Abstract
Excited electronic states created by UV excitation of the diribonucleoside monophosphates ApA, ApG, ApC, ApU, and CpG were studied by the femtosecond transient-absorption technique. Bleach recovery signals recorded at 252 nm show that long-lived excited states are formed in all five dinucleosides. The lifetimes of these states exceed those measured in equimolar mixtures of the constituent mononucleotides by one to two orders of magnitude, indicating that electronic coupling between proximal nucleobases dramatically slows the relaxation of excess electronic energy. The decay rates of the long-lived states decrease with increasing energy of the charge-transfer state produced by transferring an electron from one base to another. The charge-transfer character of the long-lived states revealed by this analysis supports their assignment to excimer or exciplex states. Identical bleach recovery signals were seen for ApA, (A)4, and poly(A) at delay times >10 ps after photoexcitation. This indicates that excited states localized on a stack of just two bases are the common trap states independent of the number of stacked nucleotides. The fraction of initial excitations that decay to long-lived exciplex states is approximately equal to the fraction of stacked bases determined by NMR measurements. This supports a model in which excitations associated with two stacked bases decay to exciplex states, whereas excitations in unstacked bases decay via ultrafast internal conversion. These results establish the importance of charge transfer-quenching pathways for UV-irradiated RNA and DNA in room-temperature solution.
Keywords: DNA excited states, DNA photodamage, electronic structure, femtosecond spectroscopy
Excited electronic states created in DNA by UV light have been studied since the 1960s but have received a great deal of attention recently because of advances in experiment and theory (1). These efforts are motivated by the desire to understand the photoreactions behind the genetic damage induced by UV light. There is also increasing interest in DNA and other arrays of π-stacked chromophores as materials for optoelectronic applications (2–4).
Striking differences have emerged between the dynamics of excited states in single bases and those in base assemblies. Excited states of single nucleobases and mononucleotides decay to the ground state primarily by ultrafast internal conversion in several hundred femtoseconds (5, 6). The additional degrees of freedom in polymeric DNA might be expected to quench singlet excited states even more rapidly, but the lifetimes actually increase dramatically. Femtosecond transient absorption measurements reveal that excited states in oligo- and polynucleotides relax in tens to hundreds of picoseconds (1, 7–10).
Although there is consensus that electronic relaxation can take place orders of magnitude more slowly in DNA polymers, contradictory assignments have been proposed for the long-lived states. Crespo-Hernández et al. (8) assigned the long-lived excited states seen in (dA)18, (dA)18·(dT)18, and (dAdT)9·(dAdT)9, to excimers formed between π-stacked bases on the same strand. This assignment is supported by their low radiative transition rates and by the red-shifted emission observed from DNA and RNA base multimers in low-temperature glasses (11) and in room-temperature solution (12, 13). However, it was not shown that excitations are localized on just two bases as is the case in an excimer or exciplex.
Other investigators have made different assignments for the long-lived states. Markovitsi et al. (14) proposed that they are Frenkel excitons delocalized over several bases. Buchvarov et al. (10) wrote that an “electronically relaxed exciton” is responsible for the ≥100-ps lifetime in (dA)n. They concluded that the exciton has a “1/e delocalization length” of 3.3 ± 0.5 bases from their analysis of pump-probe signals recorded several picoseconds after excitation. However, it is unclear what degree of delocalization Buchvarov et al. attribute to the electronically relaxed exciton responsible for the long-time signal.
In our view, the initial Frenkel excitons responsible for the strong UV absorption by DNA and RNA are not the same states seen at long times in the transient-absorption experiments (15). Instead, the initial bright states decay or trap to much longer-lived states localized on pairs of stacked bases. Recent theoretical studies provide support for this dynamic viewpoint (16, 17). Here, we report a femtosecond transient-absorption study of several RNA dinucleoside monophosphates that provides strong support for this paradigm. Long-lived excited states are observed in all dinucleosides investigated, in yields that are approximately equal to the fraction of π-stacked conformers in solution. The lifetimes are correlated with the energy of the state formed by transferring an electron from one base to its π-stacked neighbor. Finally, comparison of the signals from these minimally sized π-stacks with ones from longer sequences confirms the generality of our proposal that excited states in single-stranded nucleic acids rapidly trap to intrastrand exciplex states.
Results
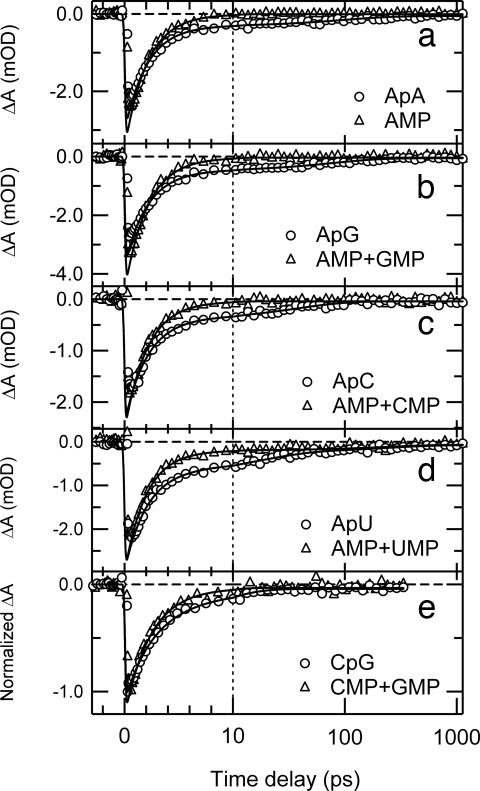
In our experiments, a femtosecond UV pump pulse abruptly creates excited electronic states in the sample under study. The ensuing dynamics are monitored by the absorption of a femtosecond probe pulse at a chosen wavelength as a function of delay time between pump and probe pulses. The ground-state population is depleted when excited states are present, and this can result in a negative absorbance change (ΔA <0) at probe wavelengths where there is strong ground-state absorption. Bleach signals are seen in all dinucleosides for probing at 252 nm after excitation at 267 nm (Fig. 1). The signals are biphasic, and nonlinear least-squares fitting reveals a dominant decay component with a time constant of ≈2 ps and a weaker decay characterized by a time constant of between 10 and 100 ps (Table 1). This extends the earlier finding of picosecond relaxation in ApA (figure 18 in ref. 1) to additional nucleobase dimers. The τ2 values for ApA and ApG were linked during fitting because the signals were kinetically indistinguishable within experimental uncertainty.
Fig. 1.
Transient absorption signals of dinucleosides at 252 nm (circles) after photoexcitation at 267 nm. (a) ApA. (b) ApG. (c) ApC. (d) ApU. (e) CpG. Signals of equimolar mixtures of the constituent mononucleotides are shown by triangles. Solid lines indicate best-fit curves obtained by global-fit analysis. The time axis in this and later figures is linear for delay times of <10 ps and logarithmic for larger values.
Table 1.
Global-fit parameters for the bleach-recovery signals probed at 252 nm
| Compound | A1/10−3 | τ1/ps | A2/10−3 | τ2/ps | A3/10−3 | τ3/ps | Offset/10−3 |
|---|---|---|---|---|---|---|---|
| ApA | −2.69 ± 0.09 | 2.05 ± 0.07 | −0.26 ± 0.03 | 105 ± 30 | — | — | −0.06 ± 0.02 |
| AMP | −3.4 ± 0.1 | ” | — | — | — | — | −0.01 ± 0.01 |
| ApG | −3.2 ± 0.1 | 2.10 ± 0.09 | −0.44 ± 0.05 | 105 ± 30 | — | — | −0.06 ± 0.03 |
| AMP + GMP | −4.5 ± 0.2 | ” | — | — | — | — | −0.04 ± 0.02 |
| ApC | −2.0 ± 0.1 | 2.1 ± 0.1 | −0.36 ± 0.05 | 45 ± 10 | — | — | −0.04 ± 0.02 |
| AMP + CMP | −2.5 ± 0.1 | ” | — | — | — | — | −0.04 ± 0.01 |
| ApU | −2.2 ± 0.1 | 2.0 ± 0.1 | −0.52 ± 0.09 | 18 ± 6 | −0.17 ± 0.07 | 240 ± 70 | −0.08 ± 0.03 |
| AMP + UMP | −2.8 ± 0.1 | ” | — | — | −0.13 ± 0.04 | ” | −0.07 ± 0.03 |
| UMP | −0.7 ± 0.1 | 2.1 ± 0.6 | — | — | −0.42 ± 0.04 | ” | −0.20 ± 0.04 |
| CpG | −0.7 ± 0.2 | 2.4 ± 0.2 | −0.4 ± 0.2 | 12 ± 8 | — | — | −0.04 ± 0.01 |
| CMP + GMP | −1.2 ± 0.1 | ” | — | — | — | — | −0.03 ± 0.01 |
The fitting function was a sum of up to three exponentials, Σi Aiexp(−t/τi), plus a constant offset. Parameters marked with dashes were not used in fitting. Parameters marked with ″ were globally linked to the row above in fitting. Errors are twice the standard deviation (2σ).
Fig. 1 also shows signals recorded for equimolar mixtures of the constituent 5′-mononucleotides for each dinucleoside (hereafter referred to as “monomer mixtures”). The signals were recorded in back-to-back measurements, and concentrations were adjusted so that all solutions had equal absorbance of 1.0 at the pump wavelength. Global fitting analysis reveals that the fast decay component (τ1 in Table 1) is the same within experimental uncertainty for each dinucleoside and its corresponding monomer mixture. In the AMP and UMP mixture, an additional decay component of 240 ± 70 ps is observed. The same decay time is observed in a solution of UMP by itself, and is assigned to the 1nπ* state (6). The signature of the long-lived 1nπ* state of CMP is not observed in the AMP + CMP and GMP + CMP mixtures because of the small fraction of light absorbed by CMP at the pump wavelength and because of the small absorption cross section of the 1nπ* state at the probe wavelength.
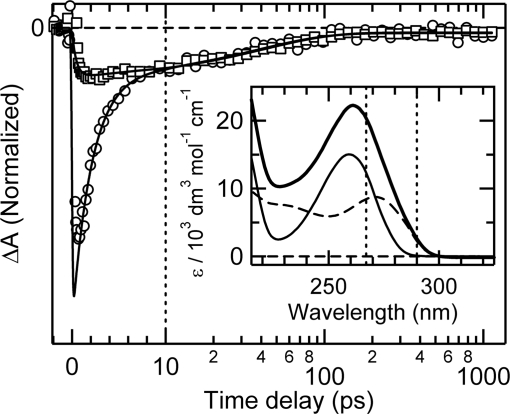
Although the UV absorption spectra of the nucleobases overlap significantly, slight shifts allow the character of the initial excited state in a dinucleoside composed of two different bases to be altered by tuning the excitation wavelength. To explore this possibility, transients were recorded for ApC at two widely separated excitation wavelengths (267 and 290 nm) with probing at 252 nm. As shown in Fig. 2, the signals are identical at delay times greater than 10 ps, but the fast decay component seen for excitation at 267 nm is completely absent at 290 nm.
Fig. 2.
Transient absorption by ApC at 252 nm after excitation at 267 nm (circles) and 290 nm (squares). The signals are normalized at 10 ps. Solid lines are best-fit curves. The Inset shows the absorption spectrum of ApC (thick line), AMP (thin line), CMP (dashed line), and the two pump wavelengths used (vertical dashed lines).
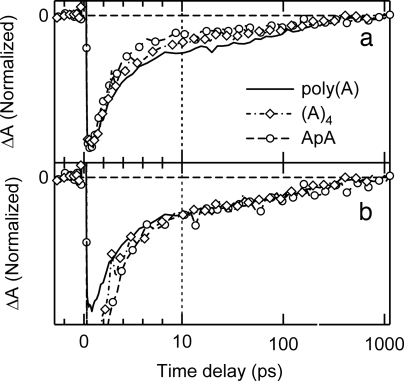
Bleach recovery signals for ApA, (A)4, and poly(A) are compared in Fig. 3. All three signals have short- and long-lived components. The ratio of the amplitude of the long-lived decay to the maximum bleach signal near time zero increases with the number of bases (Fig. 3a). When the signals are normalized at 10 ps, the decays at longer times are the same within experimental uncertainty (Fig. 3b).
Fig. 3.
Transient absorption signals of poly(A) (solid line), (A)4 (dashed-dotted line with open diamonds), and ApA (broken line with open circles) at 252 nm with photoexcitation at 267 nm. (a) The signals are normalized at the time delay at which maximal bleach is observed. (b) The signals are normalized at 10 ps after photoexcitation.
Discussion
The fast decay component in the bleach recovery signal of each dinucleoside has the same lifetime (τ ≈ 2 ps) as in the corresponding monomer mixture. Following the model first described by Crespo-Hernández et al. (8), this signal component is assigned to excited states of unstacked bases, which are assumed to decay by the same nonradiative decay pathways available to mononucleotides. Although the population returns to the electronic ground state in hundreds of femtoseconds, the bleach signals recover in aqueous solution with a characteristic time constant of ≈2 ps, which is assigned to the rate-limiting step of vibrational energy transfer to the environment (6, 8, 18).
The slow picosecond component indicates that comparatively long-lived excited states are formed in these base dimers. These states are absent in equimolar mixtures of the constituent mononucleotides, proving that they form only when two bases are in close spatial proximity. Long-lived excitations in DNA and RNA homopolymers form preferentially in π-stacked base sequences (7). We argue below that the long-lived excited states seen here are formed only in π-stacked conformers after presenting evidence that the long-lived states involve interbase charge transfer.
Long-Lived Excited States Are Exciplexes.
Emission from 2-aminopurine (2AP), a structural isomer of adenine, is thought to be quenched by electron transfer with the natural bases in DNA (19, 20), although this is disputed by some workers (21, 22). The natural bases have comparable ionization energies and electron affinities as 2-aminopurine, and their lowest singlet excited states lie even higher in energy. These factors make interbase electron transfer among the natural nucleobases highly plausible. We propose that the quenching of 2AP fluorescence in DNA and the formation and decay of the long-lived excited states reported here are caused by the same phenomenon—electron transfer between π-stacked bases.
The long-lived excited states are assigned to interbase charge-transfer states in which an electron is transferred, at least partially, from one base to a π-stacked neighbor. Previously, we referred to these states as excimers and emphasized their charge transfer character (8). In the electron-transfer literature, such states are frequently referred to as exciplexes, and we will adopt this term here to underscore the expected dominance of charge-transfer configurations in the excited-state wave function. The distinction usually made between excimer and exciplex, based on whether the two interacting molecules are identical or not, is unfortunate because it obscures their common dependence on locally excited and charge-transfer configurations.
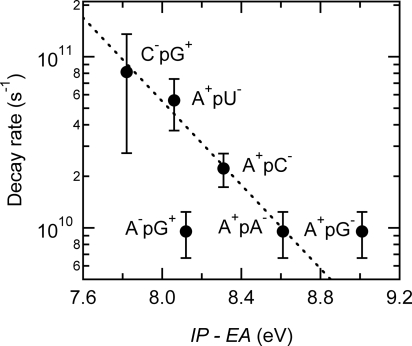
The exciplex states decay by charge recombination on the picosecond timescale. In support of this assignment, the rate of decay (1/τ2) decreases logarithmically as the thermodynamic driving force for charge recombination increases (Fig. 4). The driving force was estimated as the gas-phase ionization energy (IP) of the electron donor base minus the gas-phase electron affinity (EA) of the acceptor base (Table 2). This neglects solvation and Coulombic stabilization, but these effects, although substantial, are similar in magnitude for all base dimers. The use of gas-phase quantities provides a consistent dataset for all nucleobases and avoids issues with follow-on reactions and solvent effects on redox data (23). Importantly, the use of solution-phase redox potentials does not change the quality of the correlation in Fig. 4.
Fig. 4.
Decay rates of the long-lived states of dinucleosides plotted against the gas-phase energy of the exciplex state, IP–EA. The dashed line is a fit to guide the eye.
Table 2.
Exciplex decay rates and energetics
Correlation between the rate of decay of an excited state and IP–EA is diagnostic of electron transfer. The slowing down of charge recombination with increasing energy of the exciplex state is seen for diverse geminate radical–ion pairs (24). It is consistent with the frequent observation that highly exothermic electron-transfer reactions frequently fall in the Marcus-inverted region. However, the approximately linear correlation in Fig. 4 is also characteristic of the weak coupling limit in radiationless transitions, so we considered whether the energies of the expected Frenkel exciton states vary in the same way as the proposed exciplex states. Calculations have shown that the dipolar coupling that determines the splitting between excitonic states is no greater than several hundred cm−1 for π-stacked bases (25, 26). This is small compared with the monomer transition frequencies. Thus, A and U, which have the highest singlet energies (1), should have comparably high energy exciton states, yet ApA and ApU have radically different lifetimes. This proves that the observed rates of decay are best correlated with the energies of exciplex states and not those of Frenkel excitons.
Forward electron transfer was assumed to occur in the direction that yields the lowest-energy exciplex. For most of the dinucleosides studied, this corresponds to electron transfer from a purine to a pyrimidine base. Although pyrimidine bases are better electron acceptors than the purines (27), the latter bases combine an electron-deficient pyrimidine ring and an electron-rich imidazole ring. This could lead to electron transfer in a specific direction in purine–purine dimers like ApA and ApG because of differences in electronic coupling arising from base–base overlap (28). The directional asymmetry of electron transfer quenching of 2-aminopurine fluorescence in DNA has been extensively discussed by Barton and coworkers (29). Electron transfer in ApG is plausibly from G, the most readily oxidized base, to A, but the energy of neither the A−pG+ nor the A+pG− exciplex is well correlated with the observed rate. One possibility is that the guanine radical cation can undergo deprotonation to the solvent (30) in competition with charge recombination. Alternatively, a lower degree of charge separation in the purine–purine dimers could be responsible for the deviations in Fig. 4.
The rate of exciplex formation is not observable in these measurements because the first 10 ps of dynamics are obscured by vibrational cooling of the monomer-like excited states and by the strong two-photon absorption signal of the solvent at zero delay time. The faster initial decays of transient absorption signals at visible wavelengths (1, 8) suggest that 1 ps may be an upper bound for the time required for exciplex formation. Importantly, electron transfer between 2AP and natural DNA bases has recently been proposed to occur in <200 fs from an unequilibrated singlet excited state (31).
Influence of Excitation Wavelength on the ApC Exciplex State.
The absorption cross-section of A is 1.5 times greater than that of C at 267 nm, but at 290 nm, C absorbs >20 times more strongly than A. There is thus negligible excitation of A in ApC at 290 nm. The transient signals recorded for ApC at excitation wavelengths of 267 and 290 nm (Fig. 2) are in excellent agreement at times >10 ps, indicating that excitation of either A or C populates the same final exciplex state. Red-shifted exciplex emission from ApC at 77 K is also independent of excitation wavelength (32).
The short-time signals from ApC are strikingly different for the two excitation wavelengths. At a probe wavelength of 252 nm, the short-time vibrational cooling signal from CMP is extremely weak but is readily observable in AMP (6). The absence of any fast signal decay at 290 nm allows two important conclusions to be reached. First, there is no transfer of excitation energy to unstacked A bases. Second, because the monomer-like signals are not detectable when pumping at 290 nm and probing at 252 nm, the signals are attributable solely to collective excitations in π-stacked bases. The absence of a fast component indicates that these excited states evolve comparatively slowly with no ultrafast branching to the electronic ground state. The slow and fast signals are thus assignable to different excited-state populations and can be used to estimate exciplex yields as we now show.
Exciplex Quantum Yields.
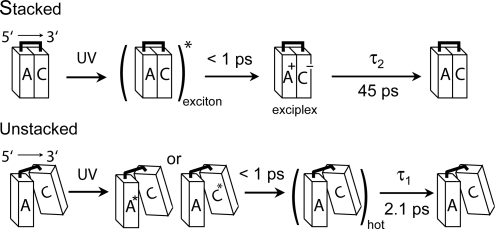
The small separation of 3.4 Å between stacked bases facilitates charge-transfer quenching of excited 2AP (29, 33). It is also responsible for the excimer/exciplex fluorescence seen from excited dinucleosides at 77 K (34). The slow decay component observed in UV-excited DNA and RNA polynucleotides is attenuated at elevated temperatures that disrupt base stacking (7). This suggests that long-lived excited states are formed only in regions where the bases are well stacked (8). Although a wide range of conformations are thermally accessible for adjacent bases in single-stranded DNA (35), we analyze our results with a two-state model in which bases are either stacked or unstacked (Fig. 5). A two-state model has been shown to explain CD and temperature-dependent UV/vis absorption spectra of DNA base multimers (36).
Fig. 5.
Kinetic model for excited-state decay in DNA. Fundamentally different relaxation pathways are postulated for stacked and unstacked conformers as illustrated for the dinucleoside ApC. The bases in π-stacked structures are in van der Waals contact, and the resulting electronic coupling produces an exciton state upon UV excitation. This initial exciton decays on an ultrafast timescale to an exciplex or charge-separated state, which subsequently decays to the electronic ground state with a time constant τ2. When bases are poorly stacked, a local excited state is produced that decays on an ultrafast time scale to a hot ground state that thermalizes with time constant τ1.
As discussed above, the wavelength-dependent transients from ApC reveal that the short- and long-time signals arise from distinct excited-state populations. The τ2 decay is assigned to excitations that result from electronic coupling between π-stacked bases, and the τ1 decay is assigned to monomer-like relaxation by unstacked bases. The signal from unstacked dinucleoside conformers is assumed to be identical in amplitude and dynamics to the signal that would be observed if the bases were present in the same concentrations as mononucleotides. Comparison of the amplitude of the short-lived dinucleoside signal with the short-lived signal amplitude from an equimolar mixture of the constituent nucleotides provides a measure of the extent of monomer-like decay. In this case, the quantum yield of long-lived excited states, fe, can be calculated from back-to-back measurements on equal absorbance solutions by using Eq. 1,
In this equation, A1d and A1m are the amplitudes of the short-lived component in the dinucleoside and monomer mixtures, respectively, and h is the hypochromicity of the dinucleoside at the pump wavelength.
We estimated fe (see Table 3) from the fit parameters in Table 1 for the adenine-containing dinucleosides using Eq. 1. Hypochromicity values at λmax from ref. 37. were used with the assumption that they differ insignificantly from h at 267 nm. The exciplex yield is very similar for all five dinucleosides even though τ2 varies over a wide range. For ApU and AMP + UMP, the sum of A1 and A3 was used in place of A1 in Eq. 1 to account for the 1nπ* state of UMP. The degree of base stacking has been estimated for many dinucleosides by NMR (38, 39) and CD spectroscopy (40–42). As summarized in Table 3, the exciplex yields calculated with Eq. 1 are comparable to or slightly less than the fraction of stacked bases estimated by NMR, whereas CD experiments provide slightly higher estimates. These differences could indicate that the two techniques are sensitive to different conformational characteristics. Although the reduction to just two conformational states, stacked or unstacked, can be criticized, a reasonable conclusion is that most excitations in stacked bases decay to exciplexes (8).
Table 3.
Hypochromicity and exciplex yields for various dinucleosides compared with the fraction of stacked bases determined by NMR and CD spectroscopy
Exciplex Lifetimes Are Independent of the Number of Stacked Bases.
Longer base stacks in base multimers could lead to delocalization of excitations over more than two bases (10, 25). Increasing excited state delocalization could lead to length-dependent signals, but identical dynamics are seen in ApA, (A)4, and poly(A) from 10 ps onwards (Fig. 3). This is strong evidence that long-lived excited states in runs of adenine bases are the same ones observed in ApA. We conclude that the initial excitons, which may exhibit length-dependent delocalization (10), decay to a common trap state localized on a pair of nearest-neighbor bases no later than a few picoseconds after excitation. As discussed above, this decay is likely to occur on the femtosecond time scale.
An important question is whether an exciplex can migrate along the DNA helix perhaps in response to backbone fluctuations induced by the environment. This raises important parallels with charge transport in DNA (19, 28, 43–46). Instead of motion of a single electron or hole, exciplex diffusion requires motion of a bound electron–hole pair. Even though the exciplex states live for many picoseconds, they are nonetheless short-lived compared with the lifetimes of nucleobase radicals (43), and this will further constrain the achievable distances. The identical signals seen at times >10 ps for ApA and poly(A), which contains hundreds of adenine bases, suggest that exciplex migration, at least in A tracts, can be neglected at room temperature or involves too small a fraction of the total exciplex population to be detectable in our experiments.
Conclusions
DNA damage by UV light is governed by relaxation pathways for excess electronic energy: Excited states either evolve to photoproducts or decay harmlessly to the electronic ground state. This work shows that the long-lived excited states seen in longer DNA model systems (7–10, 13) are formed in stacks of just two bases. The good correlation between the measured lifetimes and the driving force for charge recombination supports assignment to exciplex states created by interbase charge transfer. Long-lived signals in ApA are dynamically indistinguishable from ones in longer homoadenine sequences, indicating that a common exciplex state spanning two stacked bases is formed in all systems. Exciplexes are not formed directly but are populated rapidly from initial Frenkel excitons, which are radiatively brighter. This model, which is supported by a recent computational study (17), readily explains the dramatic differences between transient absorption and fluorescence experiments (14, 15). It also underscores the importance of both Frenkel excitonic and charge-transfer interactions in assemblies of π-stacked chromophores (8, 16, 47).
Although the precise effects of base pairing must still be elucidated, the high yields of exciplex formation in base-stacked dinucleosides suggest that decay to exciplex states is the dominant decay channel in general nucleic acids. The photostability of the DNA double helix is thus built on fundamentally different nonradiative decay mechanisms than the ones found in single bases (1, 48, 49). A central conclusion from this study is that relaxation pathways for excess electronic energy in DNA are determined primarily by its π-stacked architecture and not by the photophysical properties of its building blocks. Schreier et al. (50) provided evidence recently that thymine dimer formation in DNA is determined largely by conformational properties at the instant of excitation. It is apparent that DNA photophysics and photochemistry are controlled to a great extent by the π-stacking interactions that in large measure define DNA structure. The sensitivity of excited-state dynamics to conformational properties could therefore lead to methods for probing rapidly evolving structures in nucleic acids.
Methods
Transient absorption signals were recorded by using the amplified output of a femtosecond Ti:sapphire laser (Mira/Legend; Coherent at 50 fs, 800 nm, 1 kHz), which was divided to generate pump and probe pulses. One portion of the output was converted to the third harmonic (267 nm) and used as pump light in most experiments. The other portion was passed through optical parametric amplifiers (OPerA with SFG UV and UV/VIS options; Coherent) to produce pulses at 252 or 290 nm. The probe light transmitted through a sample was dispersed with a monochromator and detected by a photomultiplier tube. Changes of probe-light intensity were detected by a lock-in amplifier referenced to an optical chopper that modulated the pump beam. The angle between pump and probe-light polarizations was 54.7° to measure reorientation-free signals.
Five dinucleoside monophosphates were used for the transient-absorption experiments. The free acid of ApA and the ammonium salt of ApG were purchased from Sequoia Research Products. The free acid of ApC and CpG were purchased from Sigma–Aldrich. The ammonium salt of ApU was purchased from MP Biomedicals. The 5′-monoribonucleotides AMP, GMP, CMP, and UMP, and the potassium salt of poly(A) were purchased from Sigma–Aldrich. The ammonium salt of (A)4 was purchased from Midland Certified Reagent Company. All samples were used without further purification. Compounds were dissolved in a pH 7 phosphate buffer solution, and all measurements were carried out at room temperature. Solutions did not contain added salt, and the total ionic strength was determined solely by the phosphate buffer. The low ionic-strength conditions used in our experiments are not expected to influence the stacking–unstacking equilibrium or other conformational properties because dinucleosides have a single charged-phosphate group. Varying the ionic strength between 0.01 and 1.0 had no effect on the stacking equilibrium for ApA (36). Control experiments on ApA and ApU confirmed that identical signals were observed with and without added 0.1 M NaCl. Samples were held between CaF2 (front) and fused silica (back) windows separated by 1.2 mm and rotated at several hundred revolutions per minute. All sample solutions were prepared to have an optical density of 1 at the pump wavelength as determined by a commercial UV/vis spectrometer (Lambda 25; PerkinElmer). Photodegradation of the samples during experiments was periodically monitored by recording UV/vis spectra and were replaced with fresh ones if significant spectral changes were observed.
Fit functions were convoluted with a Gaussian with a full-width at half-maximum of 230 fs. This value was determined from the two-photon absorption signal measured for UV pump and probe wavelengths in pure water. Because the lifetime of the short-lived component for each dinucleoside was very similar to the lifetime observed for the equimolar mononucleotide mixture, these lifetimes were linked during global fitting. All reported uncertainties are twice the standard deviation (2σ).
Acknowledgments.
This research was supported by National Institutes of Health Grant R01 GM64563. Measurements were performed in Ohio State's Center for Chemical and Biophysical Dynamics, with equipment funded by the National Science Foundation and the Ohio Board of Regents.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Crespo-Hernández CE, Cohen B, Hare PM, Kohler B. Ultrafast excited-state dynamics in nucleic acids. Chem Rev. 2004;104:1977–2019. doi: 10.1021/cr0206770. [DOI] [PubMed] [Google Scholar]
- 2.Lewis FD. DNA molecular photonics. Photochem Photobiol. 2005;81:65–72. doi: 10.1562/2004-09-01-ir-299.1. [DOI] [PubMed] [Google Scholar]
- 3.Delos Santos GB, Lewis FD. Electronic interactions between π-stacked chromophores in nonsymmetric tertiary naphthyl di- and polyarylureas. J Phys Chem A. 2005;109:8106–8112. doi: 10.1021/jp052406+. [DOI] [PubMed] [Google Scholar]
- 4.Steckl AJ. DNA—A new material for photonics? Nature Photonics. 2007;1:3–5. [Google Scholar]
- 5.Pecourt J-ML, Peon J, Kohler B. DNA excited-state dynamics: Ultrafast internal conversion and vibrational cooling in a series of nucleosides. J Am Chem Soc. 2001;123:10370–10378. doi: 10.1021/ja0161453. [DOI] [PubMed] [Google Scholar]
- 6.Hare PM, Crespo-Hernández CE, Kohler B. Internal conversion to the electronic ground state occurs via two distinct pathways for pyrimidine bases in aqueous solution. Proc Natl Acad Sci USA. 2007;104:435–440. doi: 10.1073/pnas.0608055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crespo-Hernández CE, Kohler B. Influence of secondary structure on electronic energy relaxation in adenine homopolymers. J Phys Chem B. 2004;108:11182–11188. [Google Scholar]
- 8.Crespo-Hernández CE, Cohen B, Kohler B. Base stacking controls excited-state dynamics in A·T DNA. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 9.Kwok W-M, Ma C, Phillips DL. Femtosecond time- and wavelength-resolved fluorescence and absorption spectroscopic study of the excited states of adenosine and an adenine oligomer. J Am Chem Soc. 2006;128:11894–11905. doi: 10.1021/ja0622002. [DOI] [PubMed] [Google Scholar]
- 10.Buchvarov I, Wang Q, Raytchev M, Trifonov A, Fiebig T. Electronic energy delocalization and dissipation in single- and double-stranded DNA. Proc Natl Acad Sci USA. 2007;104:4794–4797. doi: 10.1073/pnas.0606757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisinger J, Guéron M, Shulman RG, Yamane T. Excimer fluorescence of dinucleotides, polynucleotides, and DNA. Proc Natl Acad Sci USA. 1966;55:1015–1020. doi: 10.1073/pnas.55.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plessow R, Brockhinke A, Eimer W, Kohse-Höinghaus K. Intrinsic time- and wavelength-resolved fluorescence of oligonucleotides: A systematic investigation using a novel picosecond laser approach. J Phys Chem B. 2000;104:3695–3704. [Google Scholar]
- 13.Markovitsi D, Gustavsson T, Talbot F. Excited states and energy transfer among DNA bases in double helices. Photochem Photobiol Sci. 2007;6:717–724. doi: 10.1039/b705674e. [DOI] [PubMed] [Google Scholar]
- 14.Markovitsi D, et al. Molecular spectroscopy: Complexity of excited-state dynamics in DNA. Nature. 2006;441:E7. doi: 10.1038/nature04903. [DOI] [PubMed] [Google Scholar]
- 15.Crespo-Hernández CE, Cohen B, Kohler B. Molecular spectroscopy: Complexity of excited-state dynamics in DNA (reply) Nature. 2006;441:E8. doi: 10.1038/nature04903. [DOI] [PubMed] [Google Scholar]
- 16.Bittner ER. Lattice theory of ultrafast excitonic and charge-transfer dynamics in DNA. J Chem Phys. 2006;125 doi: 10.1063/1.2335452. 094909/094901–12. [DOI] [PubMed] [Google Scholar]
- 17.Santoro F, Barone V, Improta R. Influence of base stacking on excited-state behavior of polyadenine in water, based on time-dependent density functional calculations. Proc Natl Acad Sci USA. 2007;104:9931–9936. doi: 10.1073/pnas.0703298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middleton CT, Cohen B, Kohler B. Solvent and solvent isotope effects on the vibrational cooling dynamics of a DNA base derivative. J Phys Chem A. 2007;111:10460–10467. doi: 10.1021/jp0740595. [DOI] [PubMed] [Google Scholar]
- 19.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 20.Wan C, Fiebig T, Schiemann O, Barton JK, Zewail AH. Femtosecond direct observation of charge transfer between bases in DNA. Proc Natl Acad Sci USA. 2000;97:14052–14055. doi: 10.1073/pnas.250483297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynisson J, Steenken S. One-electron reduction of 2-aminopurine in the aqueous phase. A DFT and pulse radiolysis study. Phys Chem Chem Phys. 2005;7:659–665. doi: 10.1039/b417343k. [DOI] [PubMed] [Google Scholar]
- 22.Hardman SJO, Thompson KC. Influence of base stacking and hydrogen bonding on the fluorescence of 2-aminopurine and pyrrolocytosine in nucleic acids. Biochemistry. 2006;45:9145–9155. doi: 10.1021/bi060479t. [DOI] [PubMed] [Google Scholar]
- 23.Crespo-Hernández CE, Close DM, Gorb L, Leszczynski J. Determination of redox potentials for the Watson–Crick base pairs, DNA nucleosides, and relevant nucleoside analogues. J Phys Chem B. 2007;111:5386–5395. doi: 10.1021/jp0684224. [DOI] [PubMed] [Google Scholar]
- 24.Gould IR, Ege D, Moser JE, Farid S. Efficiencies of photoinduced electron-transfer reactions: Role of the Marcus inverted region in return electron transfer within geminate radical-ion pairs. J Am Chem Soc. 1990;112:4290–4301. [Google Scholar]
- 25.Bouvier B, Gustavsson T, Markovitsi D, Millie P. Dipolar coupling between electronic transitions of the DNA bases and its relevance to exciton states in double helices. Chem Phys. 2002;275:75–92. [Google Scholar]
- 26.Czader A, Bittner ER. Calculations of the exciton coupling elements between the DNA bases using the transition density cube method. J Chem Phys. 2008;128 doi: 10.1063/1.2821384. 035101–035112. [DOI] [PubMed] [Google Scholar]
- 27.Steenken S. Purine bases, nucleosides, and nucleotides: Aqueous solution redox chemistry and transformation reactions of their radical cations and e− and OH adducts. Chem Rev. 1989;89:503–520. [Google Scholar]
- 28.Jortner J, Bixon M, Voityuk AA, Roesch N. Superexchange mediated charge hopping in DNA. J Phys Chem A. 2002;106:7599–7606. [Google Scholar]
- 29.O'Neill MA, Barton JK. Effects of strand and directional asymmetry on base–base coupling and charge transfer in double-helical DNA. Proc Natl Acad Sci USA. 2002;99:16543–16550. doi: 10.1073/pnas.012669599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuimova MK, et al. Monitoring the direct and indirect damage of DNA bases and polynucleotides by using time-resolved infrared spectroscopy. Proc Natl Acad Sci USA. 2006;103:2150–2153. doi: 10.1073/pnas.0506860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan C, Xia T, Becker H-C, Zewail AH. Ultrafast unequilibrated charge transfer: A new channel in the quenching of fluorescent biological probes. Chem Phys Lett. 2005;412:158–163. [Google Scholar]
- 32.Guéron M, Shulman RG, Eisinger J. Energy transfer in dinucleotides. Proc Natl Acad Sci USA. 1966;56:814–818. doi: 10.1073/pnas.56.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neill MA, Barton JK. DNA charge transport: Conformationally gated hopping through stacked domains. J Am Chem Soc. 2004;126:11471–11483. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 34.Koudelka J, Augenstein L. Importance of the microenviroment surrounding a chromophore in determining its spectroscopic behavior. Photochem Photobiol. 1968;7:613–617. [Google Scholar]
- 35.Martinez JM, Elmroth SKC, Kloo L. Influence of sodium ions on the dynamics and structure of single-stranded DNA oligomers: A molecular dynamics study. J Am Chem Soc. 2001;123:12279–12289. doi: 10.1021/ja0108786. [DOI] [PubMed] [Google Scholar]
- 36.Powell JT, Richards EG, Gratzer WB. Nature of stacking equilibriums in polynucleotides. Biopolymers. 1972;11:235–250. doi: 10.1002/bip.1972.360110118. [DOI] [PubMed] [Google Scholar]
- 37.Warshaw MM, Tinoco I., Jr Optical properties of sixteen dinucleoside phosphates. J Mol Biol. 1966;20:29–38. doi: 10.1016/0022-2836(66)90115-x. [DOI] [PubMed] [Google Scholar]
- 38.Lee C-H, Ezra FS, Kondo NS, Sarma RH, Danyluk SS. Conformational properties of dinucleoside monophosphates in solution: Dipurines and dipyrimidines. Biochemistry. 1976;15:3627–3639. doi: 10.1021/bi00661a034. [DOI] [PubMed] [Google Scholar]
- 39.Ezra FS, Lee C-H, Kondo NS, Danyluk SS, Sarma RH. Conformational properties of purine–pyrimidine and pyrimidine–purine dinucleoside monophosphates. Biochemistry. 1977;16:1977–1987. doi: 10.1021/bi00628a035. [DOI] [PubMed] [Google Scholar]
- 40.Brahms J, Maurizot JC, Michelson AM. Conformational stability of dinucleotides in solution. J Mol Biol. 1967;25:481–495. doi: 10.1016/0022-2836(67)90200-8. [DOI] [PubMed] [Google Scholar]
- 41.Warshaw MM, Cantor CR. Oligonucleotide interactions. IV. Conformational differences between deoxy- and ribodinucleoside phosphates. Biopolymers. 1970;9:1079–1103. doi: 10.1002/bip.1970.360090910. [DOI] [PubMed] [Google Scholar]
- 42.Johnson NP, Schleich T. Circular dichroism studies of the conformational stability of dinucleoside phosphates and related compounds in aqueous neutral salt solutions. Biochemistry. 1974;13:981–987. doi: 10.1021/bi00702a023. [DOI] [PubMed] [Google Scholar]
- 43.Lewis FD, et al. Direct measurement of hole transport dynamics in DNA. Nature. 2000;406:51–53. doi: 10.1038/35017524. [DOI] [PubMed] [Google Scholar]
- 44.Schuster GB. Long-range charge transfer in DNA: Transient structural distortions control the distance dependence. Acc Chem Res. 2000;33:253–260. doi: 10.1021/ar980059z. [DOI] [PubMed] [Google Scholar]
- 45.Conwell EM. Charge transport in DNA in solution: The role of polarons. Proc Natl Acad Sci USA. 2005;102:8795–8799. doi: 10.1073/pnas.0501406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giese B, Amaudrut J, Kohler AK, Spormann M, Wessely S. Direct observation of hole transfer through DNA by hopping between adenine bases and by tunnelling. Nature. 2001;412:318–320. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann M, et al. The lowest energy Frenkel and charge-transfer excitons in quasi-one-dimensional structures: Application to MePTCDI and PTCDA crystals. Chem Phys. 2000;258:73–96. [Google Scholar]
- 48.Serrano-Andrés L, Merchán M, Borin AC. Adenine and 2-aminopurine: Paradigms of modern theoretical photochemistry. Proc Natl Acad Sci USA. 2006;103:8691–8696. doi: 10.1073/pnas.0602991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satzger H, et al. Primary processes underlying the photostability of isolated DNA bases: Adenine. Proc Natl Acad Sci USA. 2006;103:10196–10201. doi: 10.1073/pnas.0602663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schreier WJ, et al. Thymine dimerization in DNA is an ultrafast photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orlov VM, Smirnov AN, Varshavsky YM. Ionization potentials and electron-donor ability of nucleic acid bases and their analogues. Tetrahedron Lett. 1976;4377:4378. [Google Scholar]
- 52.Li X, Cai Z, Sevilla MD. DFT calculations of the electron affinities of nucleic acid bases: Dealing with negative electron affinities. J Phys Chem A. 2002;106:1596–1603. [Google Scholar]