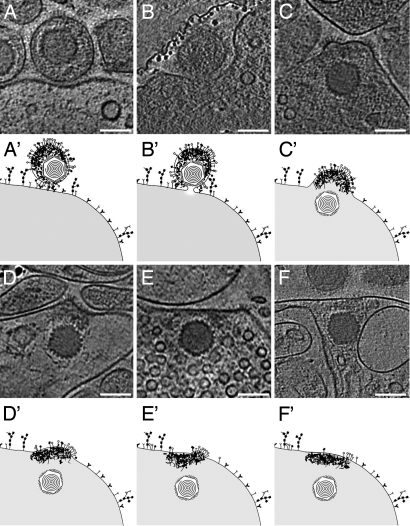
Summarizing the data based on 51 tomograms, we present a gallery of characteristic slices through the tomograms and corresponding schematic drawings for the six major steps into which we divided the entry process (Fig. 3 A–F and A′–F′). Of the 66 viral entry intermediates, we allocated 45 structures to the steps A–F, comprising 15, 1, 4, 5, 9 and 11 events, respectively. These numbers support the notion that the fusion pore represented the most short-lived intermediate. Twenty-one structures represented heterogeneous intermediates between state A and B (Fig. S1).
Fig. 3.
Structural dynamics of HSV-1 during entry into synaptosomes. Different steps during HSV-1 entry into synaptosomes, for details refer to text. (A–F) Slices of 2.7 to 12.8 nm thickness of various tomographic reconstructions from HSV-1 entry intermediates; (A′ and F′) represent the corresponding schematic drawings. (A and A′) The virus attached to the plasma membrane by interaction of specific glycoproteins with cellular receptors. (B and B′) Fusion pore formation close to the capsid proximal side of the asymmetric virion (see Movie S3 and Fig. 4 for 3D representation). A part of the carbon film is present in the upper left. (C and C′) The capsid entered the cell, and the viral membrane was integrated into the cellular membrane while keeping its curvature, being most prominent in patches studded with glycoprotein spikes. (D and D′) The membrane was partly bent, and most of the tegument remained at the entry site. (E and E′) Glycoproteins were spread along the plasma membrane. (F and F′) The membrane curvature returned to a more flattened, regular organization, and the glycoproteins were removed from the surface. (Scale bars, 100 nm.)
The first morphological stage was virus attachment to the plasma membrane (Fig. 3A). We observed a prominent gap between the viral and the cellular membrane. The width of this gap (≈20 nm) corresponded to the length of the longest glycoprotein spikes protruding from the virus. This primary interaction of the virus with the host cells involved a large contact area, apparent as an indent of the plasma membrane. Next, the viral and synaptosomal membranes came close together, and a fusion pore formed (Fig. 3B). The pore enlarged and allowed mixing of the viral content with the cytosol (Fig. 3C). The viral envelope, which had been integrated into the cell membrane, initially maintained its curvature. Subsequently, and consistent with earlier reports (10, 19), the capsid and potentially some bound inner tegument advanced into the cytosol, whereas the outer tegument remained attached to the membrane (Fig. 3D). Akin to entry into cells, the capsid was not surrounded by any densities assignable to tegument proteins. The curvature of the former viral membrane decreased (Fig. 3E) and the number of glycoproteins and the amount of tegument at the entry site declined (Fig. 3F) until no respective densities remained.
Intermediate Glycoprotein Conformations During Pore Formation and Dilation.
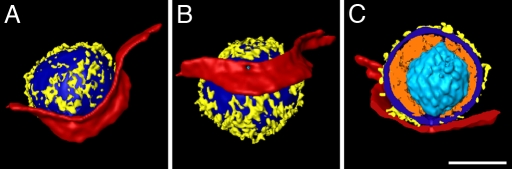
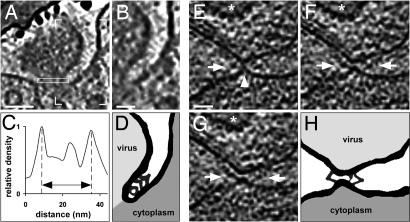
Three-dimensional renderings of the short-lived fusion pore (Fig. 3B) are presented in Fig. 4 and in Movie S3. The virion pole where the membrane is in close proximity to the capsid (1) was located close to the pore opening and can be seen through the pore neck from inside the cell (Fig. 4B), whereas most of the tegument mass was distant to the fusion pore. The pore diameter of ≈25 nm (Fig. 5C) reflected a state after the frequent reversions to the hemifusion state known as “flickering” (20), and thus the beginning of lateral pore expansion. Detailed analysis of the region between the viral and cell membranes next to the pore revealed bent V- or Y-shaped densities connecting the two membranes and possibly represented glycoprotein complexes undergoing conformational changes (Fig. 5 A, C, and D). In HSV-1 inoculated Vero cells, another example of V/Y-shaped structures was captured next to an early contact site just before pore formation, perhaps corresponding to hemifusion as indicated by the elongated neck shape and a pointed membrane contact (Fig. 5 E–H). In both stages, early contact and pore dilation, the V/Y-complexes had an arm length of up to ≈15 nm. The component of the V/Y-complex proximal to the viral membrane was in both cases curved, whereas the segment connecting to the plasma membrane was straight. The major difference between early contact and pore dilation was that the angles between the glycoprotein segment and the plasma membrane decreased from ≈50° to ≈40° concomitant with a change from a tapered contact of the viral and plasma membrane to a more parallel arrangement. Moreover, the shape of the “hinge” connecting the two components of the complex changed from a globular structure of ≈5 nm in diameter to an elongated one of ≈8 nm in length. A small structure close to the potential lipid mixing region connecting the two different membranes was observed in both states.
Fig. 4.
Detailed analysis of a fusion pore. (A–C) Surface renderings of the volume from Fig. 3B in different orientations (see Movie S3 for complete tomogram). (A) Rendering of the virus with parts of the synaptosomal plasma membrane in the same orientation as in the tomographic slice shown in Fig. 3B and Movie S3. (B) The virus was rotated vertically for ≈60° to allow a better view into the fusion pore. (C) Cut-away view of all features apart from the capsid; the orientation was changed to provide a perpendicular view of the pore. Colors used are light blue (capsid), orange (tegument), yellow (glyco proteins), dark blue (viral membrane), and red (cell membrane). Cellular elements of the synaptosome were excluded from rendering for clarity. (Scale bar, 100 nm.)
Fig. 5.
Glycoprotein bridges are formed prior and next to the fusion pore. (A) A 2.7-nm-thick slice from the volume presented in Fig. 4. Continuous V- and Y-shaped structures are localized between the two membranes. (B) Zoom into the area marked by box corners in A. (C) Line plot of pixel intensities from the pore cross-section (see rectangle in A). The pore diameter of this entry intermediate was ≈25 nm as measured between the maxima for the membrane. The gray values within the pore are higher than those of the surrounding vitreous ice. (D) Schematic drawing of membrane and protein structures from A with emphasis on connective densities and membranes. (E–G) Subsequent 2.7-nm-thick slices from a tomogram of an inoculated, nocodazole-treated Vero cell. The capsid of the virion is marked by a star. The arrowhead (E) highlights the contact between the plasma membrane and the viral envelope. The arrows in (E–G) point to prominent protein complexes between the two membranes and close to the contact site. (H) Schematic drawing of the connecting densities and membrane structures in F and G. (Scale bars; A, 40 nm; B and E, 20 nm.)
Discussion
In this study, we monitored the native structural dynamics during HSV-1 entry into adherent host cells and synaptosomes in unprecedented detail. We captured several intermediates, including the very short-lived events of pore formation and dilation by imaging single virions during the entry process in the frozen hydrated state. By only using high quality virus inocula (see Materials and Methods), we consider it most reasonable that the virions captured here at the plasma membrane represented functional intermediates in the cell entry of HSV-1 that were suspended from further progression into the cytosol by sudden freezing. Thus, they provide valuable, physiologically relevant in situ snapshots of the fusion dynamics.
In adherent cells, cytosolic capsids were located between actin bundles, which appeared to enclose the capsids (Fig. 1). Viruses are known to modulate the host's cytoskeleton and associated pathways (21, 22). Rearrangement of the actin cytoskeleton during entry via endocytosis was reported for HSV-1 (23). Thus, we suppose that the actin bundles had reorganized upon viral stimulation to facilitate capsid entry and to remove the dense cytoskeletal barrier. The morphological appearance of the cortex—bundles and filaments—implies that there was no local actin depolymerization around the capsid. However, the actin network from adherent cells at this resolution has so far only been visualized in intact filopodia (24). A direct control of the cytoskeleton organization from a cell at exactly the same area before virus entry is not feasible with the technique used here. To evaluate this aspect in full detail, the potential players in the respective signaling pathways need to be identified.
The viral membrane dynamics changed its curvature into extremely bended structures during the entry process (Fig. 3). We consider this a native feature because in cryo electron microscopy, in contrast to electron microscopy of fixed samples, membranes are preserved in their native state. The curvature was possibly stabilized by the distinct lipid contents of this domain (25) and by clusters of the glycoprotein spikes, which are present in the highly curved regions (Fig. 3 C–E) and which interact with the tegument.
Some of the glycoprotein spikes must represent the postfusion state of the glycoproteins gH/gL and gB. However, most glycoproteins were not recruited to the fusion site, and resembled those seen on the virus (1). Our data (Figs. 3–5) support the notion that only a small number of glycoprotein spikes is sufficient to catalyze HSV-1 cell entry. We did not detect any particular structure indicative for a large multisubunit complex formed by numerous spikes and receptors. Glycoproteins were densely clustered on both extracellular virions and the plasma membrane after fusion so that any postfusion conformation complexes would have been difficult to detect.
The glycoproteins and the tegument remained at the plasma membrane after the capsid was released into the cytosol (Figs. 1–3). This implies a continued contact of the tegument proteins with the cytoplasmic tails of the viral glycoproteins after fusion. During the subsequent events (Fig. 3), the membrane curvature flattened and the amount of glycoproteins and the tegument density decreased. The glycoproteins perhaps diffused away and were recycled or degraded until finally no glycoproteins remained at the entry site; the tegument possibly dissociated.
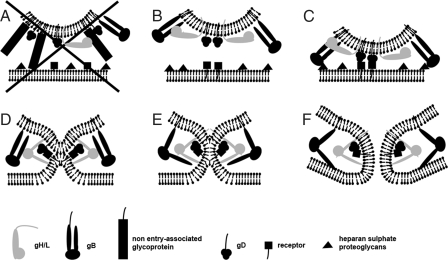
The HSV-1 virion has an asymmetric structure, characterized by a noncentric localization of the capsid. We propose that this asymmetric structure of the virion is related to different functions and results in two functional poles (Fig. S2). One pole of the virion is associated with virus assembly during secondary envelopment at cytoplasmic membranes and is characterized by a dense “cap” of tegument proteins (1). Some of these tegument proteins interact with the cytosolic tails of various glycoproteins (26, 27). These interactions impede lateral movement of glycoproteins in the membrane and maintain dense clusters of glycoprotein spikes on this pole. Bulky, non-entry-associated glycoprotein spikes in close proximity to gD may act as spacers and prevent binding of gD to its cellular receptor (Fig. 6A). On the opposite virion pole, where the capsid almost contacts the viral envelope, tegument material is sparse and, as such, are glycoprotein spikes. The larger spacing between the latter prevents sterical hindrance, and this pole may present the entry-associated glycoproteins in a functional arrangement (Fig. 6B). Such a preference for the capsid proximal side of the viral membrane to participate in the fusion pore was, in fact, observed in our study (Figs. 4 and 5). Therefore, we assign the entry function to this pole, thereby introducing the concept of a functional bipolar and structurally asymmetric virion.
Fig. 6.
Schematic model for the fusion mechanism based on the identified intermediates. (A) The bulky glycoprotein spikes in close proximity to gD, which are not involved in cell entry, act as spacers. Thus, gD cannot bind to its specific receptor, and the fusion process is not initiated. (B) Fusion can only be initiated if the contact of the entry-associated glycoprotein gD with its cellular receptor is not sterically hindered. (C) Binding of gD to its receptor might be paralleled or preceded by nonessential interactions of gB with heparan sulfate proteoglycans. (D) After receptor binding, gD undergoes a conformational change and interacts with gH/gL. This induces gH/gL to also change conformation and to flip out one segment that binds to the plasma membrane. This connects the two membranes, brings them close together and enables lipid mixing (E). Glycoprotein B is recruited to the gD/gH/gL complex by the conformational switch of gH/gL and binds to the plasma membrane (F). The fusion pore forms stabilized by gB.
Based on the current view (4, 5) of the sequence of events in HSV-1 fusion, we suggest a refined model for the fusion mechanism of HSV-1, which is compatible with the observed glycoprotein intermediates during pore formation and dilation (Fig. 5). At the “entry pole,” gD may bind to its cellular receptor (Fig. 6C) and subsequently undergo the necessary conformational changes (28) that activate gH/gL (Fig. 6D). Like viral class I fusion proteins, gH contains hydrophobic alpha-helices (29). The heterodimer gH/gL perhaps changes conformation, and one segment binds to the plasma membrane. This connects the two membranes, pulls them together and enables lipid mixing as shown for mutants lacking gB (30). Based on the lower molecular weight of the gH/gL heterodimer compared with the gB homotrimer, we suggest allocating the small structure close to the lipid mixing region of the viral and the plasma membrane (Fig. 5 D and H) to a gD-gH/gL complex (31, 32). The conformational switch of gH/gL then recruits gB to the gD-gH/gL complex. The observed V/Y-complexes in Fig. 5 had an arm length of ≈15 nm, thus possibly representing gB (33). The gB, a class III fusion protein, has features of both class I and class II fusion proteins and shares structural homologies with the class III fusion protein glycoprotein G of vesicular stomatitis virus (7). The x-ray structures of the pre- and postfusion states of the latter lead to an assignment of putative conformational changes and intermediates (34, 35). Accordingly, we suggest analogous to the V/Y-shaped structures (Fig. 5) that gB might flip out one segment that binds to the plasma membrane forming the elongated neck like structure (Fig. 6E). Finally, the fusion pore forms and expands facilitated by gB (Fig. 6F).
The suggested assignments of glycoproteins are mainly based on single structures derived from the tomograms presented in Fig. 5. Because cryo electron microscopy of biological material provides low-contrast images with a low signal to noise ratio, but with the advantage of avoiding artifacts caused by fixation, dehydration, and staining, these structures do not stand out very strongly against the background. However, the detected glycoprotein intermediate structures were very similar, although they originated from experiments with two different host systems, synaptosomes and Vero cells. Therefore, we think that these intermediates are representative for the HSV-1 fusion mechanism.
In the case of one fusion effector, crystal structures of truncated viral glycoproteins at different pH have revealed distinct pre- and postfusion conformations, and suggested a fusion mechanism by deducing hypothetical intermediate structures and molecular dynamics for all three classes of viral fusion proteins (9). However, membrane fusion during the entry of herpesviruses or poxviruses and during intracellular membrane traffic requires the interaction of several effectors (36, 37). Here, we present the first direct observation of several intermediates in virus fusion during cell entry in a native environment at close to molecular resolution, i.e., snapshots of a multicomponent fusion machinery in action.
Materials and Methods
Purification of HSV-1.
Extracellular HSV-1 virions (wild type strain 17+) were produced from African green monkey kidney (Vero) cells (CCL-81; American Type Culture Collection) or human foreskin fibroblasts (HFF) cells (kindly provided by Prashant Desai, University of Baltimore, Baltimore, MD) and purified and characterized by plaque titration and real-time PCR with previous DNase treatment as described (1, 14, 38). Viruses produced from Vero cells had a genome/pfu ratio of 6 and viruses from HFF cells of 11.
Infection of Cells Grown on Grids.
Au-grids with holey carbon films [either Quantifoil (Quantifoil) or C-flat (Protochips)] were glow-discharged for 20 s and preincubated with growth medium at 37°C and 5% CO2. Rat kangaroo kidney cells (PtK2: CCL-56; American Type Culture Collection), HFF cells, or Vero cells were grown on the grids in their respective growth medium: alpha-minimum essential medium for PtK2 and HFF and D-MEM for Vero (both Invitrogen) supplemented with 10% FCS (Sigma-Aldrich), 100 units/ml penicillin, and 100 μg/ml streptomycin (both Invitrogen) for 15 h. After pretreatment with 50 μM nocodazole (Sigma-Aldrich) for PtK2 and 7 μM nocodazole for HFF and Vero for 2 h, the cells were cooled on ice and infected with 106 pfu of HSV-1 produced from Vero cells per 25 μl of medium and grid. Virus binding to the cells was allowed for 2 h on ice; the samples were then warmed up to 37°C for 2–5 min. After adding 3 μl of colloidal gold particles (15 nm, protected with BSA), excess fluid was absorbed with filter paper. The samples were vitrified by plunge freezing in liquid ethane and stored in liquid nitrogen. Cells incubated without nocodazole were treated likewise.
Preparation of Inoculated Synaptosomes.
Isolation of synaptosomes from the forebrain of a 6-week-old male Wistar rat was adapted from Leenders et al. (39). After harvesting the F4 fraction from the 15%/23% interface of a four step (23%, 15%; 10%, 3%) Percoll gradient (Sigma-Aldrich), the synaptosomes (typical diameter ≈0.5 μm) were centrifuged for 15 min at 15,000 × g and 4°C and resuspended in 1 ml of RPMI 1640 w/Glut 25 mM Hepes medium (Invitrogen) containing 0.1% BSA (Sigma–Aldrich). The synaptosomes from 1 brain were activated at 37°C for 30 min and inoculated on ice with 109 pfu HSV-1 produced from HFF cells. Binding was allowed for 60 min on ice. Washing was carried out by centrifugation steps of 5 min at 3,500 × g and 4°C. The subsequent incubation times (1, 2, 5, 10, 30, or 60 min) and temperatures (10°C, 18°C, 25°C, 37°C or on ice) were varied for individual experiments. Finally, 3 μl of sample were applied to holey carbon films on Cu-grids (Quantifoil; glow-discharged for 20 s), 3 μl of colloidal gold particles (10 nm, protected with BSA) were added, and vitrified as described above.
Cryo electron Tomography.
Data were collected on a Philips CM300 FEG and a Tecnai Polara (FEI) transmission electron microscope, both equipped with a GATAN GIF 2002 postcolumn energy filter (Gatan), and images were collected with a 2K × 2K Multiscan CCD camera (Gatan). The microscopes were operated at 300 kV and a final magnification of 43,974 and 37,248 giving a pixel size of 0.68 nm and 0.81 nm at the specimen level, respectively. Tilt series were collected covering a minimum angular range of 120°, with an angular increment of 2°, 2.5°, or 3°. Defocus was measured along the tilt axis after each tilt and automatically maintained at −12 μm ± 0.5 μm. The total electron dose received at the specimen level was kept between 50 and 100 electrons/Å2. The exposure time was adjusted to the tilt angles by a 1/cos scheme.
Image Processing.
Alignment, reconstruction, extraction of subtomograms, and further image processing were performed using the EM (40) and TOM (41) software packages implemented in MATLAB (Mathworks). Tilted images were aligned using gold beads as fiducial markers. Three-dimensional reconstructions were calculated using weighted back projections. Visualization and segmentation was performed using Amira 4.1 (TGS) and Bsoft (42). Nonlinear anisotropic denoising (43) and Gaussian filtering was applied before segmentation. Final figure preparation and drawings were performed in PhotoShop CS2 (Adobe) and Canvas X (ACD Systems International).
Supplementary Material
Acknowledgments.
We thank C.H. Nagel for sharing his experience on inoculating synaptosomes with HSV-1; V. Lucic, G.K.W. Kong, and R. Fernández-Busnadiego for discussions and practical help with setting up of the synaptosome preparation protocol; K. Döhner for many discussions on HSV-1 cell entry experiments; I. Ibiricu and C. Hagen for support; A.C. Steven for stimulating discussions; W. Baumeister for support and encouragement; and J. A. G. Briggs and A. Leis for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Emmy-Noether-Program Grant GR1990/1-2 (to K.G.), Deutsche Forschungsgemeinschaft Priority Program 1775 Grants GR1990/2-1 (to K.G) and So403/3 (to B.S.), and European Commission New Emerging Science and Technologies Contract 012702 (to B.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801674105/DCSupplemental.
References
- 1.Grünewald K, et al. Three-dimensional structure of Herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 2.Spear PG. Herpes simplex virus: Receptors and ligands for cell entry. Cell Microbiol. 2004;6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 3.Nicola AV, Hou J, Major EO, Straus SE. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol. 2005;79:7609–7616. doi: 10.1128/JVI.79.12.7609-7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rey FA. Molecular gymnastics at the herpesvirus surface. EMBO Rep. 2006;7:1000–1005. doi: 10.1038/sj.embor.7400807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh T, et al. PILRalpha is a Herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heldwein EE, et al. Crystal structure of glycoprotein B from Herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 8.Carfi A, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 9.Weissenhorn W, Hinz A, Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming Herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Döhner K, et al., editors. Function of dynein and dynactin in Herpes simplex virus capsid transport. Mol Biol Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medalia O, et al. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 13.Lucic V, Förster F, Baumeister W. Structural studies by electron tomography: From cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 14.Döhner K, Radtke K, Schmidt S, Sodeik B. Eclipse phase of Herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J Virol. 2006;80:8211–8224. doi: 10.1128/JVI.02528-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp BG, Granzow H, Mettenleiter TC. Primary envelopment of Pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol. 2000;74:10063–10073. doi: 10.1128/jvi.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen RS, Blomberg F, Berzins K, Siekevitz P. The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J Cell Biol. 1977;74:181–203. doi: 10.1083/jcb.74.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackstone CD, et al. Biochemical characterization and localization of a non-N-methyl-D-aspartate glutamate receptor in rat brain. J Neurochem. 1992;58:1118–1126. doi: 10.1111/j.1471-4159.1992.tb09370.x. [DOI] [PubMed] [Google Scholar]
- 18.Mizoguchi A, et al. Nectin: An adhesion molecule involved in formation of synapses. J Cell Biol. 2002;156:555–565. doi: 10.1083/jcb.200103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfstein A, et al. The inner tegument promotes Herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 20.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 21.Munter S, Way M, Frischknecht F. Signaling during pathogen infection. Sci STKE. 2006;2006:re5. doi: 10.1126/stke.3352006re5. [DOI] [PubMed] [Google Scholar]
- 22.Greber UF. Signaling in viral entry. Cell Mol Life Sci. 2002;59:608–626. doi: 10.1007/s00018-002-8453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clement C, et al. A novel role for phagocytosis-like uptake in Herpes simplex virus entry. J Cell Biol. 2006;174:1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medalia O, et al. Organization of actin networks in intact filopodia. Curr Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 25.van Genderen IL, Brandimarti R, Torrisi MR, Campadelli G, van Meer G. The phospholipid composition of extracellular Herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: A tale of two membranes. Curr Opin Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 27.O'Regan KJ, Bucks MA, Murphy MA, Wills JW, Courtney RJ. A conserved region of the Herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE) Virology. 2007;358:192–200. doi: 10.1016/j.virol.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Cocchi F, et al. The soluble ectodomain of Herpes simplex virus gD contains a membrane-proximal profusion domain and suffices to mediate virus entry. Proc Natl Acad Sci USA. 2004;101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gianni T, Fato R, Bergamini C, Lenaz G, Campadelli-Fiume G. Hydrophobic alpha-helices 1 and 2 of Herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J Virol. 2006;80:8190–8198. doi: 10.1128/JVI.00504-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian RP, Geraghty RJ. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avitabile E, Forghieri C, Campadelli-Fiume G. Complexes between Herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atanasiu D, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of Herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci USA. 2007;104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stannard LM, Fuller AO, Spear PG. Herpes simplex virus glycoproteins associated with different morphological entities projecting from the virion envelope. J Gen Virol. 1987;68(Pt 3):715–725. doi: 10.1099/0022-1317-68-3-715. [DOI] [PubMed] [Google Scholar]
- 34.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the Vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 35.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the Vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 36.Chernomordik LV, Kozlov MM. Membrane hemifusion: Crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Langosch D, Hofmann M, Ungermann C. The role of transmembrane domains in membrane fusion. Cell Mol Life Sci. 2007;64:850–864. doi: 10.1007/s00018-007-6439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai P, DeLuca NA, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 39.Leenders M, G C, Sheng ZH. Multidisciplinary Approaches for Characterizing Synaptic Vesicle Proteins. Curr Protoc Neurosci. 2004 doi: 10.1002/0471142301.ns0207s28. 2004 Sep(2):Unit 2.7. [DOI] [PubMed] [Google Scholar]
- 40.Hegerl R, Altbauer A. The “EM” program system. Ultramicroscopy. 1982;9:109–116. doi: 10.1016/0304-3991(82)90233-9. [DOI] [PubMed] [Google Scholar]
- 41.Nickell S, et al. TOM software toolbox: Acquisition and analysis for electron tomography. J Struct Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.