Abstract
Improved approaches for the detection of common epithelial malignancies are urgently needed to reduce the worldwide morbidity and mortality caused by cancer. MicroRNAs (miRNAs) are small (≈22 nt) regulatory RNAs that are frequently dysregulated in cancer and have shown promise as tissue-based markers for cancer classification and prognostication. We show here that miRNAs are present in human plasma in a remarkably stable form that is protected from endogenous RNase activity. miRNAs originating from human prostate cancer xenografts enter the circulation, are readily measured in plasma, and can robustly distinguish xenografted mice from controls. This concept extends to cancer in humans, where serum levels of miR-141 (a miRNA expressed in prostate cancer) can distinguish patients with prostate cancer from healthy controls. Our results establish the measurement of tumor-derived miRNAs in serum or plasma as an important approach for the blood-based detection of human cancer.
Keywords: biomarker, miR-141, plasma, serum, prostate cancer
The development of minimally invasive tests for the detection and monitoring of common epithelial malignancies could greatly reduce the worldwide health burden of cancer (1). Although conventional strategies for blood-based biomarker discovery (e.g., using proteomic technologies) have shown promise, the development of clinically validated cancer detection markers remains an unmet challenge for many common human cancers (2). New approaches that can complement and improve on current strategies for cancer detection are urgently needed.
MicroRNAs (miRNAs) are small (typically ≈22 nt in size) regulatory RNA molecules that function to modulate the activity of specific mRNA targets and play important roles in a wide range of physiologic and pathologic processes (3, 4). We hypothesized that miRNAs could be an ideal class of blood-based biomarkers for cancer detection because: (i) miRNA expression is frequently dysregulated in cancer (5, 6), (ii) expression patterns of miRNAs in human cancer appear to be tissue-specific (7), and (iii) miRNAs have unusually high stability in formalin-fixed tissues (8–10). This third point led us to speculate that miRNAs may have exceptional stability in plasma and serum as well. We show here that miRNAs are in fact present in clinical samples of plasma and serum in a remarkably stable form. Furthermore, we establish proof-of-principle for blood-based miRNA cancer detection by using both a xenograft model system and clinical serum specimens from patients with prostate cancer. Our results lay the foundation for the development of miRNAs as a novel class of blood-based cancer biomarkers and raise provocative questions regarding the mechanism of stability and potential biological function of circulating miRNAs.
Results
Identification and Molecular Cloning of Endogenous miRNAs from Human Plasma.
Prior reports have suggested that RNA from human plasma (the noncellular component of blood remaining after removing cells by centrifugation) is largely of low molecular weight (11). We directly confirmed that human plasma contains small RNAs in the size range of miRNAs (18–24 nt) by characterizing the size of total RNA isolated from plasma by using radioactive labeling. PAGE and phosphorimaging of 5′ 32P-labeled plasma RNA demonstrated RNA species ranging from 10 to 70 nt in size, including a discernable species of size ≈22 nt characteristic of most miRNAs [supporting information (SI) Fig. S1]. The detected signal was sensitive to RNase treatment but insensitive to DNase I treatment, confirming that the signal originated from RNA (Fig. S1).
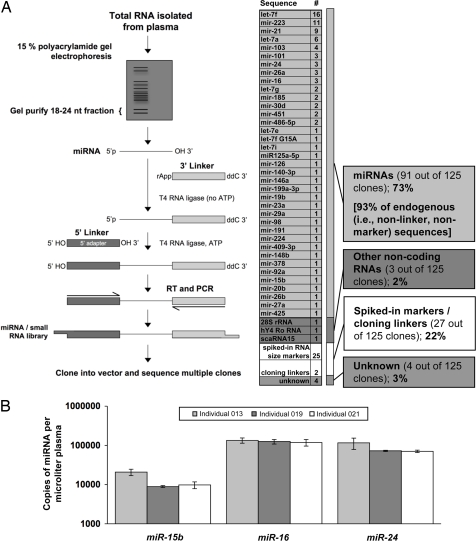
To directly determine whether miRNAs are present in human plasma, we isolated the 18- to 24-nt RNA fraction from a human plasma sample from a healthy donor (see SI Text for details on blood collection and plasma RNA isolation) and used 5′ and 3′ RNA–RNA linker ligations followed by RT-PCR amplification to generate a small RNA cDNA library (Fig. 1A). Of the 125 clones sequenced from this library, 27 corresponded to spiked-in size marker oligos or linker–linker dimers. Ninety-one of the other 98 sequences (93%) corresponded to known miRNAs, providing direct confirmation that mature miRNAs are present in human plasma and indicating that the vast majority of 18- to 24-nt plasma RNA species cloned by our protocol are indeed miRNAs (Fig. 1A). To quantitate specific miRNAs, we used TaqMan quantitative RT-PCR (qRT-PCR) assays (12) to measure three miRNAs (miR-15b, miR-16, and miR-24) in plasma from three healthy individuals. These three miRNAs, chosen to represent moderate- to low-abundance plasma miRNAs (based on the sequencing results described above), were all readily detected in the plasma of each individual at concentrations ranging from 8,910 copies/μl plasma to 133,970 copies/μl plasma, depending on the miRNA examined (Fig. 1B).
Fig. 1.
Identification of miRNAs in human plasma. (A) Cloning and sequencing of miRNAs from human plasma. The schematic diagram depicts the preparation of a small RNA library from human plasma. Briefly, the 18- to 24-nt fraction from ≈250 ng of plasma total RNA from a single donor (individual 006; described in Table S6) was isolated by PAGE. Purified miRNAs were then 3′ and 5′ligated to single-stranded oligonucleotides that contained universal primer sequences for reverse transcription and PCR. Reverse transcription and PCR generated a library of small RNA cDNA molecules that were ligated into a plasmid vector (pCR4-TOPO) and transformed into Escherichia coli. Inserts from a total of 125 individual colonies yielded high-quality sequence. Sequences were compared to a reference database of known miRNA sequences (miRBase Release v.10.1) (19) and to GenBank. Seventy-three percent of sequences corresponded to known miRNAs as shown. The next most abundant species were matches to the sequence of synthetic RNAs spiked in as radiolabeled 18- and 24-nt molecular size markers during gel isolation steps. When only endogenously derived RNA sequences are considered, miRNAs represent 93% (91 of 98) of the recovered sequences. The miRNA read designated as “let-7f G15A” denotes a sequence matching the known let-7f miRNA except for a G-to-A substitution at nucleotide position 15. (B) Quantification of representative miRNAs in normal human plasma by TaqMan qRT-PCR. The graph indicates the number of copies of each of three representative miRNAs measured in plasma obtained from three healthy individuals. In each case, values represent the average of two replicate reverse transcription reactions followed by real-time PCR. For each miRNA assay, a dilution series of chemically synthesized miRNA was used to generate a standard curve that permitted absolute quantification of molecules of miRNA/μl plasma as shown here (see Fig. S2 for standard curve plots). Values were median-normalized by using measurements of synthetic normalization controls spiked in immediately after addition of denaturing solution during RNA isolation (see SI Text for full details). The absence of amplification in reverse transcriptase-negative controls indicated that amplification was originating from an RNA template (real-time PCR plots corresponding to plasma RNA samples and negative controls are provided in Fig. S3).
Stability of Endogenous miRNAs in Human Plasma.
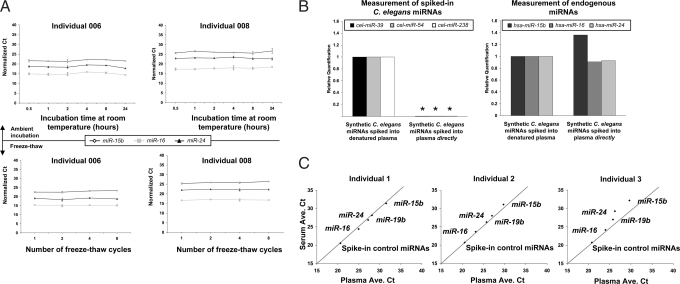
We next sought to investigate the stability of miRNAs in plasma, given that this is an important prerequisite for utility as a biomarker. Incubation of plasma at room temperature for up to 24 h (Fig. 2A Upper) or subjecting it to up to eight cycles of freeze-thawing (Fig. 2 A Lower) had minimal effect on levels of miR-15b, miR-16, or miR-24 as measured by TaqMan qRT-PCR. Given that plasma has been reported to contain high levels of RNase activity (13), we sought to determine whether the stability of miRNAs is intrinsic to their small size or chemical structure, or whether it is caused by additional extrinsic factors. We introduced synthetic miRNAs corresponding to three known Caenorhabditis elegans miRNAs (cel-miR-39, cel-miR-54, and cel-miR-238), chosen because of the absence of homologous sequences in humans, into human plasma either before or after the addition of a denaturing solution that inhibits RNase activity. RNA extraction followed by measurement of synthetic miRNAs showed that the synthetic miRNAs rapidly degraded when added directly to plasma (the time between addition of synthetic miRNAs and subsequent addition of denaturing solution was <2 min), as compared with their addition after adding denaturing solution to plasma (Fig. 2B). These results confirm the presence of RNase activity in plasma and the sensitivity of naked miRNAs to degradation. The levels of endogenous miRNAs (i.e., miR-15b, miR-16, and miR-24) were not significantly altered in any of the experimental samples (Fig. 2B), indicating that endogenous plasma miRNAs exist in a form that is resistant to plasma RNase activity.
Fig. 2.
Characterization of miRNA stability in human plasma. (A) miRNA levels remain stable when plasma is subjected to prolonged room temperature incubation or freeze-thawed multiple times. (Upper) The graphs show normalized Ct values for the indicated miRNAs measured in parallel aliquots of human plasma samples incubated at room temperature for the indicated times. The experiment was carried out by using plasma from the two different individuals noted. Normalization of raw Ct values across samples is based on the measurement of three nonhuman synthetic miRNAs spiked into each sample at known molar amounts after initial plasma denaturation for RNA isolation (described in detail in SI Text). (Lower) The graphs show normalized Ct values for the indicated miRNAs measured in parallel aliquots of human plasma samples subjected to the indicated number of cycles of freeze-thawing. Raw Ct values were normalized across samples by using the same approach as described above. (B) Exogenously added miRNAs are rapidly degraded in plasma, whereas endogenous miRNAs are stable. Three C. elegans miRNAs (chosen for the absence of sequence similarity to human miRNAs) were chemically synthesized and added either directly to human plasma (from individual 003; described in Table S6) or added after the addition of denaturing solution (containing RNase inhibitors) to the plasma (referred to as “denatured plasma”). RNA was isolated from both plasma samples, and the abundance of each of the three C. elegans miRNAs was measured by TaqMan qRT-PCR (Left), as was that of three endogenous plasma miRNAs (Right). Asterisks indicate that the abundance ratios of cel-miR-39, cel-miR-54, and cel-miR-238 added to human plasma directly, relative to addition to denatured plasma, were 1.7 × 10−5, 9.1 × 10−6, and 1.1 × 10−5, respectively and therefore too low to accurately display on the plot. (C) Abundance of miRNAs in serum and plasma collected from the same individual is highly correlated. Each plot depicts the average Ct values (average of two technical replicates) of the indicated miRNAs measured in serum and plasma samples collected from a given individual at the same blood draw. Results from three different individuals are shown. miRNA measurements were highly correlated in both sample types. Results shown for synthetic C. elegans miRNAs spiked into each plasma or serum sample (after addition of denaturing solution) demonstrate that experimental recovery of miRNAs and robustness of subsequent qRT-PCR is not affected by whether it is plasma or serum that is collected.
Comparison of miRNA Levels Between Plasma and Serum.
Given that clinical specimens of serum (the supernatant remaining after whole blood is permitted to clot) are more plentiful than plasma samples in many retrospective clinical sample repositories, we sought to determine whether miRNA measurements are substantially different in serum compared with plasma by measuring miR-15b, miR-16, miR-19b, and miR-24 in matched samples of serum or plasma collected from a given individual at the same blood draw (Fig. 2C). Measurements obtained from plasma or serum were strongly correlated, indicating that both serum and plasma samples will be suitable for investigations of miRNAs as blood-based biomarkers.
Tumor-Derived miRNAs Are Present in Plasma.
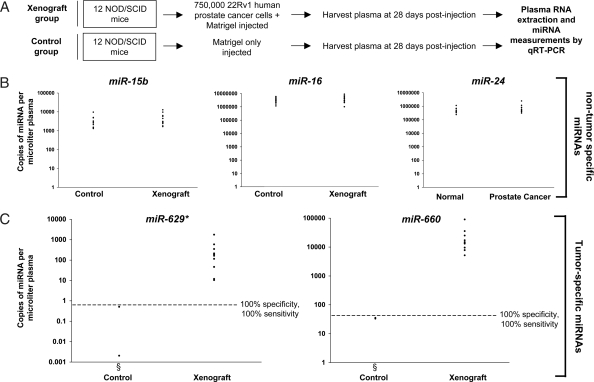
Having demonstrated that circulating miRNAs are detectable and stable in blood collected from healthy individuals, we next sought to determine whether tumor-derived miRNAs enter the circulation at levels sufficient to be measurable as biomarkers for cancer detection. We chose to study a mouse prostate cancer xenograft model system that involves growth of the 22Rv1 human prostate cancer cell line in NOD/SCID immunocompromised mice (14–17). We established a cohort of 12 mice xenografted with 22Rv1 cells injected with Matrigel and 12 control mice inoculated with Matrigel alone (Fig. 3A). Plasma was collected 28 days later (once tumors were well established), and RNA was isolated for miRNA quantitation. Because of the lack of an established endogenous miRNA control for plasma or serum, we introduced three synthetic C. elegans miRNAs (described earlier) after the addition of denaturing solution to the plasma samples and used these as normalization controls to correct for technical variations in RNA recovery (detailed in SI Text).
Fig. 3.
Tumor-derived miRNAs are detectable in plasma. (A) Schema for 22Rv1 human prostate cancer xenograft experiment. (B) MiRNAs are present in plasma of healthy control mice and their levels are not nonspecifically altered in cancer-bearing mice. Plasma levels of miR-15b, miR-16, and miR-24 were measured in 12 healthy control mice and 12 xenograft-bearing mice. The mature sequence of these miRNAs is perfectly conserved between mice and humans. Ct values were converted to absolute number of copies/μl plasma by using a dilution series of known input quantities of synthetic target miRNA run on the same plate as the experimental samples (dilution curves are provided in Fig. S2). Values shown have been normalized by using measurements of C. elegans synthetic miRNA controls spiked into plasma after denaturation for RNA isolation (details of the normalization method are provided in SI Text). (C) Tumor-derived miRNAs are detected in plasma of xenograft-bearing mice and can distinguish cancer-bearing mice from controls. Plasma levels of miR-629* and miR-660 (two human miRNAs that are expressed in 22Rv1 cells and do not have known murine homologs) were measured in all control and xenografted mice. Ct values were converted to absolute number of copies/μl plasma and normalized as described for B (see Table 5) threshold. Given that homologous miRNAs are not believed to exist in mice, the low level of signal detected for a few mice in the control group, particularly for the miR-660 assay, is likely to represent nonspecific background amplification. As expected, in the control (nontumor-bearing) mice group, qRT-PCR for miR-629* or miR-660 in plasma from most animals could not detect any appreciable signal. These points are therefore not shown on the graph, even though plasma samples from the entire group of 12 mice in the control group were studied.
We first sought to establish that endogenous (murine) miRNAs exist in mouse plasma and determine whether the presence of cancer may lead to a general increase in plasma miRNAs, whether they be tumor- or host-derived. miR-15b, miR-16, and miR-24 (which are perfectly conserved in mature sequence between human and mouse) were all readily detectable in healthy control mice (Fig. 3B) and were not expressed at substantially different levels in xenograft-bearing mice, indicating that the presence of tumor does not lead to a generalized increase in plasma miRNAs.
We next sought to identify candidate tumor-derived miRNAs for examination in plasma by first profiling the expression of 365 known miRNAs in 22Rv1 cells by using a microfluidic TaqMan low-density miRNA qRT-PCR array (Applied Biosystems). We identified two miRNAs, miR-629* and miR-660, that (i) were expressed in these cells as indicated by low cycle threshold (Ct) values and (ii) did not have known mouse homologs and therefore would be expected to be tumor-specific markers in this setting (Table S1). We next analyzed plasma samples from control and xenograft mice for the levels of miR-629* and miR-660 by TaqMan qRT-PCR. Levels of miR-629* and miR-660 were generally undetectable in the control mice, whereas they were readily detected (ranging from 10 to 1,780 copies/μl plasma for miR-629* and 5,189–90,783 copies/μl for miR-660) in all of the xenografted mice (Fig. 3C). Levels of both miR-629* and miR-660 were able to independently differentiate xenografted mice from controls with 100% sensitivity and 100% specificity. These data establish proof of the principle that tumor-derived miRNAs reach the circulation where their measurement in plasma can serve as a means for cancer detection.
To understand the basis for the wide variation in miRNA abundance observed among the different xenografted mice, we compared plasma levels of miR-629* and miR-660 with tumor mass in each mouse. Levels of these miRNAs were moderately correlated with tumor mass (Fig. S4 and see Table S7), indicating that variation in miRNA abundance across animals reflects, at least in part, the differences in tumor burden.
Tumor-Derived miRNAs in Plasma Are Not Cell-Associated.
We sought to further explore the mechanism of protection of tumor-derived miRNAs from plasma RNase activity by testing the hypothesis that they are present inside circulating tumor cells that might have escaped pelleting during primary centrifugation for plasma isolation. We took two approaches to address this hypothesis. In the first experiment, we filtered pooled plasma generated from the xenograft or control groups through a 0.22-μm filter, followed by RNA extraction from the filtrate and the material retained on the filter (referred to as the retentate). Measurement of miR-629* and miR-660 by qRT-PCR in each of the samples demonstrated that virtually all of the tumor-derived miRNAs passed through the 0.22-μm filter (Fig. S5A). As expected, tumor-derived miRNAs were essentially undetectable in all of the samples from the control group. In a second independent experiment, we subjected plasma pools from the xenograft and control groups to a series of two centrifugations (one at 2,000 × g, which should pellet any intact cells remaining after the initial centrifugation used to collect plasma, followed by another at 12,000 × g, which should pellet any large cell fragments). We assayed for miRNA expression in the starting material, any pelleted material obtained from each centrifugation, and the supernatant remaining after the 12,000 × g centrifugation. As shown in Fig. S5B, virtually all of the tumor-derived miRNA was present in the supernatant of the 12,000 × g spin. Taken together, the data indicate that tumor-derived miRNAs are not associated with intact cells or large cell fragments. These results do not exclude the possibility that circulating tumor cells or fragments of the same may have been lysed during the process of blood collection or plasma processing. Even if that is the case, however, our results show that miRNAs that may have been released are ultimately present in a stable, protected form of size much smaller than that of a typical epithelial cell.
Detection of Human Prostate Cancer Based on Measurement of a Prostate Cancer-Expressed miRNA in Serum.
We next sought to extend this approach to cancer detection in humans. We reasoned that an ideal marker would be (i) expressed by the cancer cells at moderate or high levels and (ii) present at very low or undetectable levels in plasma from healthy individuals. We established a list of likely blood-based miRNA biomarker candidates for prostate cancer by (i) compiling a list of miRNAs expressed in human prostate cancer specimens based on published miRNA expression profiling data (7, 18) and (ii) filtering out miRNAs detected in healthy donor-derived plasma in our miRNA cloning experiment (Fig. 1A) or detected on a microfluidic TaqMan qRT-PCR array analysis of plasma from a normal healthy individual (details provided in SI Text and see Table S8). This process generated a list of six leading candidates (miR-100, miR-125b, miR-141, miR-143, miR-205, and miR-296) for further investigation.
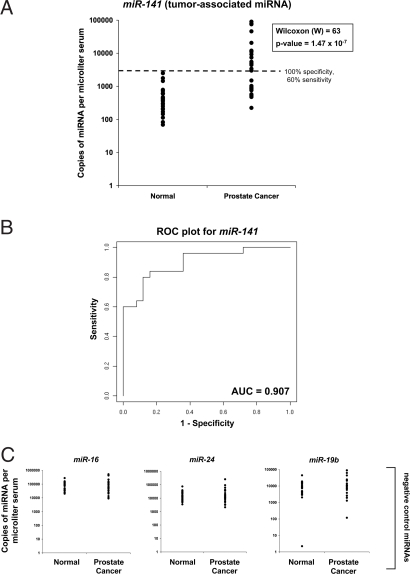
We chose to analyze these candidates in a case-control cohort of serum samples collected from 25 individuals with metastatic prostate cancer and 25 healthy age-matched male control individuals. To efficiently screen multiple miRNA biomarker candidates, we first generated two pools of serum aliquots derived from the individuals in the case and control groups, respectively. We isolated RNA from both pools and screened them for differential expression of the six candidate biomarker miRNAs by TaqMan qRT-PCR assays. Results of this screen indicated that five of six of these candidate miRNA biomarkers showed increased expression, although to varying degrees, in the prostate cancer serum pool compared with the healthy control group serum pool (Table S2). For one of the candidates (miR-205), no conclusion could be reached because miRNA levels in both pools were lower than the limit of detection of the assay (as determined by a standard curve using a dilution series of a synthetic miR-205 RNA oligonucleotide). Of all of the candidates, miR-141 showed the greatest differential expression (46-fold overexpressed) in the prostate cancer pool compared with the control pool (Table S2). We therefore focused our study on miR-141 by measuring the abundance of this miRNA in all of the individual serum samples comprising the case and control groups. Consistent with results from the analysis of pooled samples, serum levels of miR-141 were, in general, substantially higher in cancer cases compared with controls (Fig. 4A). Comparison of the two groups by a Wilcoxon two-sample test yielded W = 63 with a P = 1.47 × 10−7, confirming a significant difference in miR-141 levels between the two groups. Furthermore, serum levels of miR-141 could detect individuals with cancer with 60% sensitivity at 100% specificity (Fig. 4A). Representation of the data using a Receiver Operating Characteristic plot (Fig. 4B) reflects strong separation between the two groups, with an area under the curve (AUC) of 0.907. Comparison of miR-141 levels to prostate-specific antigen (PSA) values among the prostate cancer patients demonstrated Pearson and Spearman (rank) correlation coefficients of +0.85 and +0.62, indicating that miR-141 and PSA levels are moderately correlated (Table S3). Serum levels of nonbiomarker candidate miRNAs miR-16, miR-19b, and miR-24 were not significantly different between cases and controls, supporting the notion that miR-141 is specifically elevated in prostate cancer, as opposed to reflecting a nonspecific, generalized increase in serum miRNA levels in the setting of cancer (Fig. 4C). Taken together, the results extend to human cancer the concept that circulating miRNAs can serve as markers for cancer detection.
Fig. 4.
Detection of human prostate cancer by serum levels of tumor-associated miRNA miR-141. (A) Serum levels of miR-141 discriminate patients with advanced prostate cancer from healthy controls. Serum levels of the prostate cancer-expressed miRNA miR-141 were measured in 25 healthy control men and 25 patients with metastatic prostate cancer (clinical data on subjects is provided in Table S3). Ct values were converted to absolute number of copies/μl serum by using a dilution series of known input quantities of synthetic target miRNA run simultaneously (on the same plate) as the experimental samples (dilution curves are provided in Fig. S2). Values shown have been normalized by using measurements of C. elegans synthetic miRNA controls spiked into plasma after denaturation for RNA isolation (details of normalization method are provided in SI Text). The dashed line indicates a 100% specificity threshold. (B) Receiver Operating Characteristic (ROC) plot. The data shown in A were used to draw the ROC plot shown. (C) Serum levels of nontumor-associated miRNAs are not substantially different between patients with prostate cancer and controls. Serum levels of miR-16, miR-24, and miR-19b were measured as negative controls as they are not expected to be cancer-associated in the serum. Absolute quantification of miRNAs and data normalization were carried out as described for A.
miR-141 Is an Epithelial-Associated miRNA Expressed by Several Common Human Cancers.
miR-141 is a member of an evolutionarily conserved family of miRNAs that includes, in humans, miR-141, miR-200a, miR-200b, miR-200c, and miR-429 (19). The expression of zebrafish homologs of this family, when studied by in situ hybridization, was found to localize to various epithelial tissues (20). To gain more insight into the potential biological role of miR-141, we explored the large miRNA expression profiling dataset generated by Lu et al. (7), who profiled a diverse range of human cancer types. Consistent with findings from the zebrafish studies, the expression of miR-141 was tightly associated with expression in epithelial samples compared with nonepithelial samples (Fig. S6), and miR-141 was expressed in a wide range of common epithelial cancers including breast, lung, colon, and prostate.
To determine the relative expression of this miR-141 specifically between the epithelial and stromal compartments of prostate tissue, both comparatively between the two cell types and relative to all other known miRNAs within a cell type, we generated small RNA libraries from primary cultures of human prostate epithelial and stromal cells and subjected them to massively parallel sequencing (detailed in SI Text). We found that miR-141 (and two of its family members, miR-200b and miR-200c) was readily detected in the prostate epithelial cell dataset but strikingly absent in the prostate stromal cells (Table S4). In fact, of all miRNAs in this analysis, miR-141 and miR-200b were the two most overexpressed in prostate epithelial cells relative to prostate stromal cells (Table S4). Taken together, the data are consistent with the notion that miR-141 is an epithelial-restricted miRNA that can be detected in the circulation as a prostate cancer biomarker.
Discussion
Our Results Establish That Tumor-Derived miRNAs, Detected in Plasma or Serum, Can Serve as Circulating Biomarkers for Detection of a Common Human Cancer Type.
Although there is a long history of investigation of circulating mRNA molecules as potential biomarkers (21), blood-based miRNA studies are in their infancy. Recently, Chim et al. (22) reported the detection by qRT-PCR of miRNAs of presumed placental origin in the plasma of pregnant women, and Lawrie et al. (23) reported detecting elevations in miRNAs in serum from lymphoma patients. Beyond confirming the early reports, our study yielded (i) a more comprehensive view of plasma miRNAs by direct cloning and sequencing from a plasma small RNA library, (ii) unique results on miRNA stability that provide a firm grounding for further investigation of this class of molecules as blood-based cancer biomarkers, and (iii) evidence that tumor-derived miRNAs can enter the circulation even when originating from an epithelial cancer type (as compared with hematopoietic malignancies like lymphoma). Most importantly, our study of miR-141 in prostate cancer patients demonstrates that serum levels of a tumor-expressed miRNA can distinguish, with significant specificity and sensitivity, patients with cancer from healthy controls.
The available data indicate that miR-141 is expressed in an epithelial cell type-specific manner in a range of common human cancers. Given this, we speculate that it could have value in the setting of detecting cancer recurrence for cancer types for which clinically validated blood biomarkers are lacking (e.g., lung cancer, breast cancer, etc.). We also anticipate that advances in miRNA qRT-PCR assay design and assay optimization, and the application of alternative miRNA quantitation strategies, will substantially improve the approach and will likely be needed to detect cancer at lower tumor burdens (i.e., early-stage disease). It is likely that other blood-based miRNA markers that are specific for particular cancer types will be discovered. The results presented here establish the foundation and rationale to motivate future global investigations of miRNAs as circulating cancer biomarkers for a variety of common human cancers.
Our Study Raises Intriguing Questions Regarding the Mechanism of miRNA Stability in Plasma and Potential Biological Roles of Circulating miRNAs.
The remarkable stability of miRNAs in clinical plasma samples raises important and intriguing questions regarding the mechanism by which miRNAs are protected from endogenous RNase activity. One tantalizing hypothesis is that they are packaged inside exosomes that are secreted from cells. Exosomes are 50- to 90-nm (24), membrane-bound particles that have been reported to be abundant in plasma (25) and that have recently been shown (in cell culture studies) to contain miRNAs (26). Our results on filtration (through a 0.22-μm filter) and differential centrifugation of plasma containing tumor-derived miRNAs are certainly consistent with this hypothesis (Fig. S5). Alternative explanations include protection via association with other molecules (e.g., in a RNA–protein complex) or modifications of the miRNAs that make them resistant to RNase activity.
The high abundance of many circulating miRNAs also raises provocative questions regarding their potential biological role as extracellular messengers mediating short- and long-range cell–cell communication, reminiscent of the similar role played by small RNAs that spread within the organism in plants and C. elegans (27, 28). Additional studies will be needed to explore these exciting hypotheses.
Advantages and Potential of miRNAs as Blood-Based Cancer Biomarkers.
The availability of powerful approaches for global miRNA characterization and simple, universally applicable assays for quantitation (e.g., qRT-PCR) suggests that the discovery–validation pipeline for miRNA biomarkers will be more efficient than traditional proteomic biomarker discovery–validation pipelines, which typically encounter bottlenecks at the point of antibody generation and quantitative assay development for validation of biomarker candidates (2). In addition, the inherent regulatory function of miRNAs makes it likely that many miRNAs expressed in tumor tissue influence the biological behavior and clinical phenotype of the tumor. As the functional roles of miRNAs in tumor biology are unraveled, we envision that blood-based miRNA biomarkers that predict clinical behavior and/or therapeutic response will be identified.
Materials and Methods
Clinical Samples.
Human plasma and serum samples from healthy donors or patients with cancer were obtained with informed consent under institutional review board-approved protocols. Samples were derived from the Pacific Ovarian Cancer Research Consortium, local healthy donors from the Seattle area, or the Department of Urology, University of Washington. Details of sample collection and processing and relevant corresponding clinical data are provided in SI Text.
RNA Isolation.
Isolation of RNA from plasma or serum was carried out using a modification of the mirVana PARIS kit (Ambion) that is described in detail in SI Text.
Mouse Xenograft Experiment.
Xenografts were established in NOD/SCID mice by s.c. injection of 7.5 × 105 22Rv1 cells per mouse. Mouse blood was collected by cardiac puncture at 28 days after injection. Additional details of cell culture, establishment of xenografts, plasma collection, and RNA isolation are provided in SI Text.
qRT-PCR.
MiRNAs were quantified by using TaqMan miRNA qRT-PCR assays with modifications as described in detail in SI Text.
Normalization of Experimental qRT-PCR Data from Plasma or Serum Using Spiked-In C. elegans Controls.
Three C. elegans miRNAs were chosen because of a lack of sequence homology to human miRNAs and absence of empiric hybridization to human miRNA probes on miRNA microarrays (data not shown). Molar concentrations to be spiked-in were derived empirically to produce Ct values comparable to those of moderately abundant miRNAs (e.g., miR-16) measured in blood from human plasma from normal healthy donors (Table S9). Synthetic versions of the C. elegans miRNAs were spiked into plasma or serum after the addition of an equal volume of 2× denaturing solution (to inhibit RNases) to the samples. Full details of the use of these controls to normalize qRT-PCR results across independent RNA isolations are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank C. Nourigat, M. Mocilac, M. Comstock, and the Fred Hutchinson Cancer Research Center NOD/SCID core facility for technical assistance with mouse xenograft experiments and F. Appelbaum, A. Geballe, J. Olson, S. Tapscott, B. Torok-Storb, and S. Collins for constructive comments on the manuscript. This work was supported in part by Pacific Ovarian Cancer Research Consortium/Specialized Program of Research Excellence in Ovarian Cancer Grant P50 CA83636 (to N.U. and M.T.), National Cancer Institute Grant 5 P01 CA085859 (to R.L.V.), Pacific Northwest Prostate Cancer Specialized Program of Research Excellence Grant P50 CA97186 (to P.S.N., M.T., and R.L.V.), Core Center of Excellence in Hematology Pilot Grant P30 DK56465 (to M.T.), and the Paul Allen Foundation for Medical Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804549105/DCSupplemental.
References
- 1.Etzioni R, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Stefani G, Slack FJ. Small noncoding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs: MicroRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Xi Y, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap-frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson PT, et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Hefnawy T, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50:564–573. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 14.Attardi BJ, et al. Steroid hormonal regulation of growth, prostate-specific antigen secretion, and transcription mediated by the mutated androgen receptor in CWR22Rv1 human prostate carcinoma cells. Mol Cell Endocrinol. 2004;222:121–132. doi: 10.1016/j.mce.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Hartel A, et al. Characterization of steroid receptor expression in the human prostate carcinoma cell line 22RV1 and quantification of androgen effects on mRNA regulation of prostate-specific genes. J Steroid Biochem Mol Biol. 2004;92:187–197. doi: 10.1016/j.jsbmb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Holleran JL, Miller CJ, Culp LA. Tracking micrometastasis to multiple organs with lacZ-tagged CWR22R prostate carcinoma cells. J Histochem Cytochem. 2000;48:643–651. doi: 10.1177/002215540004800508. [DOI] [PubMed] [Google Scholar]
- 17.Sramkoski RM, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403–409. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 18.Porkka KP, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, et al. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 21.Tsang JC, Lo YM. Circulating nucleic acids in plasma/serum. Pathology. 2007;39:197–207. doi: 10.1080/00313020701230831. [DOI] [PubMed] [Google Scholar]
- 22.Chim SS, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 23.Lawrie CH, et al. Detection of elevated levels of tumor-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 24.van Niel G, et al. Exosomes: A common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 25.Caby MP, et al. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 26.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.Hunter CP, et al. Systemic RNAi in Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 2006;71:95–100. doi: 10.1101/sqb.2006.71.060. [DOI] [PubMed] [Google Scholar]
- 28.Voinnet O. Noncell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.