Abstract
SgrS is an Hfq-binding small RNA that is induced under glucose phosphate stress in Escherichia coli. It forms a specific ribo nucleo protein complex with Hfq and RNase E resulting in translational repression and rapid degradation of ptsG mRNA, encoding the glucose transporter. Here, we report translational silencing of ptsG mRNA in a defined in vitro system. We demonstrate that SgrS and Hfq are the minimum components for translational silencing to faithfully reproduce the reaction in cells. We show that ptsG-SgrS base pairing is sufficient to cause translational repression when the ptsG mRNA is forced to base pair with SgrS without the help of Hfq. The extent of translational repression correlates with the extent of duplex formation. We conclude that base pairing itself but not Hfq is directly responsible for translational silencing and the major role of Hfq in gene silencing is to stimulate the base pairing between SgrS and ptsG mRNA. This simple mechanism is in striking contrast to miRNA action in eukaryote in which the RNA is believed to act only as a guide of protein partners.
Keywords: base pairing, in vitro reconstitution, translational repression, SgrS
Base-pairing small RNAs (sRNAs) are widespread in all organisms and are involved in the regulation of gene expression primarily at posttranscriptional levels. It is becoming clear that a major class of base-pairing sRNAs in Escherichia coli bind to the RNA chaperone Hfq (1, 2) and act by imperfect base pairing to regulate mRNA function (in most cases negatively) and the translation and stability of target mRNAs under specific stress conditions (3–5). SgrS represents Hfq-binding base-pairing sRNAs in E. coli. It is induced in response to accumulation of glucose 6-phosphate (G6P) or α-methylglucoside 6-phosphate and down-regulates the expression of ptsG encoding the glucose transporter at posttranscriptional levels (6, 7).
Studies on gene silencing mediated by SgrS in the last few years have uncovered several notable features of SgrS action that could be relevant to the functions of other Hfq-binding sRNAs. SgrS forms a specific ribonucleoprotein complex with RNase E via interactions with Hfq to cause the translational repression and rapid degradation of ptsG mRNA in an RNase E-dependent manner (8). However, the RNase E-dependent mRNA degradation is dispensable for gene silencing (9). It has been established that of the predicted 23 base pairs between SgrS and ptsG mRNA, only the 6 consecutive base pairs overlapping the ptsG Shine–Dalgarno (SD) sequence are crucial for SgrS action (10). Hfq stimulates base pairing between SgrS and the target mRNA by accelerating the rate of duplex formation (10). In addition, it has been shown that membrane localization of target mRNA facilitates the action of SgrS, presumably by affecting competition between the sRNA and ribosomes (11). A more recent fascinating discovery concerning SgrS function is that SgrS acts not only as a base-pairing RNA but also as an mRNA template, encoding a small functional protein to deal with the same metabolic stress (12).
Despite the substantial progress mentioned above, several important questions still remain to be elucidated concerning the mechanisms of SgrS action. For example, although both SgrS and Hfq are essential for translational silencing of ptsG mRNA in intact cells, it is not clear yet whether SgrS and Hfq are sufficient for translational silencing. Thus, we cannot exclude the possibility that some other factors are additionally required for translational silencing. Another important question concerns the exact role of SgrS and Hfq in translational silencing. In particular, an intriguing question is which component, SgrS (and therefore base pairing itself) or Hfq, is ultimately responsible for the translational inhibition. To address these questions and to learn more about the mechanisms of action of Hfq-binding sRNAs, we investigated translational gene silencing of ptsG mRNA by SgrS and Hfq in a defined in vitro translation system (PURESYSTEM; Post Genome Institute) (13). The present study strongly suggests that SgrS and Hfq are the minimum components and that no other factors are required for SgrS-directed translational silencing of ptsG mRNA. We also demonstrate that the base pairing itself, but not the recruitment of Hfq to the translational initiation region of ptsG mRNA, is the fundamental event for translational silencing.
Results
In Vitro Translation of ptsG Using Purified Translation Components.
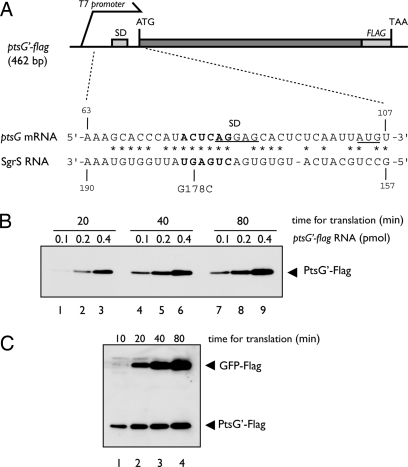
For in vitro translation of ptsG, we used a reconstituted PURESYSTEM that is composed of purified ribosome and all other factors required for translation and transcription (13). First, the protein synthesis coupled with transcription was performed by using a DNA template containing the T7 promoter and the 5′ portion of ptsG (ptsG′) encoding the N-terminal 117 aa residues of PtsG (PtsG′) (Fig. 1A). The Flag tag sequence was placed before the stop codon to allow the detection of translation products by anti-Flag antibody. The translation products were analyzed by SDS/PAGE followed by Western blotting probed with anti-Flag antibody. A significant amount of PtsG′–Flag protein was detected, indicating that the ptsG′–flag DNA fragment was efficiently transcribed and translated in vitro (data not shown). Next, the ptsG′–flag mRNA encoding PtsG′–Flag was prepared by in vitro transcription and purified through electrophoresis on a polyacrylamide gel. Then, the purified ptsG′–flag mRNA was subjected to in vitro translation assay and translation products were analyzed by SDS/PAGE followed by Western blotting. As shown in Fig. 1B, the amount of PtsG′–Flag increased with increasing amounts of RNA template and with incubation time. When both ptsG′–flag RNA and gfp–flag mRNA (internal control) of about 820 bases were translated together, the translation products from the two RNAs increased with incubation time up to 80 min (Fig. 1C).
Fig. 1.
In vitro translation of ptsG mRNA by PURESYSTEM. (A) Schematic drawing of ptsG′–flag DNA fragment. The 462-bp ptsG′–flag DNA contains the sequence for the N-terminal 117 aa residues of PtsG along with a part of its 5′ UTR. It carries also the sequence for T7 promoter before the 5′ UTR and the sequence for Flag tag peptide just before the TAA stop codon. The nucleotide sequences around translation initiation region of ptsG mRNA and the base-pairing region of SgrS are shown. The SD sequence and the initiation codon of ptsG mRNA are underlined. The asterisks correspond to nucleotides complementary to SgrS RNA (7). The bold letters represent the critical nucleotides for base pairing with SgrS (10). (B) Translation of purified ptsG′–flag RNA. Translation was carried out in 10 μl of reaction mixture by using 0.1, 0.2, and 0.4 pmol of purified ptsG′–flag RNA at 37°C. At indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting using anti-Flag antibody. (C) Mixed translation of purified ptsG′–flag and gfp–flag RNAs. Both 0.2 pmol of ptsG′–flag RNA and 0.4 pmol of gfp-flag RNA were translated together in 10 μl of reaction mixture at 37°C. At indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting using anti-Flag antibody.
Effects of Hfq and SgrS on ptsG Translation.
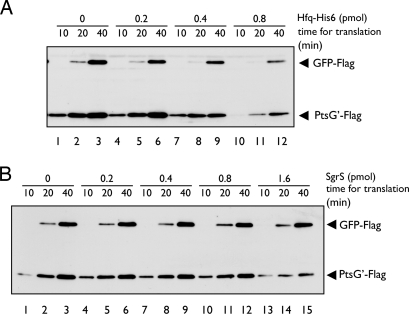
To investigate the effect of Hfq on translation, we performed the translation reaction in the presence of increasing amounts of purified Hfq–His6 for 10, 20, and 40 min (Fig. 2A). The amounts of Hfq–His6 are expressed per Hfq hexamer throughout this work. When 0.2 pmol ptsG′–flag RNA was translated along with a control gfp–flag RNA, up to 0.4 pmol Hfq–His6 inhibited translation of both RNAs only slightly (Fig. 2A, lanes 1–9). When Hfq–His6 was further increased, the amounts of translation products from both RNAs were significantly reduced (Fig. 2A, lanes 10–12). The inhibitory effect of Hfq–His6 on ptsG′–flag translation is likely to be due to nonspecific binding of Hfq to this RNA, because translation of the control gfp–flag RNA was also inhibited to the same extent. Next, we examined the effect of SgrS RNA on translation (Fig. 2B). SgrS did not affect translation of both RNAs when added at up to 0.8 pmol (lanes 1–12), whereas 1.6 pmol SgrS moderately reduced ptsG′–flag translation (lanes 13–15). The effect of SgrS on ptsG′–flag translation is specific because translation of control gfp–flag mRNA was little affected under the same condition. As shown later, the ptsG′–flag RNA could form the RNA duplex with SgrS in vitro, although slowly (see Fig. 6). Therefore, the moderate inhibition of ptsG′–flag translation by SgrS could be due to duplex formation between ptsG′–flag RNA and SgrS during the translation assay. This suggests that SgrS alone functions to inhibit the translation of target ptsG mRNA once it forms an RNA–RNA duplex with the ptsG mRNA.
Fig. 2.
Effect of Hfq or SgrS on in vitro translation. Both 0.2 pmol of ptsG′–flag RNA and 0.4 pmol of gfp-flag RNA were translated together in 10 μl of reaction mixture containing indicated amounts of purified Hfq–His6 (A) or SgrS (B) at 37°C. At indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting.
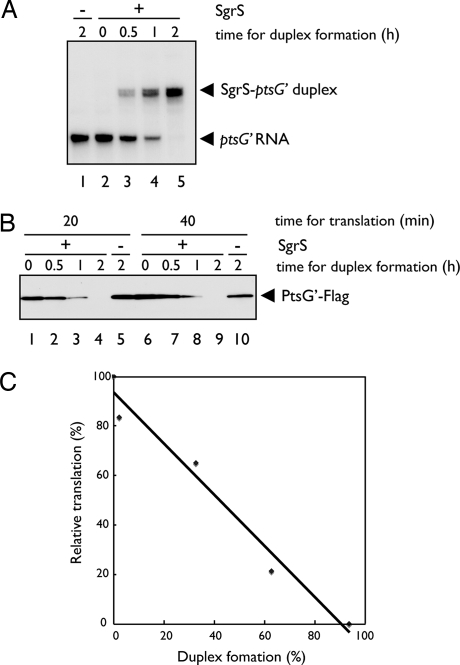
Fig. 6.
Inhibition of ptsG translation by SgrS without Hfq in vitro. (A) Formation of SgrS-ptsG′ duplex in the absence of Hfq. 32P-labeled ptsG′–flag RNA (0. 2 pmol) was incubated with SgrS RNA (0.4 pmol) in 3.5 μl of binding buffer at 37°C for the indicated times. Samples were analyzed by a gel mobility shift assay on a native polyacrylamide gel after the addition of 1 μl of RNA loading buffer. (B) In vitro translation of SgrS-ptsG mixture. 32P-labeled ptsG′-flag RNA (0. 2 pmol) was incubated with unlabeled SgrS RNA (0.4 pmol) in 3.5 μl of binding buffer at 37°C for the indicated times. Then, each sample was translated in 10 μl of reaction mixture consisting of PURESYSTEM at 37°C. At indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting. (C) Correlation between duplex formation and translational inhibition. The band signals in A and B (lanes 1–5) on the films were quantified by using Multi Gauge Ver. 3.1 software (Fujifilm) and plotted. The ptsG′ RNA band (A, lane 1) and the PtsG′–Flag band (B, lane 5) were taken as 0% duplex formation and 100% relative translation, respectively.
SgrS Efficiently Inhibits ptsG Translation in the Presence of Hfq.
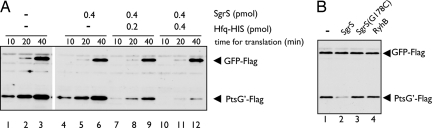
Both SgrS and Hfq are required for translational silencing of ptsG in intact cells (11). We also showed previously that Hfq dramatically stimulates duplex formation between ptsG mRNA and SgrS in vitro (10). To test the requirement for both Hfq and SgrS for translational silencing of ptsG in vitro, we carried out translation in the presence of both SgrS and Hfq. As shown in Fig. 3A, translation of ptsG′–flag mRNA was markedly inhibited when SgrS and Hfq–His6 were present together. The presence of SgrS and Hfq–His6 did not affect translation of the control gfp-flag mRNA. A mutant form of SgrS (SgrSG178C) that no longer down-regulates ptsG translation in vivo (10) was unable to inhibit ptsG′–flag translation in vitro (Fig. 3B, lane 3). A noncognate sRNA RyhB also did not affect the ptsG′–flag translation in vitro (Fig. 3B, lane 4). These results are essentially consistent with the in vivo data concerning the regulation of ptsG translation by SgrS in intact cells, indicating that in vitro, SgrS also acts to specifically inhibit ptsG translation in the presence of Hfq through base pairing. Thus, we have succeeded in faithfully reproducing the translational gene silencing of ptsG mRNA by SgrS and Hfq in vitro, using PURESYSTEM.
Fig. 3.
SgrS specifically inhibits ptsG translation in the presence of Hfq. (A) Both 0.2 pmol of ptsG′–flag RNA and 0.4 pmol of gfp-flag RNA were translated together in 10 μl of reaction mixture containing indicated amounts of purified Hfq–His6 and SgrS at 37°C. At indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting. (B) Both 0.2 pmol of ptsG′–flag RNA and 0.4 pmol of gfp-flag RNA were translated together in 10 μl of reaction mixture containing 0.4 pmol of indicated sRNA and 0.8 pmol of Hfq–His6 at 37°C. After 40 min incubation, 4.5 μl of reaction mixture was taken and subjected to Western blotting.
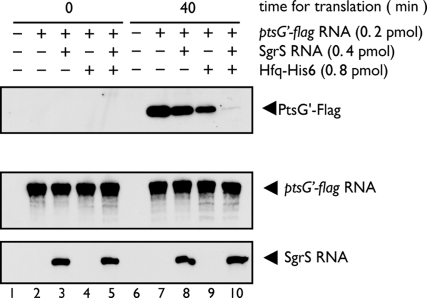
To examine whether the incubation in PURESYSTEM affects the stability of ptsG RNA, we incubated the reaction mixtures containing the indicated components for 0 min or 40 min and subjected them to both Western and Northern blot analyses. We again confirmed that the ptsG′–flag mRNA was efficiently translated when incubated without SgrS and/or Hfq for 40 min, whereas the translation was strongly inhibited in the presence of both SgrS and Hfq (Fig. 4Top, lanes 7–10). No translation product was detected at 0 min incubation (Fig. 4 Top, lanes 2–5). Northern blotting using the ptsG probe revealed that the yield of ptsG′–flag mRNA after 40 min incubation was essentially identical with that at 0 min incubation (Fig. 4 Middle). Thus, the ptsG′–flag mRNA essentially remains unchanged during the translation reaction. In addition, translation by PURESYSTEM did not affect the stability of SgrS RNA (Fig. 4 Bottom).
Fig. 4.
RNAs remain unchanged during translation reaction. Preincubation mixtures containing the indicated components (ptsG′–flag RNA, Hfq–His6, and SgrS) were incubated with PURESYSTEM mix in a final volume of 10 μl. At indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting (Top). To analyze RNAs, we took 1 μl of reaction mixture and mixed it with 100 μl of RNA buffer (20 mM sodium acetate, pH 5.5; 0.5% SDS; and 1 mM EDTA). The mixture was treated with phenol, and RNAs were precipitated with ethanol. The precipitate was dissolved in 11 μl of a sample buffer (90% formamide; 10 mM EDTA, pH 8.0; and 0. 025% bromophenol blue), and 5 μl of each sample was subjected to Northern blotting using either ptsG (Middle) or sgrS (Bottom) probes.
Direct Evidence for the Formation of an SgrS–ptsG–Hfq Ternary Complex.
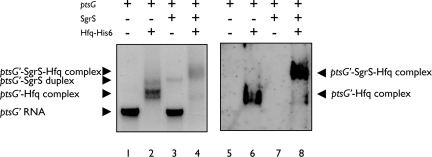
SgrS forms a ribonucleoprotein complex with Hfq and RNase E (8), and Hfq dramatically stimulates the duplex formation between ptsG mRNA and SgrS in vitro (10). Although we speculate that Hfq still associates stably with RNAs even after the duplex formation, we do not exclude the alternative possibility that Hfq may dissociate from two RNAs after the SgrS–ptsG duplex is formed. To address this question, we analyzed the complex formation between SgrS, 32P-labeled ptsG′–flag RNA, and Hfq–His6 under a mild condition (5 min at 30°C) by a gel mobility shift assay. The same gel was subjected to autoradiography (Fig. 5, lanes 1–4) and to Western blotting using anti-Hfq antibodies (Fig. 5, lanes 5–8). When the 32P-labeled ptsG′–flag RNA was incubated with SgrS alone, only a small fraction of ptsG′–flag RNA was shifted to the position corresponding to the SgrS–ptsG′ duplex (Fig. 5, lane 3). However, when the 32P-labeled ptsG′–flag RNA was incubated with Hfq–His6 alone, ptsG′–flag RNA was completely shifted, resulting in ptsG′–Hfq complexes (Fig. 5, lanes 2 and 6). Although we do not know the exact binding sites of Hfq within ptsG RNA, it should be noted that the binding of Hfq to ptsG RNA does not significantly affect ptsG translation (see Fig. 2A). When the 32P-labeled ptsG′–flag RNA was incubated with SgrS in the presence of Hfq–His6, the base pairing between SgrS and ptsG′–flag was markedly enhanced, resulting in a new band of slow mobility that is distinct from the SgrS-ptsG′ duplex (Fig. 5, lane 4). This new band apparently corresponds to the SgrS–ptsG′–Hfq-His6 ternary complex, and the existence of Hfq in the complex was confirmed by Western blotting (Fig. 5, lane 8). No SgrS-ptsG′ duplex and only a little ptsG′–Hfq–His6 complex were formed under this condition (Fig. 5, lanes 4 and 8). This strongly suggests that Hfq forms a stable ternary complex with the SgrS-ptsG′ duplex. The formation of such ternary complex may be a general property of Hfq-binding small RNAs because Zhang et al. (14) also observed bands corresponding to OxyS sRNA and its target fhlA mRNA and Hfq complex in their gel shift assay.
Fig. 5.
Complex formation between ptsG RNA, SgrS, and Hfq. 32P-labeled ptsG′–flag RNA (0. 2 pmol) was incubated with unlabeled SgrS RNA (0.4 pmol) and/or Hfq–His6 (0.1 pmol) in 3.5 μl of binding buffer at 30°C for 5 min, and 1 μl of RNA loading buffer was added. The complex formation was monitored by a gel mobility shift assay on a native polyacrylamide gel using 3 μl of each sample (lanes 1–4). One microliter of each sample was separated on the same gel and subjected to Western blotting using anti-Hfq antibody (lanes 5–8).
SgrS Can Inhibit ptsG Translation Without Hfq.
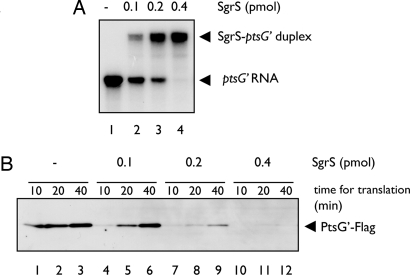
The observation that increasing amounts of SgrS cause a weak inhibition of ptsG′–flag translation in the absence of Hfq suggests that SgrS alone may be sufficient for translational inhibition of ptsG′–flag if it forms an RNA–RNA duplex with the ptsG′–flag RNA (see Fig. 2B). To test this view, we carried out a time course experiment for duplex formation by incubating a fixed amount of 32P-labeled ptsG′–flag RNA with a fixed amount of unlabeled SgrS RNA at 37°C. Approximately 50% of ptsG′–flag RNA formed a duplex with SgrS after 1 h incubation, and duplex formation was essentially completed after 2 h incubation (Fig. 6A). Thus, duplex formation between ptsG′–flag RNA and SgrS RNA proceeds quite slowly. The RNA mixtures from different incubation times were subjected to the in vitro translation assay. The inhibition of ptsG′–flag translation was shown to correlate with the incubation time, and therefore with the extent of duplex formation (Fig. 6 B and C). To accelerate duplex formation, we incubated the 32P-labeled ptsG′–flag RNA with increasing amounts of SgrS for 5 min at 70°C and then cooled the mixture down to 30°C. As expected, duplex formation was dramatically enhanced by heat treatment (Fig. 7A, lanes 1–4). The heat-treated ptsG′–SgrS mixture was subjected to in vitro translation. As shown in Fig. 7B, the translation of ptsG′–flag mRNA was reduced as the amount of free ptsG′–flag mRNA decreased. These data again imply that the ptsG′–flag mRNA base paired with SgrS is translationally inactive. We conclude that base pairing itself is sufficient and directly responsible for the translational silencing of ptsG mRNA.
Fig. 7.
In vitro translation of heat-treated ptsG′–SgrS RNA mixture. (A) Heat treatment accelerates the duplex formation between ptsG′ and SgrS. 32P-labeled ptsG′-flag RNA (0. 2 pmol) was incubated with SgrS RNA (0.4 pmol) in 3.5 μl of binding buffer at 70°C for 5 min and then cooled down to 30°C. Samples were analyzed by a gel mobility shift assay on a native polyacrylamide gel after the addition of 1 μl of RNA loading buffer. (B) In vitro translation of SgrS-ptsG′ mixture. 32P-labeled ptsG′–flag RNA (0. 2 pmol) was incubated with SgrS RNA (0.4 pmol) in 3.5 μl of binding buffer at 70°C for 5 min and then cooled down to 30°C. Then, each sample was translated in 10 μl of reaction mixture consisting of PURESYSTEM at 37°C. At the indicated times, 2.5 μl of reaction mixture was taken and subjected to Western blotting.
Discussion
The stress-induced sRNA SgrS forms a specific ribonucleoprotein complex with RNase E and Hfq, and acts through imperfect base pairing, resulting in translational inhibition and RNase E-dependent degradation of ptsG mRNA (8). However, the RNase E-dependent degradation of target mRNA is dispensable for silencing, and therefore translational inhibition is the primary event for gene silencing (9). In the present study, we addressed two questions regarding the mechanism of translational silencing: (i) are SgrS and Hfq sufficient for ptsG silencing? and (ii) which component, SgrS or Hfq, is ultimately responsible for translational silencing? The latter question is particularly intriguing because there are three possible ways by which SgrS and Hfq might, in principle, execute their function. First, SgrS (i.e., the base pairing itself) acts primarily for silencing, and the major role of Hfq is to help SgrS action by stimulating the base pairing. Second, Hfq recruited near the ribosome-binding site might be directly responsible for translational repression, and the major role of SgrS is to bring Hfq to the translation initiation site. Last, both base pairing and Hfq would act cooperatively to execute an efficient translational silencing.
In vitro reconstitution is a powerful tool in general to study the molecular mechanisms of various biological processes. We used PURESYSTEM to investigate mechanisms of translational silencing mediated by SgrS/Hfq in vitro. We first showed that PURESYSTEM allows an efficient translation of ptsG and control gfp RNAs in vitro (Fig. 1). Then, we showed that SgrS is able to efficiently inhibit ptsG translation only in the presence of Hfq, under a mild condition that is likely to mimic the in vivo situation (Figs. 2 and 3). A noncognate sRNA, RyhB, has no effect on ptsG translation (Fig. 3B). Mutant SgrS that is unable to act on the wild-type ptsG failed to repress the ptsG translation also in vitro (Fig. 3B). Thus, we could reproduce quite faithfully ptsG mRNA regulation by SgrS and Hfq in vitro. This established that both SgrS and Hfq are the minimum components and that no other factors beyond those already present in the PURESYSTEM are required for translational silencing of ptsG mRNA. The most important finding in the present study is that SgrS alone was sufficient to cause translational repression when it was forced to base pair with the ptsG RNA in the absence of Hfq by incubating the RNA mixture for a longer time (Fig. 6) or at a higher temperature (Fig. 7). Thus, Hfq is dispensable for the SgrS-mediated translational silencing if SgrS forms a duplex with ptsG mRNA. In other words, the RNA rather than protein components are directly responsible for translational repression of ptsG mRNA mediated by the SgrS–Hfq–RNase E complex. We also showed that the extent of translational repression correlates with the extent of duplex formation between ptsG mRNA and SgrS (Figs. 6 and 7). It is apparent that SgrS paired with a specific region around SD sequence prevents the binding of 16S rRNA of the ribosome, resulting in translational silencing. Our finding that the SgrS–ptsG base pairing alone is sufficient to cause translational repression is consistent with previous studies in which annealing of MicC or MicA to the cognate ompC or ompA mRNA in the absence of Hfq was shown to be able to prevent the formation of ternary complex consisting of 30S ribosome, initiator tRNA, and mRNA by toeprinting experiments (15, 16).
The present study allowed us to delineate the differential roles of each of three components of ribonucleoprotein complex in gene silencing directed by Hfq-binding sRNAs. First, the major role of sRNA is to inhibit translation of target mRNA through base pairing. Second, the major role of Hfq is to stimulate the base pairing between sRNA and its target mRNA, athough we do not know at this moment how Hfq facilitates the base pairing. In addition, Hfq also acts to stabilize sRNA and recruit RNase E to the target mRNA through sRNA. Finally, the major role of RNase E recruited to the target mRNA through a given sRNA is to destroy the base-paired mRNA and sRNA. It remains to be elucidated which regions of the two base-paired two RNAs are attacked by RNase E, which is considered to be a single-strand specific endonuclease. It should be emphasized that these three components are dependent on each other, acting as a ribonucleoprotein complex to exert their functions in intact cells. Although sRNAs have the ability to inhibit the translation of target mRNAs through base pairing, it is difficult for them to execute their function without Hfq under in vivo conditions because the rate of duplex formation between sRNAs and target mRNAs is too slow. Thus, the stimulation of base pairing by Hfq is essential for the sRNA-directed gene silencing in vivo. In addition, RNase E cannot be recruited to the target mRNA to destruct the translationally inactive mRNA without Hfq and sRNA. There is no reason to doubt that this view represents a general principle for the mechanism of Hfq-binding sRNAs in bacteria.
Concerning translational silencing of target mRNAs by bacterial sRNAs in vitro, there is a report in which the effect of RyhB on the translation of the target sodB mRNA using an S-30 system was described (17), in which exogenously added Hfq or RyhB was shown to inhibit to some extent the sodB translation. However, a trace amount of endogenous Hfq in the S-30 system would have made it difficult to test the effect of Hfq on RyhB action. We have succeeded here in faithfully reconstituting translational silencing of ptsG mRNA by SgrS and Hfq in vitro using PURESYSTEM. Recently, PURESYSTEM has been used to show translational regulation of target mRNAs by GcvB and GlmZ/Y RNAs in vitro (18, 19). It was also successfully used to reproduce elongation arrest of secM translation in vitro (20). Thus, PURESYSTEM has been proved to be a powerful tool to study molecular events related to translational regulation.
Eukaryotic sRNAs such as miRNAs and siRNAs also act to down-regulate target genes at posttranscriptional levels by decreasing translation and/or mRNA stability through base pairing with target mRNAs. This is accomplished by the formation of large ribonucleoprotein complexes, known as the RNA-induced silencing complex (RISC), before action on the target mRNAs (21–24). RISC consists of a variety of proteins such as the RNA binding protein, RNA helicase, and nuclease. Thus, bacterial sRNAs resemble eukaryotic counterparts in their functions and mechanisms of action, at least in part. However, there are some differences between the two systems regarding the mechanisms of translation initiation and the sRNA action. Most miRNAs bind to the 3′ UTR of target mRNAs, whereas in bacteria sRNA binding occurs mainly in the 5′ UTR of target mRNAs. In addition, in higher eukaryotes, it has been shown that the Ago protein also directs the repressed mRNA in the P-bodies for storage or degradation. This is apparently not the case in bacteria, in which translation is coupled to transcription. The present study has revealed an additional distinct difference between bacterial sRNAs and eukaryotic miRNAs regarding their roles. Although the mechanisms of translational inhibition mediated by RISC are controversial, a common view is that the role of miRNAs and siRNAs is to recruit or guide a diverse family of proteins specialized for silencing to the target mRNAs (21–24). In other words, protein components, rather than sRNAs, are responsible for both translational silencing and mRNA destruction. Thus, the role of base pairing of eukaryotic sRNAs is apparently different from that for bacterial sRNAs, although we do not exclude the possibility that the base pairing itself is also directly involved in translational silencing in eukaryotic cells.
Materials and Methods
DNA Fragments and Plasmids.
The 462 bp ptsG′–flag DNA fragment containing the ptsG region (+55 to +455) flanked by T7 promoter sequence and the Flag tag sequence was amplified from plasmid pTH111 (25) by PCR using primers 802 (TATTCATTAACCTTTATCGTCGTCATCT TTGTAGTCGCCAGTATCCGCCAGGTG) and 803 (GAAATTAATACGACTCACT ATAGGGACGCGTGAGAACGTAAAAAAAGC). The ptsG′–flag DNA encodes a truncated PtsG protein of 117 aa with Flag tag (PtsG′–Flag). The 844-bp gfp–flag DNA fragment containing the gfp coding region flanked by T7 promoter sequence plus an ideal SD sequence and the Flag tag sequence was amplified from a plasmid carrying the GFP coding region by first using PCR with primers 825 (AAGGAGATATACCAATGTGCG GCCGCAGTAAAGGAGA) and 826 (TATTCATTAACCTTTATCGTCGTCATCTT TGTAGTCTTTGTATAGTTCATCCA), and then universal primer and primer 826. The 121-bp ryhB DNA fragment containing the ryhB region (+1 to +85) flanked by T7 promoter sequence and the terminator sequence was amplified from plasmid pQE-RyhB by using PCR with primers 810 (GAAATTAATACGACTCACTATAGGGTCGCGATCA GGAAGA) and 811 (TATTCATTACCAGCACCCGGCTGGC). The DNA fragments containing sgrS and sgrS(G178C) were prepared as described (10). DNA fragments were purified through either polyacrylamide or agarose gel electrophoresis and used for in vitro transcription and/or translation in most cases. PCR-amplified DNA fragments were used directly as DNA templates for translation coupled with transcription in vitro.
In Vitro RNA Preparation.
The following mRNAs and sRNAs were prepared in vitro by using the DNA fragments mentioned above: ptsG′–flag RNA, gfp-flag RNA, SgrS RNA, SgrS(G178C) RNA, and RyhB RNA. In vitro transcription reactions were performed by using the CUGA7 in vitro transcription kit (Nippon Genetech). [α-32P]UTP (Amersham Biosciences) was added to the reaction mixture to generate 32P-labeled ptsG′–flag RNA. The RNA transcripts were purified on an 8% polyacrylamide gel and eluted overnight at 37°C in buffer containing 20 mM Tris·HCl (pH 7.5), 0.5 M NH4OAc, 10 mM Mg(OAc)2, 1 mM EDTA, and 0.1% SDS, followed by phenol treatment and ethanol precipitation.
In Vitro Translation Assay and Western Blotting.
Translation reaction was carried out using PURESYSTEM classic II (PURE2048C; Post Genome Institute). Unless otherwise specified, template mRNA (DNA), sRNA, and Hfq were mixed in 3.5 μl binding buffer (20 mM Tris·HCl, pH 8.0; 1 mM DTT; 1 mM MgCl2; 20 mM KCl; and 10 mM Na2HPO4–NaH2PO4, pH 8.0) and preincubated for 5 min at 30°C. Then PURESYSTEM mix was added in a final volume 10 μl, and the reaction mixture was incubated at 37°C for indicated times. The reaction was terminated by adding an equal volume of SDS/PAGE loading buffer (62.5 mM Tris·HCl, pH 6.8; 2% SDS; 10% glycerol; 5% 2-mercaptoethanol; 0.1% bromophenol blue). The samples were heated at 94°C for 5 min, subjected to a polyacrylamide–0.1% SDS gel electrophoresis, and transferred to Immobilon membrane (Millipore). The 15% polyacrylamide gel was used to detect Flag-tagged translation products. The membranes were treated with anti-Flag monoclonal antibody (Sigma). Signals are visualized by the Lumi-Light Western Blotting Substrate (Roche).
Northern Blotting.
RNA samples were resolved by electrophoresis on a 6% polyacrylamide/8 M urea in 0.5 × TBE buffer (45 mM Tris-borate, pH 8.3, and 1 mM EDTA) and blotted on to Hybond-N+ membrane (Amersham Biosciences). RNAs were visualized by using digoxigenin (DIG) reagents and kits for nonradioactive nucleic acid labeling and detection system (Roche Diagnostics). The following DIG-labeled DNA probes were prepared by PCR using DIG-dUTP: a 305-bp fragment corresponding to the 5(prime) region of ptsG and a 227-bp fragment corresponding to sgrS.
Purification of His-Tagged Hfq.
TM589 (8) harboring pQE80L–Hfq-His (10) was cultured in 200 ml of LB medium at 37°C. At OD600 = 0.2, 1 mM IPTG was added to the culture and incubation was continued for 80 min. Cells were harvested and washed with 20 ml STE (100 mM NaCl; 10 mM Tris·HCl, pH8.0; 1 mM EDTA), and suspended in 0.6 ml of 50 mM Na2HPO4–NaH2PO4; 300 mM NaCl; and 10 mM imidazole, pH 8.0. The cell suspension was treated with lysozyme (2.5 mg/ml) for 10 min at 0°C, sonicated, and centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was treated with RNaseA (2.5 mg/ml) for 10 min at 0°C and then heated at 80°C. The sample was centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was incubated with 80 μl of Ni2+–NTA agarose resin (Qiagen) for 20 min at 4°C, and Hfq–His6 protein was purified according to the manufacturer's instruction. Hfq–His6 concentration was determined by Coomassie Brilliant Blue staining. Purified Hfq–His6 was stored with storage buffer (20 mM Tris·HCl, pH 8.0; 0.1 M KCl; 5 mM MgCl2; 50% glycerol; 0.1% Tween 2; and 1 mM DTT) at −20°C.
Gel Mobility Shift Assay.
Gel mobility shift assay was performed with 0.2 pmol of 32P-labeled ptsG′–flag RNA in binding buffer (20 mM Tris·HCl, pH 8.0; 1 mM DTT; 1 mM MgCl2, 20 mM KCl; 10 mM Na2HPO4–NaH2PO4, pH 8.0). The labeled RNA fragments were incubated in the presence and absence of indicated amounts of SgrS RNA and/or purified Hfq–His6 in a 3.5-μl reaction mixture. The samples were incubated at 37°C for the indicated times; at 70°C for 5 min, followed by gradually cooling down to 30°C to promote base pairing; and then at 30°C for 5 min (Figs. 6 and 7). One microliter of loading buffer (50% glycerol, 0.1% bromophenol blue) was added, and loaded on a 4% or 5% polyacrylamide gel in 0.5× TBE containing 0.5% glycerol. The electrophoresis was carried out at 4°C. After electrophoresis, gel was dried and subjected to autoradiography. Some samples used for gel mobility shift assay (Fig. 5) were also subjected to Western blotting, as follows. First, the gel was incubated for 1 h in soaking buffer (1% SDS, 375 mM Tris·HCl, pH7.5) at room temperature and then transferred to Immobilon membrane (Millipore). The membranes were treated with anti-Hfq polyclonal antibody. Signals are visualized by the Lumi-Light Western Blotting Substrate (Roche).
Acknowledgments.
We thank Susan Gottesman for critical reading of the manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by Ajinomoto Co., Inc.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 3.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Gottesman S. In: The RNA World. 3rd Ed. Gesteland RF, Cech TR, Atkins JF, editors. New York: Cold Spring Harbor Lab Press; 2006. pp. 567–594. [Google Scholar]
- 5.Gottesman S. Micros for microbes: Non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr Opin Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 8.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto H, et al. Implication of membrane localization of target mRNA in the action of a small RNA: Mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 14.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udekwu KI, et al. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vecerek B, et al. Interaction of the RNA chaperone Hfq with mRNAs: Direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 18.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 21.Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, Belasco JG. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi H, Inada T, Postma P, Aiba H. CRP down-regulates adenylate cyclase activity by reducing the level of phosphorylated IIA(Glc), the glucose-specific phosphotransferase protein, in Escherichia coli. Mol Gen Genet. 1998;259:317–326. doi: 10.1007/s004380050818. [DOI] [PubMed] [Google Scholar]