Abstract
Vibrio cholerae, the agent of cholera, has two circular chromosomes. In bacteria that contain a single chromosome, initiation of chromosome DNA replication is mediated by DnaA, a AAA+ ATPase that unwinds the origin of replication. There is little knowledge regarding initiation of chromosome replication in bacteria with more than one chromosome. Here, we purified V. cholerae DnaA and RctB, which have been implicated in the replication of V. cholerae chromosome II, and characterized their activities in vitro. We found that RctB has origin-specific unwinding activity and can melt the origin of chromosome II (oriCIIvc) but not the origin of chromosome I (oriCIvc); conversely, DnaA promoted the unwinding of oriCIvc and not oriCIIvc. The activity of DnaA and several plasmid initiator proteins is stimulated by ATP binding. We found that RctB bound and hydrolyzed ATP even though RctB lacks any apparent ATP-binding motifs. However, we unexpectedly found that ATP inhibited the oriCIIvc binding activity of RctB, suggesting that the ATP-bound form of RctB cannot initiate replication of chromosome II. Supporting this idea, we identified an RctB mutant that does not bind ATP and found that expression of this ATP-blind RctB mutant in V. cholerae leads to significant overinitiation of chromosome II and marked inhibition of V. cholerae growth. These observations suggest that the rules that license the replication of the two V. cholerae chromosomes differ.
Keywords: initiation of replication, RctB, DnaA, cell cycle
Bacterial chromosome replication is initiated by the binding of an “initiator” protein to the origin of replication. DnaA, a conserved AAA+ ATPase, is thought to be the initiator of chromosome DNA replication in nearly all eubacteria (1). Most of our knowledge of DnaA function and regulation has been derived from studies of Escherichia coli. In this microorganism, DnaA binds to several 9-bp sequences (DnaA boxes) found in oriC (2), the origin of replication, leading to the formation of a nucleoprotein complex. Formation of this DnaA–oriC complex triggers the melting of a thermodynamically unstable A+T-rich sequence within oriC, known as the DUE (DNA Unwind Element). After unwinding of the DUE, DnaA directs loading of the DnaB helicase onto the ssDNA along with DnaC, the helicase loader (3). At this point, other components of the replisome load on to oriC and rapid bidirectional replication of the chromosome proceeds.
Only the ATP bound form of DnaA is active in initiation (4). It appears that binding of ATP to DnaA contributes to several aspects of initiation, although the underlying mechanisms are not fully understood. First, DnaA complexed with ATP has a higher affinity for certain DnaA binding sites in oriC (5). Second, it is thought that ATP-dependent interactions between DnaA molecules bound to different sites within oriC enable the formation of a higher-ordered nucleoprotein complex that is capable of melting the DUE (6). Although DnaA is an ATPase, the energy derived from ATP hydrolysis is not required for unwinding the DUE. Instead, the ATPase activity of DnaA is thought to be important for regulating its activity. Consistent with this idea, E. coli expressing an ATPase-deficient form of DnaA displays an overinitiation phenotype (7).
Not all bacteria contain a single circular chromosome like E. coli, and DnaA may not function as the initiator of replication of secondary chromosomes in such species. There is already evidence that DnaA does not function as the initiator of replication of the second chromosome in the Vibrionaceae and Photobacteriacea. The genomes in the dozens of species in these families, which include several important human and fish pathogens, all consist of two circular chromosomes (8). The origin of replication of each of the chromosomes in Vibrio cholerae, the cause of the severe diarrheal disease cholera, has been defined and characterized (9). V. cholerae chromosome I (chrI) is larger than chromosome II (chrII) and its origin of replication, oriCIvc, is similar in sequence and features to oriC. Like oriC, oriCIvc contains eight DnaA boxes, an AT-rich sequence, and a binding site for IHF. Moreover, replication of an oriCIvc-dependent minichromosome requires DnaA (9), suggesting that the process for initiating replication at oriCIvc resembles initiation at oriC. Finally, overexpression of DnaA leads to overinitiation of chrI but not chrII (10), suggesting that DnaA specifically triggers replication at oriCI.
The origin of replication of chrII, oriCIIvc, does not exhibit significant sequence similarity to oriCIvc or oriC, and it contains only one DnaA box. Instead, oriCIIvc contains many binding sites for RctB, a protein encoded by a gene located adjacent to oriCIIvc (9). Even though the predicted 658-aa RctB protein lacks similarity to any characterized initiator and does not have any known motifs, several lines of evidence suggest that this protein may act as the initiator of chrII replication. First, this protein binds to several sites within oriCIIvc (9). Second, replication of an oriCIIvc-based minichromosome requires RctB (9), and the copy number of such a minichromosome increases as the levels of RctB are raised (11). Finally, overproduction of RctB in V. cholerae promotes overinitiation of chrII and not chrI (10), suggesting that RctB specifically triggers initiation at oriCIvc.
Here, we purified RctB and characterized its activities in vitro. We found that RctB has origin-specific unwinding activity and can melt oriCIIvc but not oriCIvc. Even though RctB lacks any ATP-binding motifs, this protein bound and hydrolyzed ATP. Surprisingly, ATP inhibited the oriCIIvc binding activity of RctB, suggesting that the ATP-bound form of RctB cannot initiate replication of chrII. Supporting this idea, we identified an RctB mutant that does not bind ATP and found that expression of this “ATP blind” RctB mutant in V. cholerae leads to significant overinitiation of chrII and marked inhibition of V. cholerae growth.
Results
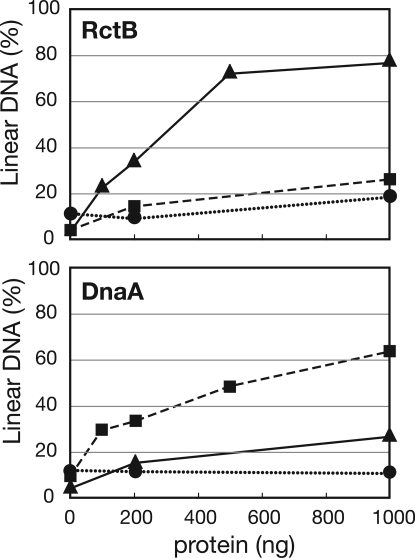
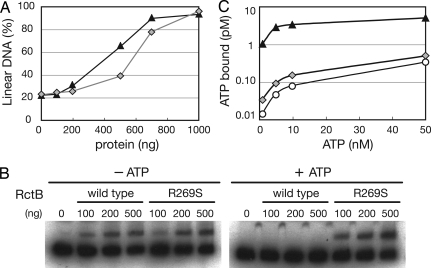
RctB and DnaA Promote the Unwinding of oriCIIvc and oriCIvc, Respectively.
Previous genetic and cytologic studies revealed that RctB and DnaA are required for the replication of oriCIIvc and oriCIvc, respectively (9, 10). To gain understanding of the biochemical activities of these proteins, we used a P1 nuclease-based assay to test whether purified RctB and/or DnaA can unwind oriCIvc and/or oriCIIvc in vitro. In this assay, V. cholerae RctB or DnaA was incubated with circular plasmid DNA substrates that harbored either oriCIvc (pOriI), oriCIIvc (pOriII), or no chromosome origin (pBR, a negative control). If the protein promotes the unwinding of an origin sequence, the DNA becomes susceptible to cleavage by the single-strand specific P1 endonuclease. The resulting linearized plasmid is then easily detected by agarose gel electrophoresis. We observed that RctB unwound the oriCIIvc-containing but not the oriCIvc-containing substrate in a concentration-dependent manner (Fig. 1), demonstrating that RctB can indeed function as the initiator of oriCIIvc-based replication. In contrast, DnaA unwound the oriCIvc-containing substrate but had no activity on the oriCIIvc-containing substrate (Fig. 1), indicating that DnaA can function as the initiator of oriCIvc-based replication. DnaA + RctB did not exhibit greater oriCIIvc-unwinding activity than RctB alone (data not shown), suggesting that although DnaA is required for the in vivo replication of an oriCIIvc-based replicon (9), this protein does not contribute to melting oriCIIvc to initiate replication of chrII. Together these observations support the idea that the two V. cholerae chromosomes rely on distinct initiators for their replication: DnaA is the initiator of chrI replication and RctB is the initiator of chrII replication.
Fig. 1.
OriCIvc and oriCIIvc unwinding activity of RctB and DnaA. Various concentrations of RctB (Upper) or DnaA (Lower) were added to reactions that contained 150 fmol of either pBR322 (●), pOriI (■), or pOriII (▲). P1 nuclease was subsequently added. Densitometric scanning of ethidium bromide-stained gels was used to determine the proportion of linearized plasmid molecules. The data presented are representative of at least three experiments.
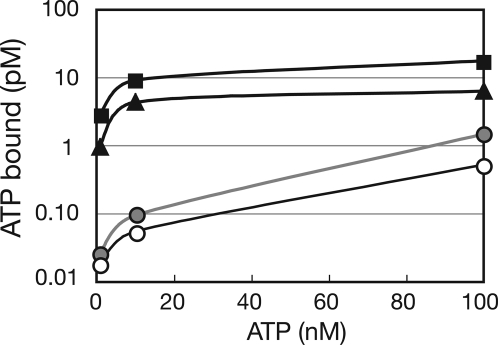
RctB and DnaA both Bind and Hydrolyze ATP.
The predicted amino acid sequence of V. cholerae DnaA contains Walker A and B motifs and thus, like E. coli DnaA, V. cholerae DnaA is expected to bind and hydrolyze ATP. In contrast, RctB lacks motifs suggestive of nucleotide binding or hydrolysis. We used a filter-binding assay to test whether V. cholerae DnaA and RctB bind ATP. Both initiator proteins bound ATPα-P32 with similar Kd (DnaA, 43 nM; RctB, 39 nM) (Fig. 2). The affinity of DnaA and RctB for ATP is comparable to that reported for DnaA purified from E. coli (4) or Mycobacterium tuberculosis (12). We also used the same assay to test whether either of the two V. cholerae initiator proteins bound UTP, CTP, or GTP but neither RctB nor DnaA had detectable affinity for these triphospho-nucleotides (data not shown). However, we found that ADP could compete with ATP for binding to RctB and DnaA [supporting information (SI) Fig. S1], suggesting that both proteins can bind ADP in addition to ATP (Fig. S1).
Fig. 2.
ATP binding by RctB and DnaA. A total of 80 nM of DnaA (■), RctB (▲), CpxR (gray circles), a protein that is not known to bind ATP, or no protein (○) were incubated with different concentrations of P32-αATP, and the bound ATP was detected by using a filter binding assay. A representative experiment of three experiments is shown.
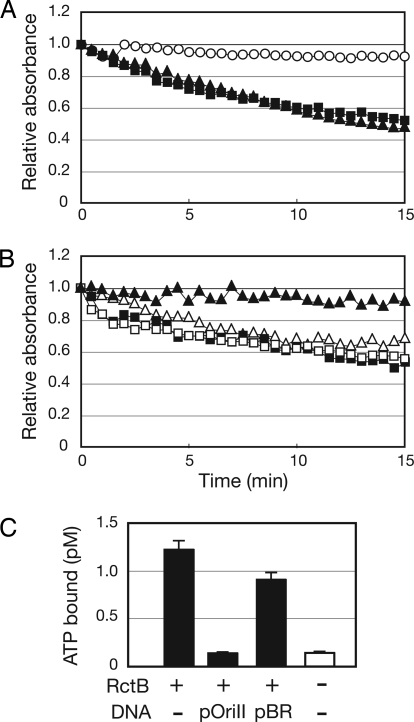
To test whether RctB and DnaA can hydrolyze ATP, we used an assay in which the conversion of ATP to ADP + Pi is linked to the oxidation of NADH to NAD+ (detected as a decrease in absorbance at 340 nm) (13). Both RctB and DnaA proved capable of hydrolyzing ATP (Fig. 3A). The calculated kcat values of ATP hydrolysis for DnaA and RctB were similar (kcat RctB, 0.41 min−1; kcat DnaA, 0.67 min−1) and in the same range as has been reported for DnaA derived from other bacteria (12).
Fig. 3.
Hydrolysis of ATP by RctB and DnaA. (A) Spectrophotometric assay of generation of ADP after incubation of 10 μM DnaA (■), 10 μM RctB (▲), or no protein (○) with ATP. (B) Influence of pOriCII DNA (triangles) or pOriCI DNA (squares) on ATP hydrolysis by DnaA (open symbols) or RctB (closed symbols). (C) Inhibition of RctB binding of ATP by pOriCII DNA. A total of 80 nM of RctB was incubated with 1 nM of P32-αATP in the presence or absence of 180 ng of pBR or pOriCII DNA. A control experiment without RctB and DNA shows the background level of P32-αATP binding to the filter. The results shown in A and B are representative of at least three experiments, and the data in C represent the average and standard deviation derived from three experiments.
oriCIIvc DNA Inhibits RctB from Binding ATP.
Supercoiled DNA has been reported to stimulate the ATPase activity of E. coli DnaA in a sequence-independent fashion (4). We tested whether supercoiled plasmids influenced ATP hydrolysis by V. cholerae DnaA or RctB. Neither pOriI, pOriII, or pBR appreciably altered DnaA's hydrolysis of ATP (Fig. 3B and data not shown). However, the ATPase activity of RctB was inhibited by DNA in a sequence-specific fashion: pOriII almost completely abolished ATP hydrolysis by RctB, whereas pOriI and pBR did not alter RctB's hydrolysis of ATP (Fig. 3B and data not shown). These observations suggest that the oriCIIvc-bound form of RctB is unable to hydrolyze ATP. The oriCIIvc-mediated block in the ATPase activity of RctB appears to occur at the level of RctB binding to ATP. The presence of pOriII decreased RctB binding of ATP to near background levels, whereas pBR had little effect on RctB binding of ATP (Fig. 3C). In aggregate, these observations suggest that RctB binding to oriCIIvc renders the initiator unable to interact with ATP.
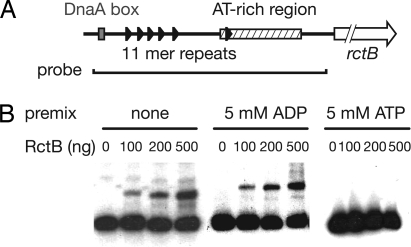
ATP Inhibits RctB Binding to oriCIIvc.
Because oriCIIvc DNA inhibited RctB binding of ATP, we explored whether ATP inhibited RctB binding to oriCIIvc. In these experiments, RctB was preincubated with ATP or ADP and then electrophoretic mobility-shift assays (EMSAs) were done to test RctB binding to a DNA probe derived from the oriCIIvc region (Fig. 4A). Premixing RctB with ATP abolished RctB binding to oriCIIvc DNA, suggesting that ATP negatively regulates RctB's ability to function as an initiator. In contrast, premixing RctB with ADP did not appear to alter RctB binding to oriCIIvc (Fig. 4B). Together, these two observations suggest the possibility that hydrolysis of ATP bound to RctB could relieve ATP-mediated inhibition of RctB binding to oriCIIvc, thereby enabling RctB to initiate replication at oriCIIvc.
Fig. 4.
Influence of ATP on RctB binding to oriCIIvc. (A) Schematic representation of the oriCIIvc region, AT-rich region (hatched area), DnaA box (gray rectangle), 11-mer repeats (black triangles), and rctB (arrow). The line below the map represents the probe used for the EMSA experiments. (B) Autoradiographs of EMSA experiments. The indicated molar quantities of RctB were preincubated for 10 min with either ATP or ADP. Afterward, the radioactive probe was added to the reaction.
An RctB Mutant That Does Not Bind ATP Promotes Overinitiation at oriCIIvc.
In a recent screen for high copy number variants of an oriCIIvc-based plasmid, we identified an RctB mutant that mediates high-copy oriCIIvc-based replication. Ordinarily, a minichromosome containing oriCIIvc, rctB, and a gene conferring resistance to kanamycin replicates with a very low copy number in E. coli (Fig. S2 and data not shown). In contrast, pYB286, which harbors a single nucleotide mutation in rctB codon 269, leading to a substitution of serine for arginine (R269S), replicates at a much higher copy number (Fig. S2). Quantitative PCR analyses revealed that the copy number of pYB286 is >100 times greater than that of plasmids lacking the R269S mutation (data not shown). To gain further understanding of the activity and regulation of RctB, we explored how the R269S RctB mutant mediates high copy oriCIIvc-based replication.
We first tested whether the enhanced copy number of pYB286 could be attributed to elevated oriCIIvc unwinding activity by this protein. As was found with wild-type (WT) RctB, the mutant R269S RctB proved capable of unwinding oriCIIvc (Fig. 5A) but did not promote formation of linear pBR (data not shown). Although the P1 nuclease-based assay is only semiquantitative, comparison of the oriCIIvc unwinding activity of R269S RctB and WT RctB did not suggest that the mutant protein had elevated oriCIIvc opening activity (Fig. 5A). Thus it seems unlikely that the elevated copy number of pYB286 can be explained by enhanced oriCIIvc unwinding by R269S RctB.
Fig. 5.
oriCIIvc unwinding and binding of ATP and oriCIIvc by R269S RctB. (A) P1 nuclease cleavage assay on oriCIIvc DNA by WT RctB (triangles) and RctB R269S (gray diamonds). (B) EMSA of RctB and R269S RctB binding to the oriCIIvc probe in the presence or absence of 5 mM of ATP. ATP was mixed with RctB for 10 min before the addition of the oriCIIvc probe. (C) Binding of ATP by 80 nM RctB (triangles) or 80 nM RctB R269S (gray diamonds) using the filter binding assay as in Fig. 2; ○ represent ATP binding to the filter observed without the addition of protein.
Alternatively, the elevated copy number of pYB286 could result from altered regulation of R269S RctB activity. We tested whether the oriCIIvc DNA binding activity of R269S RctB was inhibited by ATP. In the absence of ATP, the binding of oriCIIvc DNA by R269S and WT RctB were comparable; however, unlike WT RctB, the binding of R269S RctB to oriCIIvc was not inhibited by ATP (Fig. 5B). Thus, R269S RctB appears to be insensitive to the inhibitory effect of ATP that was observed with WT RctB.
The lack of ATP inhibition of R269S RctB binding to oriCIIvc is likely attributable to a severe reduction in the binding of ATP to this protein. ATP binding to R269S RctB was markedly reduced compared with the binding of ATP to WT RctB (Fig. 5C). ATP binding to R269S RctB was only marginally above the background levels observed when no protein was used (Fig. 5C). These observations suggest that Arg-269 is a crucial residue for RctB to bind ATP. Together, the findings that R269S RctB has approximately WT oriCIIvc unwinding activity but greatly reduced affinity for ATP suggests that the absence of ATP inhibition of RctB binding to oriCIIvc accounts for the high levels of R269S RctB-mediated oriCIIvc-based replication in E. coli.
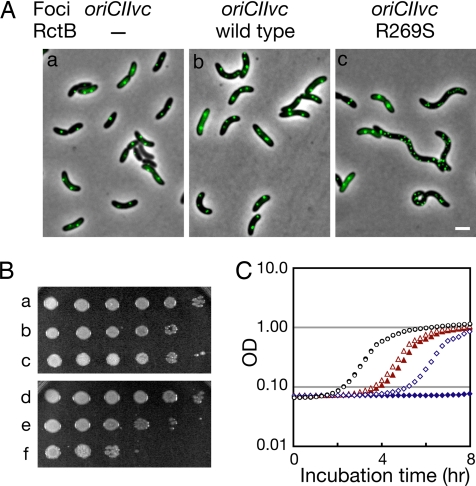
Overinitiation of Chromosome II Replication Blocks V. cholerae Growth.
We introduced plasmids containing isopropyl β-D-thiogalactopyranoside (IPTG)-inducible forms of RctB or R269S RctB into V. cholerae strain N16961 to explore the effects of these proteins on V. cholerae morphology, chromosome replication, and growth. As previously observed, overproduction of WT RctB did not significantly alter cell morphology (10). In contrast, ≈20% of cells overproducing R269S RctB exhibited filamentation (Fig. 6A), suggesting that R269S RctB blocks V. cholerae cell division. We used the fluorescent repressor-operator system (14) to evaluate the effects of overexpression of R269S RctB on the number of chrI and chrII origin regions in V. cholerae. As reported (10), overproduction of WT RctB led to a modest increase in the number oriCIIvc foci detected in most cells (Fig. 6Ab). In contrast, overproduction of R269S RctB led to a larger increase in the number of oriCIIvc foci (Fig. 6Ac). Nearly 11% of cells overexpressing R269S had six or more oriCIIvc foci, whereas <1% of cells overproducing RctB had this number of foci. Most of the cells with very large numbers (6–20 total) of oriCIIvc foci were filamentous (Fig. 6Ac). The number of foci representing the terminus of chrII (terIIvc) did not show a corresponding increase in cells overexpressing R269S; <4% of cells had ≥6 terIIvc foci (Fig. S3). Together, these observations are consistent with the idea that expression of R269S RctB leads to overinitiation of chrII replication in vivo. The effects of R269S RctB were chromosome-specific and no increase in the number of oriCIvc foci was detected in cells expressing R269S RctB (Fig. S3). The different effects of overexpression of WT vs. R269S RctB cannot be attributed to greater expression of the mutant protein, as Western blotting revealed that WT RctB was more abundant in these assays (data not shown)
Fig. 6.
In vivo characterization of V. cholerae overexpressing R269S RctB. (A) Detection by fluorescent microscopy of oriCIIvc, using TetR-YFP in an N16961 derivative that contains a tet operator array near oriCIIvc. Representative fields after 1 h of induction of TetR-YFP and RctB are shown. (Scale bar: 2 μm.) (B and C) Growth of V. cholerae N1691 with or without overexpressed RctB or R269S RctB. In B, overnight cultures of N16961 harboring a control vector (a and d), a vector containing WT rctB (b and e), or a vector containing R269S rctB (c and f) were serially diluted and spotted onto LB + chloramphenicol (5 μg/ml) with (d–f) or without (a–c) 100 μM IPTG. In C, the overnight cultures were diluted 1,000-fold into fresh media ± IPTG, and growth was monitored by absorbance at 600 nm. Control vector, no IPTG, (open black circles); control vector, + IPTG (filled black circles); WT rctB, no IPTG, (open red triangles); WT rctB + IPTG, (filled red triangles), R269S rctB, no IPTG (open blue diamonds), R269S rctB + IPTG (filled blue diamonds).
Overproduction of R269S RctB profoundly inhibited V. cholerae growth. When overnight cultures of V. cholerae strains harboring IPTG-inducible forms of RctB or R269S RctB grown in the absence of inducer were plated in the absence or presence of IPTG (Fig. 6B Upper and Lower, respectively), ≈100-fold fewer colonies arose when R269S RctB was overproduced (Fig. 6B, compare rows c and f); in marked contrast, overexpression of WT RctB did not inhibit V. cholerae growth in this assay (Fig. 6B, compare rows b and e). Furthermore, when these overnight cultures were diluted in fresh media ± IPTG, there was no detectable growth of the cells overproducing R269S RctB for the 8-h culture period (Fig. 6C, filled blue diamonds). Together with the observations described above, these findings suggest that overinitiation of chrII replication mediated by R269S RctB is extremely toxic to cells, at least in part because it blocks V. cholerae cell division.
Discussion
There is scant knowledge of the mechanisms that govern chromosome replication in bacteria that have genomes consisting of more than one chromosome. Our findings strongly support the hypothesis that the two V. cholerae chromosomes have distinct initiators that function in an origin-specific manner: DnaA initiates replication of chrI and RctB initiates replication of chrII. Because these proteins are conserved in all sequenced species of vibrio and photobacteria, it is likely that DnaA and RctB function as chrI and chrII initiators, respectively, in all members of the Vibrionaceae and Photobacteriacea. In this work, we did not focus on the regulation of V. cholerae DnaA, but given the similarity of the V. cholerae and E. coli DnaA amino acid sequences (79% identity and 83% similarity) it is possible that at least some of the mechanisms that are known to govern DnaA activity in E. coli also control DnaA activity in V. cholerae. Unlike DnaA, RctB is conserved only in vibrio and photobacteria species, and it lacks any functional motifs. Our observation that RctB, like DnaA, can bind and hydrolyze ATP, suggested that ATP might regulate RctB activity in a similar manner as described for DnaA. However, we unexpectedly found that ATP inhibited RctB binding to oriCIIvc, suggesting that ATP-RctB does not function as the initiator of chrII replication. In marked contrast, only the ATP bound form of DnaA is able to initiate replication in E. coli (4).
To our knowledge, the negative regulation of an initiator protein by ATP is without precedent among bacterial initiator proteins. Furthermore, like DnaA, the characterized eukaryotic and Archaea initiator proteins are AAA+ ATPases that are only active in their ATP-bound forms and inactivated by ATP hydrolysis (1). RctB, although an ATPase, may use ATP hydrolysis in the opposite manner as other initiators, as a positive switch enabling its melting of the origin of V. cholerae chrII. Overall, our findings are consistent with a model in which ATP binding to RctB renders it unable to bind to oriCIIvc. The reason ATP-RctB does not bind oriCIIvc is not known. However, it is unlikely that ATP and oriCIIvc compete for the same binding site on RctB, because a truncated RctB fragment binds ATP but not oriCIIvc (data not shown). Instead, it is possible that ATP acts as an allosteric inhibitor of RctB. It is tempting to speculate that a cell-cycle-dependent factor regulates hydrolysis of ATP by RctB, thereby restricting the availability of active (non-ATP-bound) RctB and preventing excessive initiation of chrII replication. While bound to oriCIIvc, RctB cannot bind ATP and is not subject to its inhibitory effects.
Our findings suggest that there must be a complex interplay between RctB, ATP, and oriCIIvc and perhaps other factors such as DnaA as well. We found that ATP inhibited RctB binding to oriCIIvc and conversely that oriCIIvc inhibited RctB binding to ATP. Unexpectedly, we did not detect significant ATP inhibition of RctB unwinding of oriCIIvc DNA using the P1nuclease assay (data not shown). This observation may suggest that the P1 assay is a more sensitive assay of RctB binding to oricIIvc than the EMSA, presumably because it reveals transient RctB-oriCIIvc interactions. In vivo, it seems likely that the relative levels and subcellular distribution of RctB, ATP, and oriCIIvc as well as additional factors determine when chrII replication begins. Future studies must define the factors that promote the formation and dissolution of the RctB–oriCIIvc complex. We did not find a role for DnaA in oriCIIvc unwinding in vitro, but DnaA is required for replication of an oriCIIvc-based replicon in E. coli (9). It is possible that DnaA promotes the loading of the replicative helicase (DnaB) on to oriCIIvc, especially because V. cholerae does not encode a homologue of DnaC, the E. coli helicase loader.
Although ATP appears to control the initiator activities of RctB and DnaA in opposite fashions, the two proteins seem to have several similar features. Both proteins are weak ATPases that may use ATP hydrolysis to regulate their activity. As for DnaA, the ATPase activity of RctB is not required for its origin-unwinding activity. We observed that WT RctB can melt oriCIIvc in the absence of ATP; furthermore, R269S RctB is able to melt oriCIIvc even though it does not bind ATP. Like R269S RctB, E. coli DnaAcos is defective for ATP binding and causes lethal overinitiation at oriC (7, 15). Overinitiation mediated by R269S RctB is expected if ATP negatively regulates RctB's interaction with oriCIIvc, whereas the reason DnaAcos mediates overinitiation at oriC is unclear.
Two studies have suggested that the replication of the two V. cholerae chromosomes is coordinated (16, 17). How could coordinated chromosome replication be achieved if ATP positively regulates oriCIvc unwinding by DnaA and negatively regulates oriCIIvc unwinding by RctB? Because we do not understand the other factors that regulate the activities of the two initiators of V. cholerae chromosome replication, it is possible that the opposite effects of ATP on DnaA and RctB do not impact the coordinated replication of the two chromosomes. Another potential explanation for this conundrum may be the proposal by Rasmussen et al. (17) that chrI and chrII do not initiate replication synchronously; instead they proposed that the two chromosomes terminate their replication synchronously. If this is the case, replication of some chrI-encoded product could provide a cue that triggers the hydrolysis of ATP bound to RctB, allowing chrII to initiate replication at a time that will ensure that the two chromosomes terminate replication synchronously. It is also possible that DnaA-ADP, generated by initiation of chrI replication, preferentially promotes initiation at oriCIIvc.
Materials and Methods
Protein Purification.
V. cholerae dnaA, rctB, and rctB R269S were subcloned into the pET28b His-Tag vector (Novagen). The DNA sequence of each construct was experimentally confirmed. After expression of these genes in E. coli strain BL21, the cells were lysed in a French pressure cell and the N-terminal His-tagged proteins were purified by using His-trap nickel columns (Amersham) and FPLC as described (18). The purity of each of these proteins was >90% as estimated by SDS-PAGE.
P1 Nuclease Cleavage Assay.
P1 nuclease cleavage of open complexes was carried out essentially as described by Bramhill and Kornberg (19). Each reaction (50 μl) contained 40 mM Hepes-KOH (pH 7.6), 8 mM magnesium acetate, 30% glycerol, 320 μg/ml BSA, 5 mM ATP, 150 fmol of supercoiled plasmid DNA, and different concentrations of RctB, RctB R269S, or DnaA. After incubation at 37°C for 10 min, P1 nuclease (1.2 units in 3 ml of 30 μM sodium acetate, pH 5.5) was added and incubated at 37°C for 30 s. P1 nuclease activity was then stopped by addition of 40 μl of stop buffer (25 mM EDTA, 1% SDS). For quantification of the fraction of the DNA that was linearized by the P1 nuclease, a portion of the reaction was electrophoresed in 0.8% TAE (40 mM Tris-acetate, 1 mM EDTA) agarose gel and then stained with ethidium bromide. Quantity One software (Bio-Rad) was used to determine the proportion of the plasmid DNA that was linearized by P1 nuclease.
ATP (ADP) Binding Assay.
Binding of RctB and DnaA to ATP and ADP was assessed with a nitrocellulose filter binding assay essentially as described by Sekimizu et al. (4). Some 80 nM of protein (RctB, DnaA, or CpxR) was incubated in 40 μl of buffer E [50 mM Tricine KOH (pH 8.25), 0.5 mM Mn acetate, 0.3 mM EDTA, 7 mM DTT, 20% glycerol, 0.007% Triton X-100] with different concentrations of P32-αATP for 15 min on ice. The reaction mixture was filtered onto a nitrocellulose membrane (Millipore HA 0.45 μM) presoaked in buffer E. The filter was washed with 10 ml of cold buffer F [50 mM Tricine KOH (pH 8.25), 0.5 mM Mn acetate, 0.3 mM EDTA, 5 mM DTT, 17% glycerol, 10 mM ammonium sulfate, 0.005% Triton X-100] and dried, and then the amount of radioactivity retained on the filter was measured in a liquid-scintillation counter.
ATP Hydrolysis Assay.
Hydrolysis of ATP was monitored by using a coupled spectrophotometric assay as described by Huang and Hackney (13). The reaction buffer contained 25 mM Tris acetate (pH 7.5), 13 mM magnesium acetate, 1.8 mM DTT, 5 mM phosphoenolpyruvate, 20 units/ml pyruvate kinase, 20 units/ml lactate dehydrogenase, 5 mM ATP, 100 μg/ml purified BSA, 10 μM of protein (RctB or DnaA), 250 ng supercoiled DNA, and 0.3 mM NADH. The 100-μl reaction was set up directly in the cuvette of a Beckman DU 530 spectrophotometer at room temperature. The reaction was initiated by addition of ATP and then monitored for 15 min.
oriCIIvc DNA Binding Assay.
Probes for electrophoretic mobility-shift experiments were amplified by PCR using chromosomal DNA prepared from V. cholerae strain N16961 as a template; P32-labeled PCR primers (5′- CACTCAGGTTGTGGATAAAC-3′, 5′-GATCCGTATCACACTTACCG-3′) were used to label the probe. The labeled probe was purified from an acyrlamide gel, and the amount of P32 incorporated into the probe was determined by liquid scintillation counting. Twenty-microliter reactions were performed by incubating 5,000 cpm of probe DNA with different concentrations of RctB in a reaction buffer containing 200 mM Tris·Hcl (pH 7.5), 10 mM EDTA, 800 mM NaCl, 12.5 μg/ml sonicated salmon sperm, and 0.1 mg/ml of BSA for 10 min at room temperature. In some experiments, RctB was preincubated in a reaction buffer containing 5 mM of ATP or ADP. The reactions were then electrophoresed in a 6% DNA retardation gel and visualized by autoradiography.
Isolation of RctB R269S.
We constructed a linear DNA molecule that consisted only of rctA-oriCIIvc-rctB and a kanamycin resistance gene. After in vitro ligation, attempts to recover a circular form of this DNA molecule after its transformation into E. coli DH5α routinely failed, presumably because rctA is a negative regulator of RctB (20). On the rare occasions when colonies arose after this transformation, they usually contained deletions in rctA (as in pYB289) or mutations in rctB. One of the mutant rctB sequences (in pYB286) was predicted to have a serine rather than an arginine at position 269 (R269S) and proved capable of mediating high copy replication of an oriCIIvc-based plasmid. The WT and R269S rctB genes were amplified by PCR and cloned into the EcoRI site of pGZ119EH (21), yielding pYB285 and pYB291, respectively. After confirming the nucleotide sequences of these plasmids, they were used for the studies shown in Fig. 6.
Microscopy.
To observe cell and nucleoid morphology in V. cholerae strains harboring plasmids expressing WT or mutant rctB, cells were grown in LB broth until OD600 ≈0.1 and then RctB was overexpressed by adding 100 μM of IPTG. After 1 h, cells were fixed with fixation solution (2.5% formaldehyde, 30 mM NaPO4, pH 7.5). Cell nucleoids were stained with 250 ng/ml of DAPI. Images were captured and processed as described (18). oriCIIvc and oriCIvc were visualized as described (10, 14, 22). The V. cholerae strain containing tetOP array at terIIvc was constructed as described (22).
Plasmids and Strains.
Plasmids and strains used in this study are listed in Table S1.
Supplementary Material
Acknowledgments.
We thank Brigid Davis and Sarah McLeod for their helpful comments on the manuscript. This work was supported by grants from the Howard Hughes Medical Institute and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803904105/DCSupplemental.
References
- 1.Zakrzewska-Czerwinska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev. 2007;31:378–387. doi: 10.1111/j.1574-6976.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- 2.Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 3.Masai H, Nomura N, Arai K. The ABC-primosome. A novel priming system employing dnaA, dnaB, dnaC, and primase on a hairpin containing a dnaA box sequence. J Biol Chem. 1990;265:15134–15144. [PubMed] [Google Scholar]
- 4.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 5.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci USA. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mott ML, Berger JM. DNA replication initiation: Mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 7.Simmons LA, Kaguni JM. The DnaAcos allele of Escherichia coli: Hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol Microbiol. 2003;47:755–765. doi: 10.1046/j.1365-2958.2003.03333.x. [DOI] [PubMed] [Google Scholar]
- 8.Okada K, Iida T, Kita-Tsukamoto K, Honda T. Vibrios commonly possess two chromosomes. J Bacteriol. 2005;187:752–757. doi: 10.1128/JB.187.2.752-757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan ES, Waldor MK. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- 10.Duigou S, et al. Independent control of replication initiation of the two Vibrio cholerae chromosomes by DnaA and RctB. J Bacteriol. 2006;188:6419–6424. doi: 10.1128/JB.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J Bacteriol. 2005;187:7167–7175. doi: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto K, Muniruzzaman S, Rajagopalan M, Madiraju MV. Modulation of Mycobacterium tuberculosis DnaA protein–adenine–nucleotide interactions by acidic phospholipids. Biochem J. 2002;363:305–311. doi: 10.1042/0264-6021:3630305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang TG, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J Biol Chem. 1994;269:16493–16501. [PubMed] [Google Scholar]
- 14.Lau IF, et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 15.Simmons LA, Breier AM, Cozzarelli NR, Kaguni JM. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol Microbiol. 2004;51:349–358. doi: 10.1046/j.1365-2958.2003.03842.x. [DOI] [PubMed] [Google Scholar]
- 16.Egan ES, Lobner-Olesen A, Waldor MK. Synchronous replication initiation of the two Vibrio cholerae chromosomes. Curr Biol. 2004;14:R501–R502. doi: 10.1016/j.cub.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26:3124–3131. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaichi Y, Fogel MA, McLeod SM, Hui MP, Waldor MK. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. J Bacteriol. 2007;189:5314–5324. doi: 10.1128/JB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 20.Venkova-Canova T, Srivastava P, Chattoraj DK. Transcriptional inactivation of a regulatory site for replication of Vibrio cholerae chromosome II. Proc Natl Acad Sci USA. 2006;103:12051–12056. doi: 10.1073/pnas.0605120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessl M, et al. Dissection of IncP conjugative plasmid transfer: Definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fogel MA, Waldor MK. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol Microbiol. 2005;55:125–136. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.