Abstract
The subseafloor marine biosphere may be one of the largest reservoirs of microbial biomass on Earth and has recently been the subject of debate in terms of the composition of its microbial inhabitants, particularly on sediments from the Peru Margin. A metagenomic analysis was made by using whole-genome amplification and pyrosequencing of sediments from Ocean Drilling Program Site 1229 on the Peru Margin to further explore the microbial diversity and overall community composition within this environment. A total of 61.9 Mb of genetic material was sequenced from sediments at horizons 1, 16, 32, and 50 m below the seafloor. These depths include sediments from both primarily sulfate-reducing methane-generating regions of the sediment column. Many genes of the annotated genes, including those encoding ribosomal proteins, corresponded to those from the Chloroflexi and Euryarchaeota. However, analysis of the 16S small-subunit ribosomal genes suggests that Crenarchaeota are the abundant microbial member. Quantitative PCR confirms that uncultivated Crenarchaeota are indeed a major microbial group in these subsurface samples. These findings show that the marine subsurface is a distinct microbial habitat and is different from environments studied by metagenomics, especially because of the predominance of uncultivated archaeal groups.
Keywords: Archaea, Chloroflexi, marine sediment, quantitative PCR
With perhaps one-third of Earth's biomass (1), the marine subsurface represents a unique, widespread, and largely understudied microbial ecosystem that influences large-scale geochemical cycles (2). It has been difficult to determine which microorganisms are responsible for most major geochemical cycles in the subsurface, including those for methane and sulfate (3, 4). Recently, a study of the isotopic fractionation of carbon pools in the subsurface led to the suggestion that new metabolisms may be possible in this unique environment, where substrates are restricted and cells may survive on incredibly long time scales (5).
The microbial groups that have been detected in the marine subsurface, especially from sediments collected on Ocean Drilling Program (ODP) Leg 201, are composed mostly of uncultivated representatives. The bacterial groups predominating clone libraries from these sediments include JS1 and Chloroflexi (4, 6, 7). Archaeal groups that predominate clone libraries include DSAG (MBGB), MCG, MBGC, and SAGMEG (4, 7–9). Most of these groups are either characteristic to the subseafloor or have discrete subsurface clades and are unique enough that they may not amplify well with traditional primers (10).
In addition to potentially providing insight into the genetic functions of subsurface organisms, a metagenomic analysis provides additional information about the members of the microbial population. This is particularly important for the sediments collected from the Peru Margin during ODP Leg 201, where previous studies have yielded widely divergent views about the prevalence of Archaea in the subsurface. Some studies suggest that Archaea are a small minority of the subsurface community (4, 11) whereas others indicate that Archaea are the majority of the population (5), and still others suggest an intermediate result (12). Methodological differences may be responsible for these disparate views, perhaps caused by differences in levels of cellular activity.
It has been proposed that metagenomic analysis yields the most accurate quantitative view of the microbial world (13). Multiple methods are available for metagenomic research and whereas all provide a direct view of the microbial population, pyrosequencing may allow the least bias because DNA is not replicated in Escherichia coli, and no specific primers are used (14). For the deep subsurface, the lack of primer discrimination is of particular importance because of the potential to discover novel microbial groups (10). Additionally, pyrosequencing is the most feasible sequencing approach to apply to deep sediment because of the extremely low DNA yields from this environment (15). Pyrosequencing has been shown to be useful and accurate in describing the microbial community found within a deep mine (16), a wooly mammoth (17), and honey bee colonies (18). Here, we use pyrosequencing to gain a fresh view of the microbiology of the ODP Leg 201 sediments, advance our understanding of this immense ecosystem, and test whether Archaea achieve a significant population at depth.
Results
DNA was extracted from the deeply buried marine sediment from ODP Site 1229 at 1, 16, 32, and 50 m below seafloor (mbsf). As expected, relatively low amounts of DNA were retrieved from the deeper sediments (15). At 1 mbsf, a total of 400 ng was extracted. At 16 mbsf, only 0.120 ng was extracted, and at 32 and 50 mbsf, 12 and 2.8 ng was extracted, respectively. Because of low yield, DNA from sediment >1 mbsf was subjected to whole-genome amplification (WGA) before sequencing (19, 20). To control for variations potentially induced by WGA (21), the 1-mbsf sample (original) was also subjected to WGA (amplified), and all five samples [1 (original), 1 (amplified), 16, 32, and 50] were prepared for pyrosequencing.
Sequencing yielded a total of 61.9 Mb of sequence from five samples at four separate horizons of Peru Margin ODP Leg 201 Site 1229 (Table 1). These data were in 622,129 reads, with average sequence length of 100 bp, a typical output for the Roche GS-20 Sequencer. This dataset was analyzed against the nonredundant database by BLASTX. Only 5.65–14.14% of the metagenome had a detectable homology (Table 1), with the lowest similarity at 16 mbsf. When searched against the protein family database (Pfam), 3.06–8.01% of reads could be classified into a protein family [Table 1 and supporting information (SI) Dataset S1]. The results from both search methods were compared, and a high level of similarity was found, typically with BLAST comparison providing a more detailed annotation than Pfam comparison, confirming the validity of BLAST comparison for general annotation. Because WGA can be prone to stochastic bias, a comparison of BLASTX results for the original (unamplified) and amplified 1-mbsf samples was made. It revealed similar complexity in the two datasets: The maximum (and average) number of matches to a single database sequence was 32 (1.411) and 31 (1.443) for original and amplified, respectively.
Table 1.
Results of pyrosequencing and metagenomics on DNA extracted from deeply buried marine sediments at ODP Site 1229
| Pyrosequenced samples |
Percentage of metagenome |
||||||
|---|---|---|---|---|---|---|---|
| ODP sample code | Depth, mbsf | Total reads | Sequence total, Mb | BLAST result | Pfam result | Annotation category | 16S rRNA gene |
| *1H1 | 1 | 107,977 | 10.7 | 13.29 | 7.10 | 5.51 | 0.04 |
| 1H1 | 1 | 125,842 | 12.5 | 11.27 | 5.83 | 4.49 | 0.01 |
| 3H2 | 16 | 135,726 | 13.5 | 5.65 | 3.06 | 2.35 | 0.04 |
| 4H5 | 32 | 168,462 | 16.8 | 5.83 | 3.28 | 2.79 | 0.04 |
| 7H1 | 50 | 84,122 | 8.4 | 14.14 | 8.01 | 6.08 | 0.02 |
*denotes original, unamplified sample.
The best match to the Pfam database was used as the identity for a rudimentary classification of sequence reads into metabolic categories, based on gene ontology (GO) terms. By using these categories, 2.35–6.08% of data were separated into an assigned category [Tables 1 and 2 and Dataset S2], and some data occupied more than one category. Detailed geochemical and lithological data available for this environment (22) suggests that metabolic genes would largely vary with depth. With depth, however, there were few observed overall changes in metabolic functional classes (Table 2). As expected, decreasing amounts of genes coding for locomotion and cell communication were seen with increasing depth. Genes coding for aromatic compound degradation were seen at higher levels <1 mbsf and those coding for phosphorous metabolism became more rare at depth (Table 2). Surprisingly, genes for methanogenesis and photosynthesis did not vary greatly with depth, averaging 1.186% and 0.05% of total genes, respectively.
Table 2.
Percentage of genes in metabolic pathways per sample as determined by comparisons of homologous proteins that have undergone GO annotation
| Annotation category | Sample depth, mbsf |
||||
|---|---|---|---|---|---|
| 1 original | 1 amplified | 16 | 32 | 50 | |
| Amino acid and derivative metabolism | 15.0 | 15.9 | 16.1 | 15.9 | 15.9 |
| Carbohydrate metabolism | 17.0 | 14.6 | 15.7 | 18.1 | 14.6 |
| Nitrogen compound metabolism | 11.6 | 11.6 | 12.3 | 12.2 | 11.4 |
| Protein metabolism | 10.2 | 9.80 | 11.2 | 9.57 | 13.5 |
| Transport | 8.86 | 9.39 | 6.93 | 8.42 | 6.89 |
| Cofactor metabolism | 7.36 | 8.30 | 6.86 | 5.66 | 6.48 |
| RNA metabolism | 5.76 | 8.06 | 7.60 | 7.93 | 9.92 |
| DNA metabolism | 7.98 | 7.86 | 8.52 | 7.74 | 6.80 |
| Vitamin metabolism | 4.06 | 4.20 | 3.97 | 3.87 | 4.77 |
| Cellular lipid metabolism | 4.89 | 3.56 | 3.44 | 3.62 | 3.56 |
| Aromatic compound metabolism* | 2.40 | 2.41 | 3.80 | 2.69 | 2.83 |
| Methanogenesis | 1.34 | 0.96 | 1.01 | 1.42 | 1.20 |
| Response to stress | 0.75 | 0.82 | 1.10 | 0.92 | 0.73 |
| Cell communication* | 0.83 | 0.74 | 0.43 | 0.43 | 0.41 |
| Phosphorus metabolism* | 0.68 | 0.67 | 0.38 | 0.50 | 0.41 |
| Sulfur metabolism | 0.23 | 0.36 | 0.14 | 0.16 | 0.25 |
| Secondary metabolism | 0.61 | 0.35 | 0.20 | 0.45 | 0.24 |
| Cell cycle | 0.18 | 0.26 | 0.14 | 0.19 | 0.12 |
| External encapsulating structure | 0.03 | 0.14 | 0.10 | 0.08 | 0.04 |
| Photosynthesis | 0.09 | 0.05 | 0.01 | 0.06 | 0.04 |
| Locomotion* | 0.04 | 0.03 | 0.00 | 0.00 | 0.00 |
| Cell division | 0.06 | 0.03 | 0.06 | 0.10 | 0.03 |
The differences between 1 original and 1 amplified have been used as a measure of “noise.” The * denotes categories with an above-noise degree of change consistent with a depth trend. More detailed annotation information may be found in Dataset S1.
As in most subsurface marine environments, the major microbial metabolic pathways in this environment are expected to be sulfate reduction and methanogenesis. At ODP Site 1229, methane is produced in the sediment column, with maximum pore-water concentrations observed at 50 mbsf (22). However, to a first order, there is no overwhelming evidence of such a metabolic change reflected in the metagenome (Table 2). Some evidence for methanogenesis at 50 mbsf is seen by the detection of the gene for methylene-5,6,7,8-tetrahydromethanopterin dehydrogenase, an essential enzyme in the methanogenesis pathway. This gene is detected more frequently at greater depths, with one detection each at 1 and 32 mbsf and four at 50 mbsf. However, despite the geochemical evidence that methanogenesis is a major subsurface metabolism at this site, no homologs of methyl-coenzyme reductase (mcrA) were seen, despite specific attempts to recognize the gene.
Based on the pore-water profiles of sulfate at ODP Site 1229, sulfate reduction is expected to be abundant at 1, 16, and 32 mbsf and absent at 50 mbsf (22). In this metagenome, at 1 mbsf, a gene for the β-subunit of sulfite reductase was detected, and at 16 mbsf, a gene similar to the α-subunit of dissimilatory sulfite reductase was found. No sulfite reductases or other marker genes for sulfate reduction were detected in the metagenome at 32 mbsf, despite the generally very high level of conservation of these genes across all branches of life (23). As expected, no such genes indicative of sulfate reduction were found at 50 mbsf.
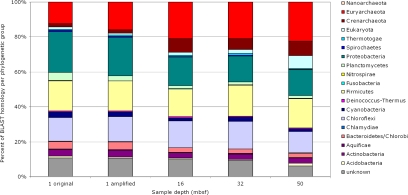
By using the most similar sequence to assign phylogenetic identity, the metagenome makes small but significant changes with depth (Fig. 1). The 1-mbsf samples are distinct from deeper samples, which have a larger number of archaeal genes and a lower number of proteobacterial genes. In the 1-mbsf samples, 22–24% of the metagenome matches proteobacterial genes and this decreases to 16%, 15%, and 15% at 16, 32, and 50 mbsf, respectively. In contrast, the abundance of euryarchaeotal genes starts at 12–16% at 1 mbsf and then increases to 21%, 21%, and 22% at 16, 32, and 50 mbsf, respectively. Three other groups also have interesting trends. The genes related to Chloroflexi and Firmicutes remain mostly constant, with Chloroflexi gene relatives ranging from 12% to 16% and Firmicutes from 16% to 18% of the total genes, independent of depth. Crenarchaeota, initially at 2% at 1 mbsf, climb to 8% at 16 mbsf, 6% at 32 mbsf, and 8% at 50 mbsf. Finally, eukaryotic gene relatives are seen throughout the sediment column. Eukaryotic gene relatives include members of the Apicomplexa, Arthropoda, Ascomycota, Basidiomycota, Chlorophyta, Chordata, Echinodermata, Microsporidia, Nematoda, Platyhelminthes, and Streptophyta. The phylum that is consistently well represented throughout all sediment depths is Ascomycota, some of which have been cultivated from these sediments (24). At 50 mbsf, Chordata help make an unexpected 5% increase in eukaryotic gene relatives, a possible result of amplification bias or contamination because the excess sequences appear similar. The other group responsible for this large increase is again the Ascomycota, the only cultured eukaryotes of this environment.
Fig. 1.
Phylogenetic identities of the BLASTX hits to metagenome against the protein nonredundant database. Shown are the percentages of the total of identifiable hits (1 original, n = 14,341; 1 amplified, n = 14,176; 16 mbsf, n = 7,670; 32 mbsf, n = 11,267; 50 mbsf, n = 11,889).
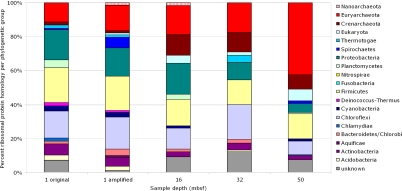
Although the phylogenetic identity of a BLAST hit provides an initial analysis of the subsurface metagenome, general metabolic genes are not exact phylogenetic markers. Additionally, many subsurface microbial groups such as JS1, DSAG, MCG, and SAGMEG have no close relatives whose genomes have been sequenced, which may cause their sequences to be assigned to a more distant relative. To get a better view of the phylogenetic distribution across the subsurface metagenome, a search specific for ribosomal protein genes was undertaken. By using matches to Pfam entries, the metagenome was analyzed specifically for a set of 24 ribosomal protein genes chosen as a subset of 31 ribosomal protein genes that have been used to provide phylogenetic identification of metagenomes (Table S1) (14, 25). An average of 92 ribosomal protein genes was found within each sample. According to the phylogenetic identity of these ribosomal proteins (Fig. 2), the major changes seen with depth are Proteobacteria decreasing with depth (from 18% at 1 (original) to 5% at 50 mbsf) and Euryarchaeota increasing with depth (from 11% at 1 (original) to 42% at 50 mbsf). Additionally, Crenarchaeota make up only 2% (original) and 1% (amplified) of the proteins seen at 1 mbsf, however, they increase to 12%, 11%, and 8% at 16, 32, and 50 mbsf. Other changes seen fall within the “noise” seen by the comparison of 1-mbsf original and amplified datasets. In these two samples, Spirochaetes increase by 5%, and Planctomycetes decrease by 5% in the amplified dataset, whereas other phylogenetic groups shift only 1–2% between samples.
Fig. 2.
Phylogenetic identities, based on matches to Pfam entries for 24 ribosomal proteins (detailed in Table S1). Shown are the percentages of the total of identifiable hits (1 original, n = 107; 1 amplified, n = 79; 16 mbsf, n = 65; 32 mbsf, n = 97; 50 mbsf, n = 130).
The results of this ribosomal protein analysis (Fig. 2), although reflecting the total gene analysis nicely (Fig. 1), were in contrast with previous studies of ribosomal RNA genes from ODP Site 1229. Previous studies detecting 16S rRNA saw high levels of Crenarchaeota and little if any signal for Euryarchaeota (4, 5). Although the discovery of a large number of Euryarchaeota could be precipitated by the primer-independent method of pyrosequencing, there is also the possibility that so few crenarchaeal sequences are known that database searches miss or misidentify these unique subsurface microbial groups (10, 26). Therefore, an in silico search for small-subunit ribosome, or 16S rRNA, gene was made to provide biomarkers for either domain. Because the 16S rRNA genes have been characterized from many environments, including these deep Peru Margin sediments (4–6, 8–11, 26, 27), they provide a more complete database from which to ascertain the phylogenetic identity of marker genes.
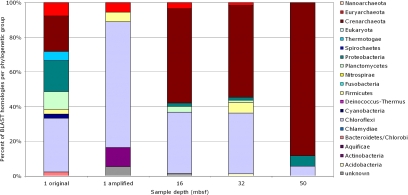
Based on BLASTN comparison, matches to small-subunit ribosomal genes often had high homology (expectancy values <1 × 10−9). Because it is difficult to define a species from a single 100-bp sequence, but not difficult to assign a microbial group, results were classified into the same groups as used in the prior analysis (Figs. 1 and 2). The 1-mbsf 16S rRNA phylogenetic profiles (Fig. 3) vary between original and amplified samples, unlike the more consistent whole-genome phylogenetic profiles (Figs. 1 and 2). Single-gene identifications are not as reliable as multigene identifications because the dataset may not be sufficiently sampled [suggested by the lack of assembly of these sequences (data not shown) and also because of the potential for single-gene amplification bias created by WGA. As such, the change between the original and amplified datasets at 1 mbsf served as a control for the potential variation or “noise” that could occur in other samples with depth.
Fig. 3.
Phylogenetic groups of subsurface metagenome, based on 16S rRNA homologous gene fragments. Shown are the percentages of the total of identifiable hits (1 original, n = 39; 1 amplified, n = 18; 16 mbsf, n = 56; 32 mbsf, n = 65; 50 mbsf, n = 17).
Despite this variation, two stark trends become apparent. First, as with the total metagenome, the overall percentage of archaeal 16S rRNA genes increases with depth, making up to 88% of the total ribosomal genes at 50 mbsf (Fig. 3). Surprisingly, these 16S rRNA genes are all from Crenarchaeota. Second, the number of Chloroflexi 16S rRNA genes decrease at 50 mbsf, going from 30% to 72% of the population at 1, 16, and 32 mbsf down to 6% at 50 mbsf. Small subunit ribosomal genes for Proteobacteria, Firmicutes, Planctomycetes, Actintobacteria, Acidobacteria, Bacteriodetes, and Thermotogae are also detected in smaller numbers and with no consistent trend with depth. Many relatives of the detected sequences had been reported during studies on Peru Margin sediments (Tables S2–S6).
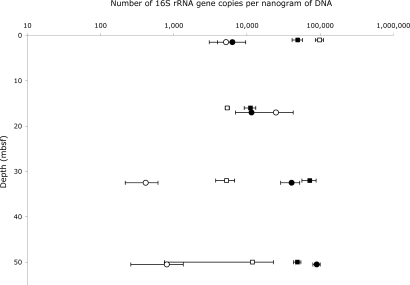
Because of the discrepancy between whole gene/ribosomal protein phylogenetic profiles and small-subunit ribosomal RNA gene detection in terms of archaeal relatedness, DNA extracted from Peru margin sediments was also analyzed by qPCR. The SYBR green method was used to detect small-subunit ribosomal genes with appropriate primer sets to specifically amplify general Bacteria and low-temperature Crenarchaeota (see SI Text). Primer sets were also tested for other microbial groups but failed to be group-specific under the test conditions. By using this approach, the number of ribosomal RNA genes in each nanogram of total DNA for both original and WGA samples was calculated at each depth studied (Fig. 4), and the relationship of Crenarchaeota with depth and the ratio of Crenarchaeota to Bacteria was determined.
Fig. 4.
Quantitative PCR data from original and amplified sediment DNAs. Crenarchaeota (771F/957R) are shown as circles, Bacteria (519F/907R) are shown as squares. Original DNAs are open symbols, RepliG amplified DNAs are filled symbols. Each data point represents the average of multiple measurements, with the standard deviation of measurements shown as error. The archaeal results have been graphed with a 0.5-m offset to distinguish data points.
Based on qPCR, the 1-mbsf samples are dominated by bacterial rRNA genes, as seen for 16S rRNA genes in the metagenome. At 16 mbsf, the microbial community seen by qPCR reflects the metagenome in that it is nearly evenly split between Crenarchaeota and Bacteria (16S rDNA in the metagenome is 53% Crenarchaeota). Additionally, there is good agreement between the original and amplified DNA samples at 1 and 16 mbsf, suggesting little if any stochastic WGA bias (in terms of small-subunit ribosomal genes) occurred at these depths. No PCR inhibition was exhibited by either of these samples.
At 32 and 50 mbsf, qPCR analysis of the small amounts of original DNA was hampered by PCR inhibitors present in the sample. To overcome this, the DNA samples were diluted and measured at different dilutions, and averages were taken for both DNA concentration measurements and qPCR results from different DNA dilutions. No inhibition of DNA measurement or PCR was observed in the amplified samples. At 32 mbsf, qPCR analysis shows Bacteria appear to be more abundant than the Crenarchaeota, although in the amplified samples, the difference between the groups is small (16S rRNA genes in the metagenome are 54% Crenarchaeota). At 50 mbsf, there is large error in the original DNA qPCR measurements, because of the aforementioned inhibition that resulted in little reproducibility in results. The WGA sample at this depth shows a larger number of crenarchaeal rRNA genes with small error, presumably from relief from inhibition. At 50 mbsf, Crenarchaeota account for 88% of 16S rRNA gene sequences in the metagenome. By qPCR, they are still dominant in the amplified DNA but by a lesser margin. Based on clone libraries made after the qPCR, most Bacteria amplified are related to the Chloroflexi genera and the Archaea are related to low-temperature Crenarchaeota, similar to the phylogenies seen in the metagenome (Fig. 3). Also consistent with the 16S rRNA genes detected in the metagenome is the increase in Crenarchaeota seen between 1 mbsf and deeper sediment horizons.
Discussion
This metagenome from deeply buried sediment allows the examination of many uncutivated groups of microorganisms. Many genes (up to 85%) in this environment are unidentifiable, with no close relatives in the nonredundant database. In contrast, tests using the genome from the Crenarchaeon Pyrobaculum showed that when good homologous genes are available for identification, at least 55% of a novel genome can be identified despite the short gene fragments generated by GS20 pyrosequencing (details in SI Text). However, it is difficult to ascertain what portion of the unidentifiable sequences are novel, chimeras, other amplification artifacts, or may be identified once closer relatives are in the genomic databases.
The analysis of identified metabolic functional genes, as sorted by GO category, showed little relevance to the geochemistry measured at ODP Site 1229. Although total euryarchaeotal genes increased in areas presumed to be responsible for methane generation, few functional methanogenesis genes were found. Additionally, sulfate reduction is rampant in shallow sediments, yet few genes were found for this process. As with previous studies of the subsurface, the potential does remain that this environment may house organisms with metabolisms that are too distinct to detect (4, 5). However, this study did not rely on primer amplified fragments, meaning that the divergence would have to be so great as to not be detected even with homologous gene detection.
The bacterial group Chloroflexi has again been shown to be a major part of the subsurface microbial community in both the metagenome and qPCR analysis. Many groups of uncultivated Chloroflexi have been found in previous subsurface studies and a wide phylogenetic group is assembling (4). The potential metabolic diversity of this group is unknown, and although many of the genes from the subsurface were very similar to those known from Dehalococcoides ethenogenes, the ribosomal sequences are still quite distinct, making metabolic assignment based on this cultivated representative unreliable.
In this metagenome and in the qPCR results, Archaea are a significant portion. However, many subsurface studies have concluded that Archaea are an insignificant portion of the subsurface community (4, 11). A new analysis of the methods used in these studies may explain some of these results, because the commonly used TaqMan qPCR primers may detect only 10% of the MCG group of Archaea commonly found in the subsurface (10). The current study also may explain the decrease of reliable detection by qPCR with depth (Fig. 4). Generally, the results suggest that as total DNA becomes scarce and the quality of that DNA decreases with depth, the qPCR estimations of the microbial populations decrease, with those for the Crenarchaeota decreasing more than those for the Bacteria. DNA quality and quantity can be improved by WGA (Fig. 4), which may yield better detection levels, but this is not a recommended solution because of potential stochastic bias in WGA. The growing evidence of archaeal importance in the subsurface (5, 9, 28, 29) makes subsurface archaeal groups important to examine in future studies.
Of these archaeal groups, this study predicts that the Crenarchaeota are a dominant phylotype of the sediment at depth, based on the focused search for small-subunit ribosomal genes in both the metagenome and by qPCR. Although this does not agree with the phylogenetic profile seen for total genes or ribosomal proteins, this does agree with previous results measuring phospholipids and rRNA (5), which showed that Crenarchaeota were the dominant active fraction of the microbial population at depth on the Peru Margin. Many of the known crenarchaeal groups of the subsurface, particularly the MBGB (DSAG) and AAG Archaea, branch quite low, based on 16S rRNA, in the crenarchaeal clade (7, 9, 10). The apparent underrepresentation of Crenarchaeota in the metagenome could be due to the large distance between these subsurface crenarchaeal groups and the nearest relatives with sequenced genomes, leading archaeal genes to be misidentified as euryarcheal or unidentified altogether. Additionally, the suggestion also remains that ribosomal sequences from Euryarchaeota, particularly methanogens, may be unusual (4). However, by using the primer-independent approach of pyrosequencing, it should be possible to detect a small subunit ribosomal gene, even in cases where conserved primer pairs fail.
An analysis of multiple metagenomes found Proteobacteria to be the consistently dominant microbial group found in the environment (13). At 1 mbsf, the homologous genes of the subsurface metagenome are predominantly proteobacterial, as in these other environments. The dominance of Bacteria at 1 mbsf has also been seen through fluorescent in situ hybridization (FISH) of fixed sediments from ODP Site 1229 (5). However, at depth, the subeafloor metagenome becomes a unique dataset, with Archaea becoming a dominant group based on ribosomal sequence analysis (Figs. 2 and 3). It was also shown by FISH, rRNA, and lipid analysis that Archaea, in particular Crenarchaeota, are abundant at depth at this site (5). Archaea have also been suggested by both 16S studies and lipid analyses to be the most abundant microbial domain in other subsurface marine sediments (28, 29). Crenarchaeota have often been found as dominant microorganisms in deep ocean waters through PCR-based and microscopic studies (30, 31). Archaea have also been shown by 16S rRNA clone libraries to be dominant in limited subsurface ecosystems (32, 33) and acid mine drainage (34).
In contrast to other metagenomic studies of terrestrial and pelagic environments, which show Archaea to be in minimal numbers, these metagenomic results, along with SYBR-green-based qPCR, show high numbers of Archaea at depth. The influence of Archaea on geochemical cycles, other than methane generation, has largely been ignored in many estimates of global activity. As Leininger et al. (35) and Wuchter et al. (36) determined, Crenarchaeota may be the dominant contributors to nitrification in both terrestrial and pelagic environments, and although archaeal genes for ammonia oxidation have been found in other environments, none have been detected in the subsurface metagenome. However, because the majority of the metagenome did not match database sequences, little information about the dominant metabolic functions can be deduced for these sites. The metagenome results presented here clearly illustrate the need for a higher number of crenarchaeal or subseafloor genome sequences to be in the databases. The microbial populations in the marine subseafloor are distinctly different from many surface and pelagic environments, and until further information becomes available for comparison, the true diversity and function of these interesting microbes will likely remain hidden.
Materials and Methods
DNA Extraction.
Sediment samples were collected on-ship and frozen at −80°C (22). Samples were shipped on dry ice to Pennsylvania State University and were kept at −80°C until analysis. Sediment cores from ODP Leg 201 Site 1229 were used in this study. Sediments from ODP Site 1229 horizons: 1H1 (1 mbsf), 3H2 (16 mbsf), 4H5 (32 mbsf), and 7H1 (50 mbsf) were placed at −20°C overnight and then aseptically scraped. The first centimeter of sediment material was discarded. Subsequent scrapings were homogenized and weighed. The MoBio UltraClean Microbial DNA kit (MoBio Laboratories) was used to perform DNA extractions, with modifications (described in SI Text).
Evaluation and Amplification of Extracted DNA.
The concentration of extracted DNA was measured by using the Quant-It PicoGreen dsDNA reagent (Invitrogen). Only the 1-mbsf sample contained enough DNA to perform direct sequencing (referred to as original). Therefore, the remaining samples were subjected to a whole-genome amplification using the REPLI-g Mini kit (Qiagen) according to the manufacturer's instructions. In addition, a dilution of 1-mbsf sample was also subjected to WGA (1 amplified). The reaction was left at 30°C for 8 h, after which a 3-min inactivation at 65°C was performed. Approximately 6 μg of product was obtained for the 1- and 16-mbsf samples, and 4 μg was obtained for the 32- and 50-mbsf samples. All amplified DNAs were diluted and checked for community composition by intergenic transcribed spacer fingerprinting by using both bacterial and archaeal primer sets as described (3). The overall community composition between samples was similar.
Preparation of DNAs for Pyrosequencing.
The original, unamplified DNA from the 1-mbsf sample was denoted “1 original.” Amplified samples were denoted “1 amplified,” “16,” “32,” and “50.” Samples were sequenced by the Pennsylvania State University Center for Genomic Analysis on a GS20 Sequencing System (454; Life Sciences) as described (17).
Sequence Analysis.
All samples were subjected to sequencing as half-plate reactions, where the Pico Titer plates were divided into two samples, resulting in a total of 61.9 Mb of sequence. Data has been deposited in the National Center for Biotechnology Information GenBank Archive (SRA001015). BLAST analyses were performed for total genes, ribosomal protein genes, and small-subunit ribosomal genes (details in SI Text).
Quantitative PCR.
Quantitative PCR was performed for both Archaea and Bacteria by using primer sets 519F/907R for Bacteria (37, 38) and 771F/957R for low-temperature Crenarchaeota (39). Control DNAs were used to make standard curves to determine the number of ribosomes detected per threshold cycle. Creation of control DNAs, preparation of DNA templates, and reaction conditions are described in SI Text.
Threshold cycles were determined for each sample, and the standard curves from both archaeal and bacterial control plasmids were used to calculate ribosomal copy number for each sample. Based on the concentration of DNA added to the well, each sample was standardized to RNA copy number per nanogram of DNA, and the average and standard deviations for all samples from each horizon was taken.
Supplementary Material
Acknowledgments.
We thank the ODP shipboard scientific party, Lynn Tomsho, Ji Qi, and Tom Canich for assistance. Samples for this research were provided by ODP [sponsored by the National Science Foundation (NSF) and participating countries under the Joint Oceanographic Institutions]. This work was supported by NSF Grants OCE 05-50601 (to J.F.B., C.H.H., S.T.F., and S.C.S.) and MO-0347475 (to J.E.B and J.F.B.); Department of Energy Grant DE-FG02-93ER20117 (to J.E.B. and J.F.B.); and by the National Aeronautics and Space Administration (NASA) Astrobiology Institute under NASA–Ames Cooperative Agreement NNA04CC06A (J.F.B., C.H.H., and J.E.B.). The GS20 facility at the Pennsylvania State University Center for Genome Analysis is funded, in part, by a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds appropriated by the legislature. J.F.B. was also supported by a NASA Postdoctoral Program Fellowship administered by Oak Ridge Associated Universities (ORAU).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All sequences are deposited into the National Center for Biotechnology Information Short Read Archive (accession no. SRA001015).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709942105/DCSupplemental.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Hondt S, Rutherford S, Spivack AJ. Metabolic activity of subsurface life in deep-sea sediments. Science. 2002;295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 3.Biddle JF, House CH, Brenchley JE. Enrichment and cultivation of microorganisms from sediment from the slope of the Peru Trench (ODP Site 1230) Proc ODP Sci Results. 2005;201:1–19. [Google Scholar]
- 4.Inagaki F, et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA. 2006;103:2815–2820. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biddle JF, et al. Novel heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster G, Parkes RJ, Fry JC, Weightman AJ. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol. 2004;70:5708–5713. doi: 10.1128/AEM.70.9.5708-5713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teske AP. Microbial community composition in deep marine subsurface sediments of ODP Leg 201: Sequencing surveys and cultivations. Proc ODP Sci Results. 2006;201:1–19. [Google Scholar]
- 8.Parkes RJ, et al. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature. 2005;436:390–394. doi: 10.1038/nature03796. [DOI] [PubMed] [Google Scholar]
- 9.Sørensen KB, Teske A. Stratified communities of active archaea in deep marine subsurface sediments. Appl Environ Microbiol. 2006;72:4596–4603. 2006. doi: 10.1128/AEM.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teske A, Sørensen KB. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 11.Schippers A, et al. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433:861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- 12.Mauclaire L, Zepp K, Meister P, McKenzie J. Direct in situ detection of cells in deep-sea sediment cores from the Peru Margin (ODP Leg 201, Site 1229) Geobiology. 2005;2:217–223. [Google Scholar]
- 13.von Mering, et al. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 14.Huson DG, Auch A, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster G, Newberry CJ, Fry JC, Weightman AJ. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: A cautionary tale. J Microbiol Methods. 2003;55:155–164. doi: 10.1016/s0167-7012(03)00140-4. [DOI] [PubMed] [Google Scholar]
- 16.Edwards RA, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poinar HN, et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;331:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 18.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 19.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinard R, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Hondt SL, Jørgensen BB, Miller DJ, et al. Distributions of microbial activities in deep subseafloor sediments. Science. 2004;306:2216–2221. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- 23.Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biddle JF, House CH, Brenchley JE. Microbial stratification of deeply buried marine sediment reflects changes in sulfate/methane profiles. Geobiology. 2005;3:287–295. [Google Scholar]
- 25.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 26.Sørensen KB, Lauer A, Teske A. Archaeal phylotypes in a metal-rich and low-activity deep subsurface sediment of the Peru Basin, ODP Leg 201, Site 1231. Geobiology. 2004;2:151–161. [Google Scholar]
- 27.Webster G, et al. Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol. 2006;58:65–85. doi: 10.1111/j.1574-6941.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- 28.Reed DW, et al. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl Environ Microbiol. 2002;68:3759–3770. doi: 10.1128/AEM.68.8.3759-3770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipp J, Hinrichs K. The deep biosphere: Quantitative and taxonomic constraints through microbial lipids. Geochim Cosmochim Acta. 2007;71:A584. [Google Scholar]
- 30.Herndl GJ, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 32.Chapelle FH, et al. A hydrogen-based subsurface microbial community dominated by methanogens. Nature. 2002;415:312–315. doi: 10.1038/415312a. [DOI] [PubMed] [Google Scholar]
- 33.Takai K, et al. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles. 2004;8:269–282. doi: 10.1007/s00792-004-0386-3. [DOI] [PubMed] [Google Scholar]
- 34.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 35.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 36.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane DJ. Nucleic Acid Techniques in Bacterial Systematics. New York: Wiley; 1991. 16S/23S rRNA sequencing; pp. 205–248. [Google Scholar]
- 38.Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 39.Ochsenreiter T, Selesi D, Bonch-Ozmolovskaya L, Quaiser A, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.