Abstract
Restriction endonucleases (REases) protect bacteria from invading foreign DNAs and are endowed with exquisite sequence specificity. REases have originated from the ancestral proteins and evolved new sequence specificities by genetic recombination, gene duplication, replication slippage, and transpositional events. They are also speculated to have evolved from nonspecific endonucleases, attaining a high degree of sequence specificity through point mutations. We describe here an example of generation of exquisitely site-specific REase from a highly-promiscuous one by a single point mutation.
Keywords: genetic recombination, protein engineering, R.Kpnl, metal ion coordination, promiscuous activity
Type II REases are part of restriction–modification (R-M) systems that protect bacterial cells against invading foreign genomes (1). The enzymes are highly sequence specific and cleave DNA at the target sites several orders of magnitude more readily than at noncanonical sequences. Because of their high sequence specificity they play crucial role in DNA manipulation and characterization. Of the 3,700 Type II REases identified from different bacterial species so far, only 262 specificities have been characterized (2). The growth in the applications of REases in DNA manipulations warranted engineering of these enzymes possessing novel specificities, and numerous efforts have been undertaken toward this goal. These attempts to engineer REases with altered specificities were largely unsuccessful because of the high degree of sequence specificity already attained through specific contacts to the bases by different amino acid residues from various structural segments of the enzyme. This would imply that changing the specificity may require modification of several of the contacts with the bases and the phosphate backbone.
In nature, evolution of sequence specificity in R-M systems is achieved through genetic recombination, gene duplication, replication slippage, and transposition (3–8). Genetic recombination occurring in nature to reassort target recognition domains (TRDs) leading to evolution of sequence specificity was demonstrated in Salmonella Type I R-M systems (3, 4). Later the reassortment of TRDs has been used to generate Type I REase with different specificity in the laboratory (5). Generation of new sequence specificity by TRD swapping was demonstrated in a Type II REase recently (9). Point mutations were also considered to contribute in evolving sequence specificity, but no examples have been documented so far (1, 10).
In this study, we provide evidence for point mutations having a role in evolving sequence specificity in a REase. Although R.KpnI is a typical Type IIP REase, it exhibits prolific promiscuous cleavage. In the presence of Mg2+, the most common metal ion required for REases, R.KpnI cleaves plasmid DNA at variety of noncanonical sites in a robust fashion, generating extensive DNA fragmentation. We describe here a single point mutant, which carries out DNA cleavage in an extremely site-specific manner, confirming the importance of point mutations in evolving sequence specificity.
Results and Discussion
Two Modes of DNA Cleavage by R.KpnI in the Presence of Mg2+.
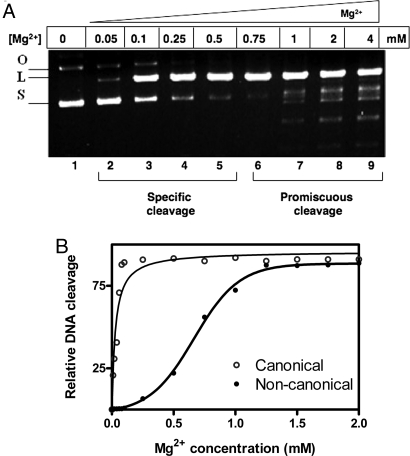
R.KpnI is a highly-promiscuous REase in Mg2+ catalyzed reactions unlike most other REases (11, 12). In Fig. 1A, the characteristic cleavage pattern by R.KpnI is shown in a range of Mg2+ concentrations. At lower concentrations of the metal ion (50–500 μM), only cleavage of the cognate sequence was observed (lanes 2–5). However, at concentrations of Mg2+ >500 μM, promiscuous cleavage was evident (lanes 6–9). The specific and promiscuous modes of cleavage by R.KpnI depending on the Mg2+ concentration were substantiated by analysis of the cleavage of oligonucleotides. The oligonucleotide substrate containing the canonical sequence was cleaved efficiently by R.KpnI even at 20 μM Mg2+. In contrast, noncanonical sequences were efficiently cleaved only at >500 μM Mg2+ [Fig. 1B and supporting information (SI) Fig. S1]. These results implied that R.KpnI exhibits two different metal activation profiles—one for canonical and the other for noncanonical DNA substrates. The enzyme follows a hyperbolic metal activation profile for the canonical substrate and a sigmoidal pattern for the noncanonical substrate (Fig. 1B). These data were subjected to Hill plot analysis (Fig. S2; and see SI Text), and the Hill coefficient for the metal ion binding is presented in Table 1. Hill plot analysis revealed the binding of additional metal ion to the enzyme and its role in the cleavage of noncanonical sequences. Thus, the recruitment of an additional metal ion near to or distant from the active site of the enzyme appeared to be necessary to induce promiscuous cleavage by the enzyme. If that is the case, mutation of the residues involved in the second site metal ion coordination should abolish promiscuous activity, resulting in a highly sequence-specific enzyme. We tested this hypothesis.
Fig. 1.
Two distinct modes of DNA cleavage by R.KpnI. (A) R.KpnI (15 nM) was incubated with pUC18 DNA (1 μg), and the reaction was initiated with different concentrations of Mg2+ as indicated. Lane 1, no metal ion; lanes 2–5, 50–500 μM Mg2+; lanes 6–9, 0.75–4 mM Mg2+. The products were subjected to electrophoresis on 1% agarose gel. S, L, and O indicate the respective position of the supercoiled, linear, and open circular forms of the plasmid. (B) Graphical representation of DNA cleavage profile by R.KpnI (15 nM) with increasing concentrations of Mg2+ (0.01–2 mM) using canonical and noncanonical oligonucleotide substrates.
Table 1.
Hill coefficients for Mg2+ binding
| Substrate | [Mg]0.5, μM | n |
|---|---|---|
| -GGTACC- | 20 | 1.7 |
| -GaTACC- | 600 | 3.5 |
[Mg]0.5, concentration of Mg2+ that yields 50% cleavage; n, Hill coefficient.
Identification of the Secondary Metal-Binding Site in R.KpnI.
We had demonstrated in our previous studies that R.KpnI belongs to the HNH superfamily and follows the single metal ion mechanism for catalysis (13). H149 of R.KpnI acts as a general base and is essential for the catalysis. The residues D148 and Q175 are involved in metal ion coordination. Mutations at these metal-binding residues affected Mg2+-mediated DNA cleavage and thus are important for the primary catalysis. To identify the secondary metal ion-binding site, the sequence of R.KpnI was analyzed. The analysis revealed the presence of a putative PD148x14D163xK165 motif in R.KpnI, a sequence motif involved in metal ion coordination and catalysis in most REases. In the linear sequence arrangement of R.KpnI this motif appears to be a typical PD… D/ExK motif similar to other REases and could be a candidate for second metal ion binding. To analyze whether the D163 of the putative motif has any role in secondary metal binding, the residue was targeted for mutational analysis. The mutant enzymes (D163A, D163E, D163N and D163I) were overexpressed and purified. The mutant proteins were soluble and showed comparable characteristics to those of R.KpnI and CD studies showed no structural perturbations (Fig. S3).
High-Fidelity DNA Cleavage by Mutant R.KpnI.
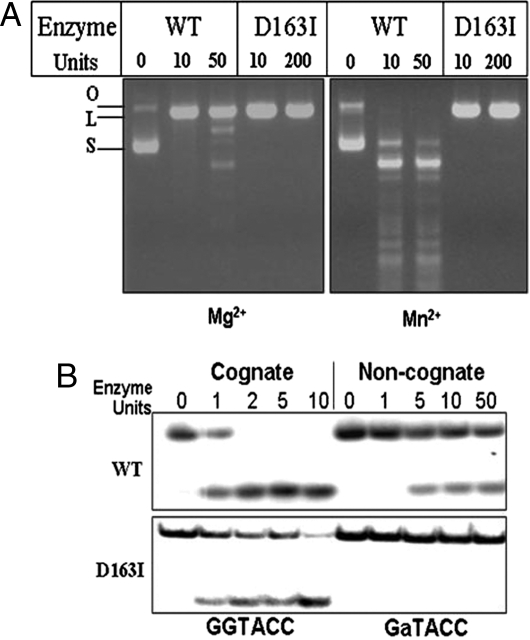
DNA cleavage by the D163 variants of R.KpnI was analyzed in the presence of 2 mM Mg2+ by using pUC18 DNA as a substrate. All of the mutant enzymes showed DNA cleavage activity in the presence of Mg2+ (Table 2), indicating that D163 was not required for the primary catalysis. Among the mutants, D163I showed specific activity comparable to that of WT R.KpnI (Table 2). In reactions carried out with saturating Mg2+ or Mn2+ (2 mM) concentrations, D163I did not show any promiscuous behavior even at very high enzyme concentrations (Fig. 2A). Oligonucleotide substrate containing the most preferred noncanonical site (-GaTACC-) was refractile to cleavage by the mutant enzyme, without compromising the efficiency of cleavage of canonical site (Fig. 2B). We have examined other residues in the predicted motif (PD148x14D163xK165). Mutation in the K165 residue also abolished promiscuous activity of R.KpnI (Fig. S4). However, the specific activity of K165A is lower than that of WT R.KpnI unlike D163I and hence was not subjected to further analysis (Fig. S4).
Table 2.
Specific activities of WT and D163 variants
| Enzyme | Specific activity (units/mg of protein) |
|---|---|
| WT | 1.2 × 106 |
| D163A | 0.4 × 105 |
| D163E | 1.1 × 104 |
| D163N | 1.3 × 104 |
| D163I | 0.8 × 106 |
Fig. 2.
Effect of D163I mutation of R.KpnI on DNA cleavage specificity. (A) pUC18 DNA (1 μg) was incubated with various concentrations of WT or mutant D163I enzyme for 1 h at 37°C in the presence of 5 mM Mg2+or Mn2+ and analyzed on 1% agarose gel. S, L, and O indicate the respective positions of the supercoiled, linear, and open circular forms of the plasmid. (B) Labeled oligonucleotides (0.2 pmol) containing canonical (-GGTACC-) or noncanonical (-GaTACC-) sequences were incubated with various concentrations of mutant D163I for 30 min at 37°C in the presence of 5 mM Mg2+. The cleavage products were analyzed on 12% urea–acrylamide gel.
D163 Is Important for Additional Metal Ion Coordination and Promiscuous Cleavage.
Substitution of I for D at position 163 led to the loss of promiscuous activity and resulted in high-fidelity R.KpnI (Fig. 2 A and B). The metal-dependency experiments showed that both WT and D163I have comparable metal activation profiles with the canonical substrate, indicating that mutation did not affect the primary metal binding and canonical DNA cleavage (Fig. 3A). When DNA cleavage patterns with the WT and D163I mutant were compared, the mutant enzyme showed only site-specific DNA cleavage irrespective of the metal ion concentrations (Fig. 3B). The residue D163 thus appears to be involved in second metal ion coordination, which is required for imparting promiscuous DNA cleavage. Fluorescence and near-UV CD spectral analysis revealed the absence of any major tertiary structural changes in D163I protein (Fig. S5).
Fig. 3.
Metal-dependent DNA cleavage by D163I. R.KpnI or D163I (15 nM) was incubated with 0.2 pmol of labeled oligonucleotides containing canonical sequence (-GGTACC-) or pUC18 DNA (1 μg). The reaction was initiated with various concentrations of Mg2+ as indicated, and the products were separated by electrophoresis on 12% urea–acrylamide gel or 1% agarose gel. S, L, and O indicate the respective positions of the supercoiled, linear, and open circular forms of the plasmid. (A) Graph depicting the activation profile of WT and D163I at Mg2+ concentrations ranging from 0.01 to 1 mM. (B) pUC18 DNA cleavage profiles with increasing concentrations of Mg2+ (0.05–5 mM).
The Mutant Exhibits Similar Kinetic Behavior to That of WT R.KpnI.
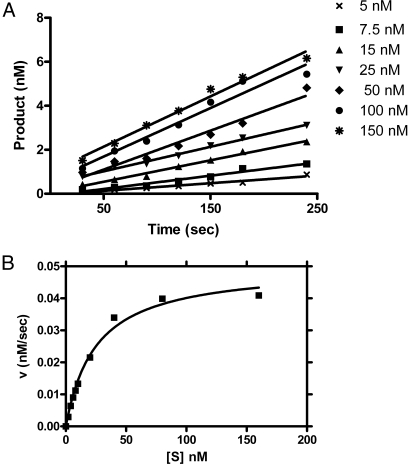
The steady-state kinetics studies (Fig. 4A) revealed that the product release rates of D163I was similar to R.KpnI and followed the Michaelis–Menten kinetics (Fig. 4B). The kinetic constants (KM and Vmax) derived from Lineweaver–Burk plots indicated that the mutant enzyme has comparable specificity constant (kcat/KM) with Mg2+ or Mn2+ (Table 3). The turnover number for the mutant was not vastly different from that of R.KpnI, implying that the mutant is equally efficient in catalysis. Kinetic parameters could not be determined with noncanonical substrates even at very high concentrations of the mutant enzyme and metal ions, because the mutation had annulled the promiscuous activity of R.KpnI without significantly altering the primary catalysis.
Fig. 4.
Steady-state kinetic analysis of D163I with canonical substrate (GGTACC) in the presence of Mg2+. D163I (1 nM) was incubated with canonical substrate in the assay system. Reactions were initiated by the addition of 5 mM Mg2+ and stopped by adding loading dye containing 10 mM EDTA. (A) Rate of DNA cleavage with different concentrations (5–150 nM) of canonical substrate in the reaction buffer. (B) A representative Michaelis–Menten plot. The graphs were obtained by plotting the rates of DNA cleavage at various concentrations of oligonucleotide substrate y using GraphPad Prism Version 4.
Table 3.
Kinetic constants of WT and D163I mutant for canonical and noncanonical substrates
| DNA substrates and cofactors |
KM (M) |
kcat (s−1) |
kcat/KM (M−1s−1) |
|||
|---|---|---|---|---|---|---|
| WT | D163! | WT | D163I | WT | D163I | |
| Duplex I (-GGTACC-) | ||||||
| Mg2+ | 23 × 10−9 | 25 × 10−9 | 0.071 | 0.0334 | 3.1 × 106 | 1.33 × 106 |
| Mn2+ | 22 × 10−9 | 22 × 10−9 | 0.078 | 0.0328 | 3.5 × 106 | 1.33 × 106 |
| Duplex II (-GaTACC-) | ||||||
| Mg2+ | 71 × 10−9 | ndc | 0.0026 | ndc | 3.7 × 104 | ndc |
| Mn2+ | 115 × 10−9 | ndc | 0.015 | ndc | 1.3 × 105 | ndc |
| Duplex III (-GGTtCC-) | ||||||
| Mg2+ | 169 × 10−9 | ndc | 0.0024 | ndc | 1.4 × 104 | ndc |
| Mn2+ | 112 × 10−9 | ndc | 0.013 | ndc | 1.2 × 105 | ndc |
WT, wild type; D163I, mutant; ndc, no detectable cleavage.
Type II REases are highly sequence-specific compared with other classes of nucleases. In the early days of discovery of R-M systems, it has been suggested that sequence specificity in this group of enzymes could have evolved through genetic recombination or mutations (1, 10). Although a role for genetic recombination was established by analyzing the natural recombinants and engineering in the laboratory (3–5), no example is known for point mutation in achieving sequence specificity. Subsequently, replication slippage, transpositional events, gene duplication, etc., were also shown to have a role in evolution of sequence specificity (6, 8, 14). Engineering REases in the laboratory with new sequence recognition has proven to be a difficult task. Initially, the TRD swapping has been successfully applied to alter the DNA sequence specificity of Type I REases (5, 7). Combinatorial reassortment of TRDs between Type IIB REases has been used more recently to generate a REase with novel DNA specificity (9). Yet another approach to obtain new specificity was to construct hybrid proteins that contain the DNA-binding and/or catalytic modules from different sources. For example, isolated catalytic domains of Type IIS REases (R.FokI and R.BmrI) were fused with specific DNA-binding domains of transcriptional regulators, to generate chimeric endonucleases with altered specificities (15–17). A selection system (methylation activity-based selection) has also been developed to produce REases with new specificity (18). This method appears to be of limited application because it can only be used on bifunctional type IIG REases (19). The bifunctional REase (R.Eco57I) has been engineered into a monofunctional MTase by cleavage-center disruption. The mutants showing altered DNA modification specificity were selected based on their ability to protect predetermined DNA targets. Later, the cleavage center was reconstituted to that of WT enzyme (18). These strategies are, however, difficult to be adopted to single domain-containing Type IIP REases.
Rational protein design and directed evolution approaches have been used to generate REases with altered substrate specificity based on the structural information. Although several mutant enzymes have been isolated, they have only limited substrate preference (20–24). An example is the directed evolution of R.BstYI endonuclease (5′-RGATCY-3′) that resulted in genetic selection of two R.BstYI variants with increased substrate specificity toward 5′-AGATCT-3′ and discrimination against 5′-GGATCC-3′ (25). In these cases, no major changes were observed in the cleavage characteristics of the enzyme. The conversion of R.KpnI from a highly-promiscuous to a very specific REase by a single amino acid change reported here highlights the role of mutations in evolving sequence specificity. This study thus provides an example for engineering specificity with a single point mutation.
Finally, R.KpnI is peculiar in retaining robust promiscuous cleavage despite being a typical Type IIP REase in all other characteristics. This property of the enzyme has led us to suggest that it is evolutionarily stuck in the process of attaining a high degree of sequence specificity (12). In nature, retaining the promiscuity in the cleavage characteristics of the enzyme may provide certain advantage for the organism. However, it is conceivable that some strains of the Klebsiella pneumoniae may have evolved the enzyme with high specificity through point mutations.
Materials and Methods
Site-Directed Mutagenesis.
R.KpnI mutants (D163A, D163E, D163N, D163I and K165A) were generated by site-directed mutagenesis using the megaprimer inverse PCR method (26). The oligonucleotides used in this study are listed in Table S1. Expression plasmid pETRK encoding the WT kpnR gene was used as a template (27). Oligonucleotide primers carrying the respective amino acid codon substitutions were used for mutagenesis. After confirming the mutation by sequencing, the mutant REases were expressed in Escherichia coli BL26(F–ompT hsdSB(rB-mB-)gal dcm Δlac(DE3)nin5 lacUV5-T7gene1) containing M.KpnI and purified as described (28). Purified enzymes were dialyzed against buffer containing EDTA, followed by EDTA-free buffer to remove traces of bound metal ions (12). For all of the assays, enzymes (WT and mutants) were diluted 1,000-fold in EDTA-free reaction buffer.
DNA Cleavage and Steady-State Kinetic Assays.
R.KpnI and its mutants were incubated in 10 mM Tris·HCl (pH 7.4), 5 mM 2-mercaptoethanol, with increasing concentrations of MgCl2 for 1 h at 37°C and 1 μg of pUC18 DNA containing a single site for R.KpnI (29). The cleavage products were analyzed on 1% agarose gels. Steady-state kinetic experiments were carried out as described (12). The kinetic parameters were determined by fitting the change in the velocity with substrate concentration to the double reciprocal (1/v versus 1/[S]) Lineweaver–Burk plot by using GraphPad Prism Version 4.
Supplementary Material
Acknowledgments.
We thank K. P. Gopinathan for critical reading of the manuscript and the Department of Biochemistry, Indian Institute of Science, for fluorescence spectrometry and CD analysis. M.S. and K.V. are Senior and Junior Research Fellows, respectively, of the Council of Scientific and Industrial Research, Government of India.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804974105/DCSupplemental.
References
- 1.Arber W, Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—Enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuller-Pace FV, Bullas LR, Delius H, Murray NE. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci USA. 1984;81:6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagaraja V, Shepherd JC, Bickle TA. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985;316:371–372. doi: 10.1038/316371a0. [DOI] [PubMed] [Google Scholar]
- 5.Fuller-Pace FV, Murray NE. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci USA. 1986;83:9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauster R. Evolution of type II DNA methyltransferases: A gene duplication model. J Mol Biol. 1989;206:313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- 7.Gubler M, Braguglia D, Meyer J, Piekarowicz A, Bickle TA. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992;11:233–240. doi: 10.1002/j.1460-2075.1992.tb05046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meister J, et al. Macroevolution by transposition: Drastic modification of DNA recognition by a type I restriction enzyme following Tn5 transposition. EMBO J. 1993;12:4585–4591. doi: 10.1002/j.1460-2075.1993.tb06147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urbanaviciene JS, et al. Generation of DNA cleavage specificities of type II restriction endonucleases by reassortment of target recognition domains. Proc Natl Acad Sci USA. 2007;104:10358–10363. doi: 10.1073/pnas.0610365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 11.Chandrashekaran S, Saravanan M, Radha DR, Nagaraja V. Ca2+-mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease. J Biol Chem. 2004;279:49736–49740. doi: 10.1074/jbc.M409483200. [DOI] [PubMed] [Google Scholar]
- 12.Saravanan M, Vasu K, Kanakaraj R, Rao DN, Nagaraja V. R.KpnI, an HNH superfamily REase, exhibits differential discrimination at non-canonical sequences in the presence of Ca2+ and Mg2+ Nucleic Acids Res. 2007;35:2777–2786. doi: 10.1093/nar/gkm114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci USA. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan SH, Bao Y, Ciszak E, Laget S, Xu SY. Catalytic domain of restriction endonuclease BmrI as a cleavage module for engineering endonucleases with novel substrate specificities. Nucleic Acids Res. 2007;35:6238–6248. doi: 10.1093/nar/gkm665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rimseliene R, Maneliene Z, Lubys A, Janulaitis A. Engineering of restriction endonucleases: Using methylation activity of the bifunctional endonuclease Eco57I to select the mutant with a novel sequence specificity. J Mol Biol. 2003;327:383–391. doi: 10.1016/s0022-2836(03)00142-6. [DOI] [PubMed] [Google Scholar]
- 19.Roberts RJ, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanio T, Jeltsch A, Pingoud A. On the possibilities and limitations of rational protein design to expand the specificity of restriction enzymes: A case study employing EcoRV as the target. Protein Eng. 2000;13:275–281. doi: 10.1093/protein/13.4.275. [DOI] [PubMed] [Google Scholar]
- 21.Schottler S, Wenz C, Lanio T, Jeltsch A, Pingoud A. Protein engineering of the restriction endonuclease EcoRV—Structure-guided design of enzyme variants that recognize the base pairs flanking the recognition site. Eur J Biochem. 1998;258:184–191. doi: 10.1046/j.1432-1327.1998.2580184.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhu ZY, Zhou J, Friedman AM, Xu SY. Isolation of BsoBI restriction endonuclease variants with altered substrate specificity. J Mol Biol. 2003;330:359–372. doi: 10.1016/s0022-2836(03)00595-3. [DOI] [PubMed] [Google Scholar]
- 23.Alves J, Vennekohl P. In: Restriction Endonucleases. Pingoud AM, editor. Berlin: Springer; 2004. pp. 393–412. [Google Scholar]
- 24.Zhang P, Bao Y, Higgins L, Xu SY. Rational design of a chimeric endonuclease targeted to NotI recognition site. Protein Eng Des Sel. 2007;20:497–504. doi: 10.1093/protein/gzm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samuelson JC, Xu SY. Directed evolution of restriction endonuclease BstYI to achieve increased substrate specificity. J Mol Biol. 2002;319:673–683. doi: 10.1016/S0022-2836(02)00343-1. [DOI] [PubMed] [Google Scholar]
- 26.Kirsch RD, Joly E. An improved PCR mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrashekaran S, Babu P, Nagaraja V. Characterization of DNA binding activities of over-expressed KpnI REase and modification methylase. J Biosci. 1999;24:269–277. [Google Scholar]
- 28.Saravanan M, Vasu K, Ghosh S, Nagaraja V. Dual role for Zn2+ in maintaining structural integrity and inducing DNA sequence specificity in a promiscuous endonuclease. J Biol Chem. 2007;282:32320–32326. doi: 10.1074/jbc.M705927200. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.