Abstract
Visual pigments are G-protein-coupled receptors that provide a critical interface between organisms and their external environment. Natural selection has generated vertebrate pigments that absorb light from the far-UV (360 nm) to the deep red (630 nm) while using a single chromophore, in either the A1 (11-cis-retinal) or A2 (11-cis-3,4-dehydroretinal) form. The fact that a single chromophore can be manipulated to have an absorption maximum across such an extended spectral region is remarkable. The mechanisms of wavelength regulation remain to be fully revealed, and one of the least well-understood mechanisms is that associated with the deep red pigments. We investigate theoretically the hypothesis that deep red cone pigments select a 6-s-trans conformation of the retinal chromophore ring geometry. This conformation is in contrast to the 6-s-cis ring geometry observed in rhodopsin and, through model chromophore studies, the vast majority of visual pigments. Nomographic spectral analysis of 294 A1 and A2 cone pigment literature absorption maxima indicates that the selection of a 6-s-trans geometry red shifts M/LWS A1 pigments by ~1500 cm−1 (~50 nm) and A2 pigments by ~2700 cm−1 (~100 nm). The homology models of seven cone pigments indicate that the deep red cone pigments select 6-s-trans chromophore conformations primarily via electrostatic steering. Our results reveal that the generation of a 6-s-trans conformation not only achieves a significant red shift but also provides enhanced stability of the chromophore within the deep red cone pigment binding sites.
Visual pigments are responsible for mediating vision and color discrimination. These pigments are members of the G-protein-coupled receptor (GPCR)1 family, having seven transmembrane α helices as the core structure. The visual pigments in the retina are classified according to sequence similarity into mid- and long-wavelength sensitive, M/LWS (>510 nm); short-wavelength sensitive set 1, SWS1 (350–450 nm); short-wavelength sensitive set 2, SWS2 (440–461 nm); rhodopsins, RH1 (470–500 nm), and rhodopsin-like cone pigments, RH2 (460–510 nm) (1, 2) (Figure 1). These pigments contain a protein called opsin and a retinal chromophore, which is a derivative of vitamin A. The chromophore in vertebrate visual pigments is either 11-cis-retinal (an A1 retinal) or 11-cis-3,4-dehydroretinal (an A2 retinal) chromophore (Figure 2). The A2 retinal extends the π system by one double bond inside the ring and absorbs at longer wavelengths than the A1 retinal (3, 4).
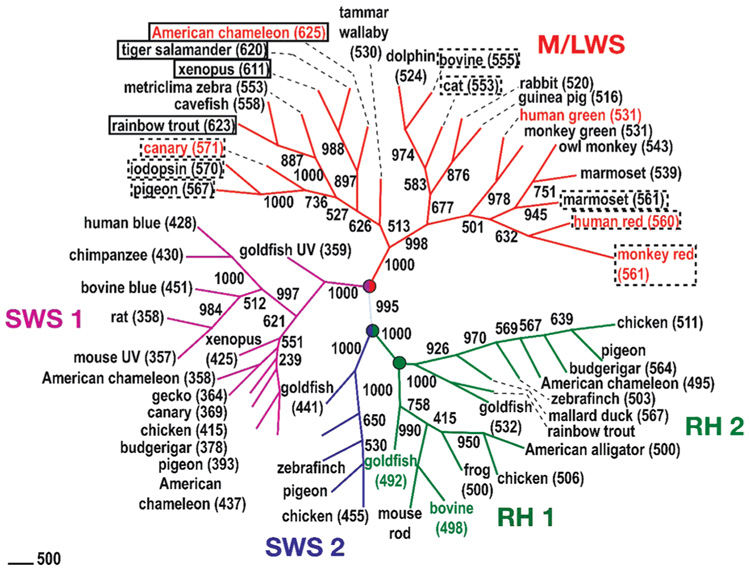
FIGURE 1.
This study’s premise is that many of the deep red cones are bathochromically shifted to higher wavelength by adopting a 6-s-trans conformation of the 11-cis-retinal chromophore. The figure places solid rectangles around those pigments for which A2 6-s-trans conformations are predicted, with dashed lines indicating more tentative assignments for the A1 6-s-trans pigments. The phylogenetic tree shown here is constructed from aligned sequences by neighbor-joining methods through ClustalW. The distances of the sequences are estimated on the basis of the Dayhoff PAM matrix by Prodist of the PHYLIP program (version 3.65). The numbers along the branches indicate the clustering percentage obtained from 1000 bootstrap resamplings. The values enclosed in parentheses are the observed absorption maxima of the pigments, when available. Classification nodes are marked with circles, and the seven pigments modeled in this study are labeled in color.
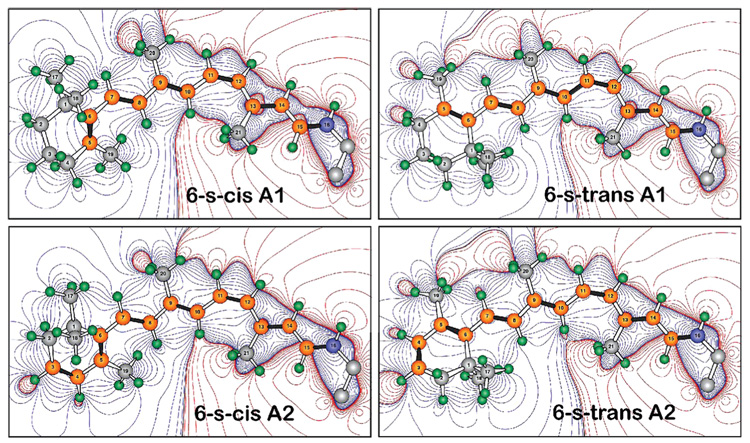
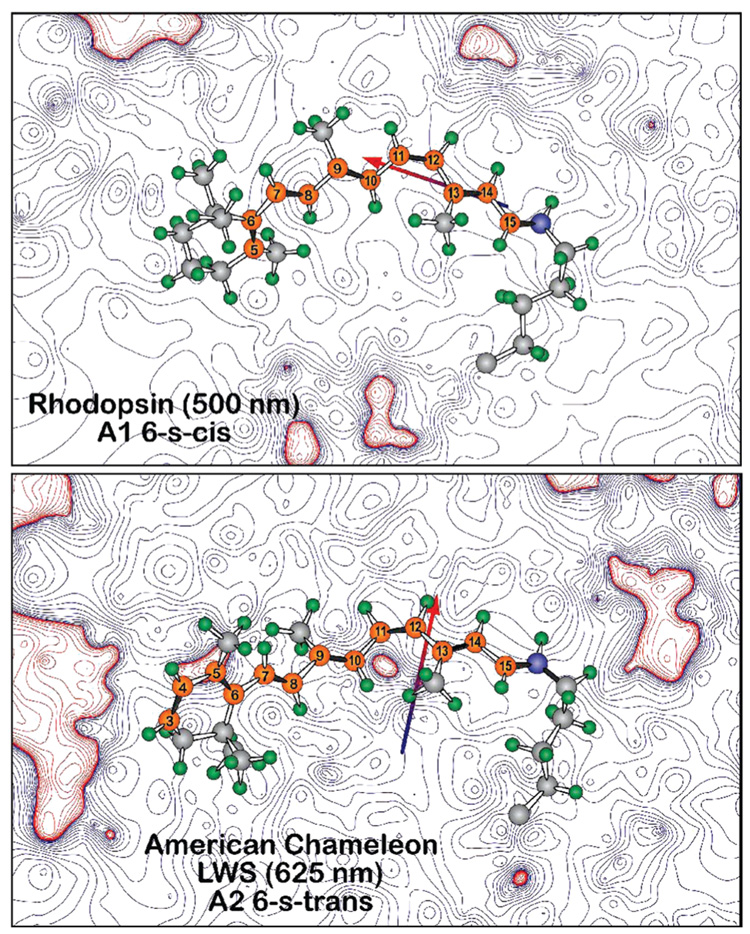
FIGURE 2.
Vertebrate visual pigments select either A1 retinal or A2 (3,4-dehydroretinal) 11-cis-retinal as the bound chromophore. A2 retinal has an additional double bond in the β-ionone ring, which red shifts the absorption spectrum. The contours describe the differential electrostatic field based on the point-charge Mulliken population [B3LYP/6-31G(d)]. The red contours indicate areas of positive charge, and the blue contours indicate areas of negative charge relative to the chromophore as a whole, which carries a net positive charge. Note that the 6-s-trans conformers move positive charges toward the ring relative to the 6-s-cis conformers.
Spectral tuning of visual pigments allows vertebrates to adjust visual acuity to suit their environment. Because red—green light is preferentially transmitted near the surface of the water (5), most freshwater fish and amphibians select an A2 retinal chromophore to extend visual sensitivity to longer wavelengths (2, 6). A variety of tuning mechanisms relevant to both A1 and A2 chromophores have been proposed. Electrostatic chromophore—protein interactions represent the most extensively studied mechanisms because the protein-bound chromophore is positively charged (7–13). The exceptions are the UV-cone pigments, which have a deprotonated Schiff base chromophore (14–16). The absorption spectrum of the chromophore is also sensitive to the conformation of the chromophore, and it has long been known that the selection of a 6-s-trans chromophore conformation will red shift the chromophore (11). The origin of this red shift is the enhanced planarity of the π system of the 6-s-trans conformation, which decreases the energy of the lowest unoccupied molecular orbital (11). Indeed, a 6-s-trans ring conformation was first proposed by Chen and co-workers as a spectral tuning mechanism in iodopsin (λmax = 571 nm) (17). More recently, Makino and co-workers tested this hypothesis by incorporating locked 6-s-cis,9-cis retinal analogues into bleached salamander, squirrel, monkey red, and monkey green cone pigments (18). Their study, however, concluded that the differences in λmax between the red- and green-absorbing pigments do not depend upon the conformation of the C6–C7 bond of the retinal. We revisit this conformational mechanism in the present paper by examining different chromophore binding site models and homology models of M/LWS pigments. We demonstrate that deep red cones preferentially select a 6-s-trans conformer and that this selection is directly responsible for as much as a 100 nm red shift in the A2 red cone pigments. The evidence for A1 6-s-trans pigments is less compelling but warrants additional study.
Although the M/LWS pigments contain a protonated Schiff base and high binding site similarity across species, the reported absorption maxima of the vertebrate M/LWS opsins populate distinct regions of the spectrum (Figure 3). The A2 pigments provide the most dramatic example, with the vast majority near 532 or 620 nm, with only a small number in between. The A1 pigments populate three differentiated regions, with maxima at 520, 533, and 564 nm, and are a more complicated group to analyze. Our initial structure–function studies of these groupings sought differences in the amino acid residues in or near the binding site. We found no systematic changes in binding site residues that could account realistically for the dramatic differentiation depicted in Figure 3. Other studies have confirmed the lack of obvious residue-based mechanisms (18). We propose that the regional differences are due to chromophore conformer selection, a more subtle wavelength mechanism that can be triggered by modest changes in the binding site, as we explore below.
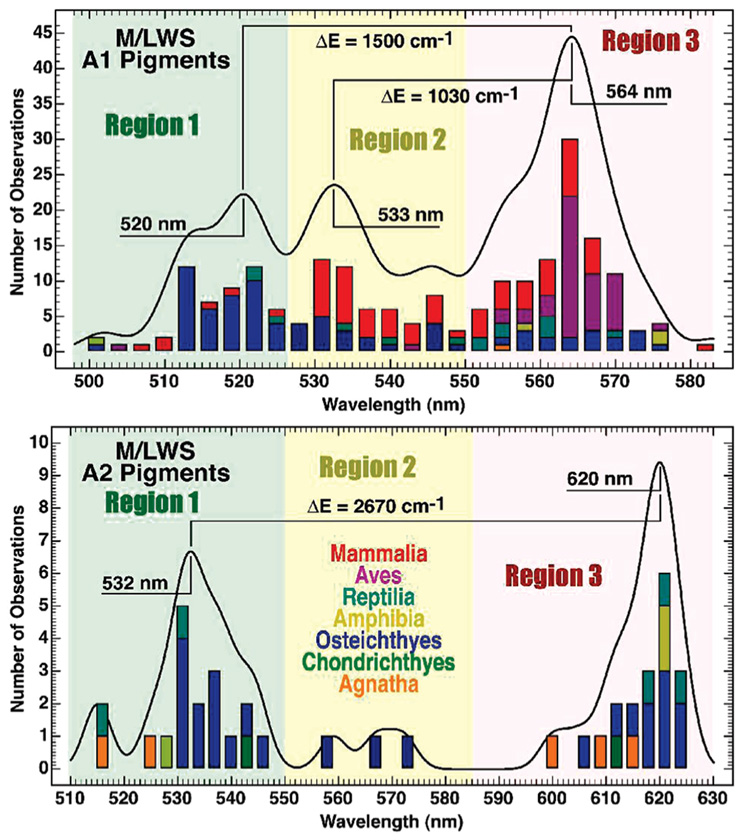
FIGURE 3.
Histogram analysis of 294 M/LWS pigments reveals the distribution of absorption maxima into two or three regions of the visible spectrum for A1 (top) and A2 (bottom) pigments. We refer to those red cones with absorption maxima in region 3 as “deep red” cones and propose that these pigments have 6-s-trans chromophores. The literature data and associated references are listed in Tables S1 and S2 of the Supporting Information.
MATERIALS AND METHODS
Chromophore Binding Site Models
The isolated chromophore models that we used to examine the effect of 6-s-cis versus 6-s-trans conformation on the electronic properties of the isolated chromophores are shown in Tables S3 and S4 of the Supporting Information. The ground-state geometries of the models were minimized by DFT (B3LYP/6-31G(d)) methods in Gaussian 03 (19). The excited-state (spectroscopic) properties were calculated using the MNDO-PSDCI theory including full single- and double-configuration interaction within the chromophore π system (15, 20–22).
Homology Modeling of M/LWS Pigments of Different Vertebrates
We constructed homology models of the following seven proteins referenced via accession number: P14592 (American chameleon red, 625 nm), CAB91996 (canary red, 571 nm), AAC12941 (dolphin green, 524 nm), AAA49169 (goldfish green, 532 nm), NP_000504 (human green, 531 nm), NP_06445 (human red, 560 nm), and AAD40324 (monkey red, 560 nm). The sequences of M/LWS pigments of different vertebrates were aligned against the sequence of rhodopsin [Protein Data Bank (PDB) code 1U19] (23) using the BLOSUM62 scoring matrix in MathScriptor (www.mathscriptor.org). Hydrogen atoms were added, and an initial minimization was executed in CAChe (Fujitsu, Inc., Beaverton, OR) by MM2 methods (24, 25). All hydrogen atoms were minimized fully prior to optimization of the whole protein, and the backbone was locked during these minimizations. To create models with a 6-s-trans retinal chromophore, the dihedral angle of the ring was altered to change the ring conformation of the retinal.
The models were further optimized by adopted basis Newton–Raphson (ABNR) methods, with harmonic constraints on the backbone and dihedral constraints on the chromophore polyene chain using CHARMM (26, 27). The parameters for the chromophore were adapted from the optimized retinal structure by Hermone and Kuczera (28). The system was gradually heated from 0 to 300 K over an interval of 20 ps, followed by 50 ps of simulation at 300 K to equilibrate the system. All molecular dynamics (MD) simulations were carried out for 2 ns, with a time step of 0.001 ps, assuming a dielectric constant of 1.0. A nonbonded cutoff of 15 Å with a nonbonding smoothing function applied between 11 and 14 Å was used for all calculations. All MD simulations were carried out using the Charmm22 parameter proteins and TIP3P waters (26, 27).
Monkey red models were also generated on the basis of the human red pigments. We included in this study both 9-cis and 11-cis chromophores to better understand the relationship between this theoretical investigation and the previous experimental study by Makino et al. (18).
RESULTS
Chromophore Binding Site Models
The ground- and excited-state properties of the 10 sets of binding site models were calculated, and their properties are summarized in Tables S3 and S4 in the Supporting Information. The molecular diagram that is shown to the left of the data (Tables S3 and S4 in the Supporting Information) shows the geometry of the chromophore with the lowest energy conformation. A few of our models contain a protonated Schiff base without a negative counterion, while the remainder contains a negatively charge species near the Schiff base present either as a carboxylate ion group or a perchlorate ion. We have also constructed a binding site model with a total charge of −1 by introducing a carboxylate ion group and a model with a total charge of +1 by adding an iminium ion near the polyene chain of the retinal. In general, protonated Schiff bases without negative counterions select the 6-s-trans. Most others select 6-s-cis.
Homology Models
To examine the sequence similarity among M/LWS pigments, a number of vertebrate pigments were aligned with respect to the rhodopsin crystal structure (PDB code 1U19). As observed in previous studies, we identified no charged residues within 5 Å of the chromophore other than the primary counterion (29, 30). We also examined the residues at positions 164, 261, and 269 (rhodopsin numbering). A hydroxyl-containing residue at these positions has been shown to red shift the absorption maxima (30, 31). However, we observe that many of the region 1 pigments also contain Tyr, Thr, and Ser at these positions, indicating that these particular residues are not the primary source of the red shift. A summary of our findings for selected region 1 and 3 pigments is shown in Tables S5 in the Supporting Information. We conclude that serine at position 164 may play a small role in enhancing the region 1–3 shift, but this residue is not a primary source of the significant red shift. The mechanism that we propose here is the selection of a 6-s-trans chromophore conformation in the deep red (region 3) pigments. Because this selection would be a discontinuous conformational change, a large shift in the nomographic spectral distribution would be anticipated. That feature is observed for the A2 chromophores (Figure 3).
DISCUSSION
The first question that we seek to answer is whether the spectral shifts observed in the histograms of Figure 3 are consistent with 6-s-cis to 6-s-trans conformational changes of the chromophore. To explore this question, we prepared a set of 10 11-cis-retinal Schiff-base chromophores with various covalent substituents that mimic the extremes of electrostatic environments observed within the visual pigments. All but 1 of the 10 models were protonated, and a few involved isolated protonated Schiff bases without counterions. Carboxylate ion groups and perchlorate ions were used to represent negatively charged ions, while iminium ions were introduced to represent positively charged ions near the chromophore. The models are shown in Tables S3 and S4 in the Supporting Information along with the calculated results. The ground-state geometries were minimized using density functional methods [B3LYP/6-31G(d), Gaussian 03, Gaussian Corp., Pittsburgh, PA], and the photo-physical properties were calculated using MNDO–PSDCI theory (see the Materials and Methods). The MNDO–PSDCI method has been found to be very reliable for calculating the absorption properties of retinal chromophores, both isolated and protein-bound (15, 22). The transition energies into the low-lying strongly allowed excited state are summarized in the histogram of Figure 4. The wavelength shift for our A1 models is comparable to the shift for isolated chromophores observed by Honig and co-workers (11). The model chromophore studies predict a 6-s-cis to 6-s-trans average bathochromic shift of ~1680 cm−1 for A1 chromophores and ~1970 cm−1 for A2 chromophores (Figure 4), which are to be compared to the observed region 1–3 bathochromic shifts of ~1500 and ~2670 cm−1, respectively (Figure 3). The good agreement for the A1 chromophores is encouraging and provides support for our hypothesis. The calculated shift for the A2 chromophores is smaller than the observed shift by roughly 30%. We provide one possible explanation for the larger-than-calculated observed shift based on binding site electrostatics below. Nevertheless, the qualitative agreement is encouraging and provides support for our proposal that the bathochromic shift between regions 1 and 3 is associated with a 6-s-cis to 6-s-trans conformational change.
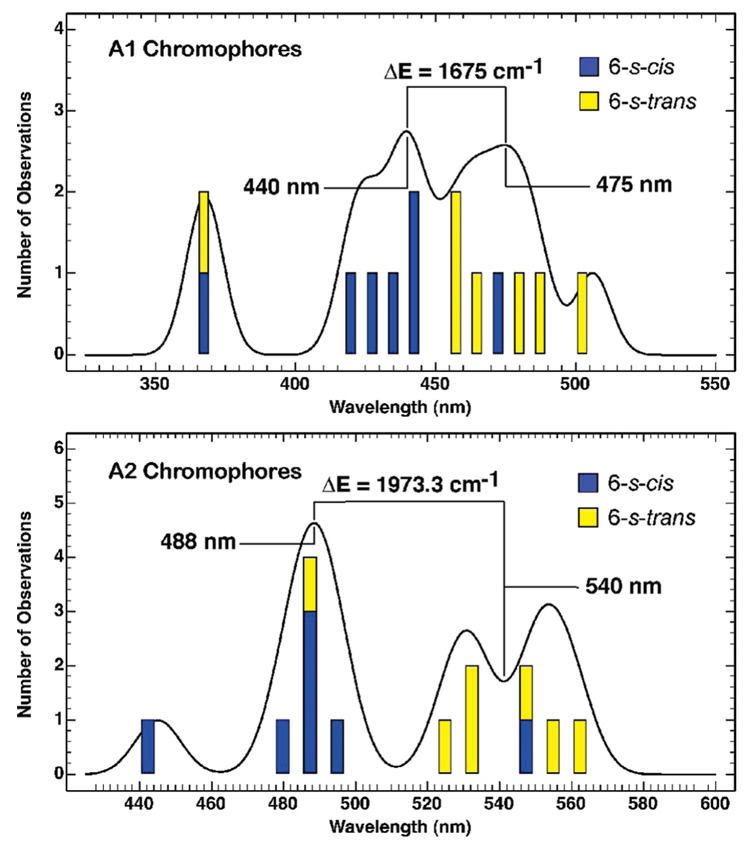
FIGURE 4.
Histogram of the transition energies of the 10 model chromophores calculated using the MNDO-PSDCI theory including full single- and double-configuration interaction within the π system. The structures and the data are listed in Tables S3 and S4 in the Supporting Information. The calculations confirm that the 6-s-trans conformers are on average red-shifted relative to the 6-s-cis conformers. A histogram analysis reveals that the difference in the transition wavelength between the 6-s-trans and the 6-s-cis conformers is similar to that observed between regions 1 and 3 of the experimental A1 and A2 histograms (Figure 3).
The next step is to explore the energetics of the conformer selection process. The availability of a crystal structure for rhodopsin (23, 32) coupled with the significant protein homology within the visual pigment family (33) makes homology modeling a viable approach. Our homology modeling methods and procedures are identical to those used previously to study the UV cone pigments (15) and include 2 ns molecular dynamics using the Charmm22 force field. The calculations indicate that the pigments that fall under region 3 of our histogram (Figure 3), such as American chameleon red, human red, and canary red, select a 6-s-trans conformation as shown by lower s-trans energies (Table 1). This finding supports the hypothesis of Chen and co-workers on iodopsin, a pigment that has an 80% sequence identity with the human red cone opsin. Iodopsin, an A1 region 3 pigment, was proposed by Chen and co-workers to contain a 6-s-trans ring conformation as part of its spectral tuning mechanism (17). The pigments that fall under region 1 of our histogram (dolphin green and goldfish green) gave lower s-cis energies, indicating that region 1 pigments select a 6-s-cis ring conformation. However, the energetic difference for the A1 deep red cone is within the error margins of these calculations. The studies on monkey red demonstrate the importance of carrying out long-term (2 ns) molecular dynamics prior to minimization. After 1 ns, the binding site retains a preference for a 6-s-cis chromophore, and only after 2 ns does the 6-s-trans chromophore achieve comparative preference. However, a key observation is that these calculations do not predict energy differences larger than the intrinsic error in such procedures. Hence, the theoretical results are illustrative but not authoritative.
Table 1.
Comparison of the Calculated Energies of Rhodopsin and Six Cone Pigments Containing 6-s-cis versus 6-s-trans Chromophoresa
| energy (Charmm) (kcal/mol) | ||
|---|---|---|
| models | 6-s-cis | 6-s-trans |
| rhodopsin (498 nm) | −7213.735 | −7053.548 |
| A1-containing opsins | ||
| dolphin green (524 nm) | −6217.510 | −6088.184 |
| human green (531 nm) | −7072.558 | −6734.553 |
| human red (560 nm) | −6919.252 | −7131.425 |
| canary red (571 nm) | −5127.131 | −5159.844 |
| A2-containing opsins | ||
| goldfish green (532 nm) | −6262.406 | −6099.316 |
| American chameleon red (625 nm) | −6263.410 | −6345.979 |
Energies following 2 ns molecular dynamics of rhodopsin (1U19)-based homology models (see the text).
We also created monkey red models with a 9-cis chromophore. Our calculations predict that the monkey red binding site with a 9-cis chromophore preferentially selects a 6-s-cis conformation (Table 2). This result provides insight into the Makino study (18), which indicated that the 9-cis,6-s-cis conformer is selected. However, we can find no discrete protein–chromophore interactions that are responsible for selecting 6-s-cis in the 9-cis chromophore but 6-s-trans in the 11-cis chromophore. Rather, we believe this selection process is primarily electrostatic and global to the entire binding site (see the discussion below). Both 9-cis and 11-cis chromophores are very stable in the binding site, and in the case of monkey red, the 9-cis chromophore is calculated to be more stable. This surprising result deserves further study. However, for the purposes of the present study, we conclude that our results and conclusions are consistent with the experimental results reported in Makino et al. (18).
Table 2.
Comparison of the Calculated Energies of Monkey Red Homology Models Containing 6-s-cis and 6-s-trans A1 Retinal Chromophores
| energy (Charmm) (kcal/mol) | ||
|---|---|---|
| models | 6-s-cis | 6-s-trans |
| monkey red (560 nm)a | −7229.297 | −7086.673 |
| monkey red (560 nm)b | −6908.607 | −7127.620 |
| monkey red (9-cis)b | −10811.176 | −10508.343 |
The model is based on the human red model that has been run with a 1 ns molecular dynamics. The structure was optimized by ABNR methods, and the energy was calculated (see the text).
The same as in a but after 2 ns molecular dynamics.
Although our calculations on both the A1 and A2 chromophores provide support for the selection of a 6-s-trans geometry in deep red cone pigments, the evidence is stronger for the A2 pigments for two reasons. The first reason is based on the observation that there are very few pigments found between regions 1 and 3 in the A2 histogram (Figure 3). This observation suggests that a single discrete mechanism is responsible for the spectral shift, and the lack of a residue-based source for this shift suggests that the chromophore may be involved. The 6-s-cis and 6-s-transconformations are separated by a relatively large barrier (> 10 kcal/mol in the protein), and hence, these two conformations will not be rapidly interconverting. The protein will then relax to accommodate the conformation to yield a stable absorption band characteristic of the lower energy conformation. This recipe will produce the type of histogram that we observe for the A2 pigments (Figure 3). The second reason that the A2 pigment assignment is more compelling derives from an analysis of the molecular dynamics calculations. During dynamics, the protein equilibrates to provide enhanced stabilization of whatever chromophore is inserted and the 6-s-trans conformer requires about 400 ps to stabilize in the deep red A2 pigments but often more than 1.6 ns for the deep red A1 pigments. We conclude that ambient temperature differentiation is more efficient in the A2 pigments, and thus, our modeling is more reliable for these pigments. Conversely, the A1 studies are less certain in terms of both the histograms (high occupation of region 2) and in the requirement that conformational equilibration requires long-term dynamics.
We cannot identify any discrete protein—chromophore interactions within the binding sites of these proteins that are responsible for preferential selection of the 6-s-trans conformer. Rather, we conclude that the 6-s-trans conformer is selected via the combination of two electrostatic effects, which are summarized in Figure 5. This figure compares the electrostatic fields in the plane of the chromophores in rhodopsin (which selects 6-s-cis) and chameleon red (which selects 6-s-trans). The atomic charges associated with the positively charged chromophore have been neglected, and hence, the binding site displays a net negative charge. In the case of rhodopsin, a majority of the negative charge is concentrated near the protonated Schiff base. In contrast, the negative charge in chameleon red is more evenly distributed, which means that there is a net shift of negative charge toward the ring. An examination of Figure 2 provides a good perspective on why this result will enhance stabilization of the 6-s-trans conformation, which has a charge distribution that shifts more chromophore positive charge toward the ring. Dipole–dipole interactions are also important. In American chameleon red, the net dipole moment of the binding site is oriented orthogonal to the chromophore, whereas in rhodopsin, the dipole moment of the binding site is along the axis of the chromophore. The former situation preferentially stabilizes the 6-s-trans conformer, and the latter preferentially stabilizes the 6-s-cis conformer. We refer to the above effects as electrostatic steering and note that an increasing negative charge near the ring will also enhance the red shift associated with the low-lying strongly allowed 1Bu+-like state. This follows from the fact that, upon excitation, negative charge is shifted toward the Schiff base. This charge shift will enhance the stability of the excited state relative to the ground state when the negative charge is shifted toward the ring. This observation may explain why the region 1–3 separation is about 30% larger for the A2 pigments than that calculated on the basis of the isolated models.
FIGURE 5.
Electrostatic contours in the plane of the chromophores of bovine rhodopsin (1U19) and a 6-s-trans homology model of American chameleon based on 1U19. The contours are based on Mulliken charges from a single SCF PM3 Mozyme calculation on a Charmm-based structure following 2 ns molecular dynamics. The contours are associated with the protein residues, ignoring the chromophore charges. Note that, in both cases, the chromophore is bathed in an electrostatic field that is predominantly negative (blue contours). However, the dipole moment of the binding site is nearly orthogonal between the two proteins, with the rhodopsin binding site favoring a 6-s-cis conformer (top) and the American chameleon binding site (bottom) favoring a 6-s-trans conformer.
What remains to be explained is what conformation is selected in those A1 pigments that occupy region 2 of Figure 3. We do not have an answer to this question. Homology calculations on two pigments in this region were ambiguous, and it is possible that a mixture of 6-s-cis and 6-s-trans conformers are present in some of these pigments.
Supplementary Material
SUPPORTING INFORMATION AVAILABLE Over 200 absorption maxima of various vertebrate red cone pigments used to generate the histograms shown in Figure 3, as well as a summary of the quantum mechanical calculations. This information is available free of charge via the Internet at http://pubs.acs.org.
Footnotes
This work was supported in part by the National Institutes of Health (NIH, GM-34548) and the National Science Foundation (NSF, BES-0412387). Computational facilities were supported by the Harold S. Schwenk Sr. Distinguished Chair in Chemistry at the University of Connecticut.
Abbreviations: GPCR, G-protein-coupled receptor; M/LWS, mid-and long-wavelength sensitive; DFT, density functional theory; MNDO–PSDCI, molecular neglect of differential overlap with partial single- and double-configuration interaction; BLOSUM62, block substitution matrix 62; BLAST, basic local alignment and search tool; CAChe, computer-aided chemistry; MM2, molecular mechanics 2; ABNR, adopted basis Newton–Raphson; CHARMm, chemistry at Harvard molecular mechanics; MD, molecular dynamics.
REFERENCES
- 1.Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog. Retinal Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dartnall HJ, Lythgoe JN. The spectral clustering of visual pigments. Vision Res. 1965;5:81–100. doi: 10.1016/0042-6989(65)90057-x. [DOI] [PubMed] [Google Scholar]
- 4.Harosi FI. An analysis of two spectral properties of vertebrate visual pigments. Vision Res. 1994;34:1359–1367. doi: 10.1016/0042-6989(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 5.Reckel F, Melzer RR, Parry JWL, Bowmaker JK. The retina of five atherinomorph teleosts: Photoreceptors, patterns and spectral sensitivities. Brain, Behav., Evol. 2002;60:249–264. doi: 10.1159/000067191. [DOI] [PubMed] [Google Scholar]
- 6.Provencio I, Loew ER, Foster RG. Vitamin A2-based visual pigments in fully terrestrial vertebrates. Vision Res. 1992;32:2201–2208. doi: 10.1016/0042-6989(92)90084-v. [DOI] [PubMed] [Google Scholar]
- 7.Nakanishi K, Balogh-Nair V, Arnaboldi M, Tsujimoto K, Honig R. An external point-charge model for bacteriorhodopsin to account for its purple color. J. Am. Chem. Soc. 1980;102:7945–7947. [Google Scholar]
- 8.Kropf A, Hubbard R. The mechanism of bleaching rhodopsin. Ann. N.Y. Acad. Sci. 1958;74:266–280. doi: 10.1111/j.1749-6632.1958.tb39550.x. [DOI] [PubMed] [Google Scholar]
- 9.Oseroff AR, Callender RH. Resonance Raman spectroscopy of rhodopsin in retinal disk membranes. Biochemistry. 1974;13:2443–2448. doi: 10.1021/bi00717a027. [DOI] [PubMed] [Google Scholar]
- 10.Mathies R, Freedman TB, Stryer L. Resonance Raman studies of the conformation of retinal in rhodopsin and isorhodopsin. J. Mol. Biol. 1977;109:367–372. doi: 10.1016/s0022-2836(77)80040-5. [DOI] [PubMed] [Google Scholar]
- 11.Honig B, Greenberg AD, Dinur U, Ebrey TG. Visual-pigment spectra: Implications of the protonation of the retinal Schiff base. Biochemistry. 1976;15:4593–4599. doi: 10.1021/bi00666a008. [DOI] [PubMed] [Google Scholar]
- 12.Yoshizawa T. The road to color vision: Structure, evolution and function of chicken and gecko visual pigments. Photochem. Photobiol. 1992;56:859–867. doi: 10.1111/j.1751-1097.1992.tb09707.x. [DOI] [PubMed] [Google Scholar]
- 13.Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. Mechanisms of spectral tuning in blue cone visual pigments: Visible and Raman spectroscopy of blue-shifted rhodopsin mutants. J. Biol. Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 14.Vought BW, Dukkipatti A, Max M, Knox BE, Birge RR. Photochemistry of the primary event in short-wavelength visual opsins at low temperature. Biochemistry. 1999;38:11287–11297. doi: 10.1021/bi990968b. [DOI] [PubMed] [Google Scholar]
- 15.Kusnetzow AK, Dukkipati A, Babu KR, Ramos L, Knox BE, Birge RR. Vertebrate ultraviolet visual pigments: Protonation of the retinylidene Schiff base and a counterion switch during photoactivation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:941–946. doi: 10.1073/pnas.0305206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasick JI, Applebury ML, Oprian DD. Spectral tuning in the mammalian short-wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- 17.Chen JG, Nakamura T, Ebrey TG, Ok H, Konno K, Derguini F, Nakanishi K, Honig B. Wavelength regulation in iodopsin, a cone pigment. Biophys. J. 1989;55:725–729. doi: 10.1016/S0006-3495(89)82871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makino CL, Kraft TW, Mathies RA, Lugtenburg J, Miley ME, van der Steen R, Baylor DA. Effects of modified chromophores on the specral sensitivity of salamander, squirrel and macaque cones. J. Physiol. 1990;424:545–560. doi: 10.1113/jphysiol.1990.sp018082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb M, Cheeseman JR, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski J, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith TA, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Wallingford, CT: Gaussian, Inc.; 2003. [Google Scholar]
- 20.Shima S, Ilagan RP, Gillespie N, Sommer BJ, Hiller RG, Sharples FP, Frank HA, Birge RR. Two-photon and fluorescence spectroscopy and the effect of environment on the photochemical properties of peridinin in solution and in the peridinin–chlorophyll–protein from Amphidinium carterae. J. Phys. Chem. 2003;A 107:8052–8066. [Google Scholar]
- 21.Martin CH, Birge RR. Reparameterizing MNDO for excited state calculations using ab initio effective Hamiltonian theory: Application to the 2,4-pentadien-1-iminium cation. J. Phys. Chem. 1998;A 102:852–860. [Google Scholar]
- 22.Ren L, Martin CH, Wise KJ, Gillespie NB, Luecke H, Lanyi JK, Spudich JL, Birge RR. Molecular mechanism of spectral tuning in sensory rhodopsin II. Biochemistry. 2001;40:13906–13914. doi: 10.1021/bi0116487. [DOI] [PubMed] [Google Scholar]
- 23.Okada T, Sugihara M, Bondar A-N, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 24.Allinger NL, Burkert U. Molecular Mechanics. Washington, D.C.: American Chemical Society; 1982. [Google Scholar]
- 25.Allinger NL, Kuang J, Thomas HD. Molecular mechanics (MM2 and MM3) calculations on aliphatic and aromatic nitro compounds. THEOCHEM. 1990;68:125–148. [Google Scholar]
- 26.Brooks B, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 27.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. 1998;B 102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 28.Hermone A, Kuczera K. Free-energy simulations of the retinal cis–trans isomerization in bacteriorhodopsin. Biochemistry. 1998;37:2843–2853. doi: 10.1021/bi9717789. [DOI] [PubMed] [Google Scholar]
- 29.Lin SW, Imamoto Y, Fukada Y, Shichida Y, Yoshizawa T, Mathies RA. What makes red visual pigments red? A resonance Raman microprobe study of retinal chromophore structure in iodopsin. Biochemistry. 1994;33:2151–2160. doi: 10.1021/bi00174a023. [DOI] [PubMed] [Google Scholar]
- 30.Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red–green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 31.Chan T, Lee M, Sakmar TP. Introduction of hydroxyl-bearing amino acids causes bathochromic spectral shifts in rhodopsin. Amino acid substitutions responsible for red–green color pigment spectral tuning. J. Biol. Chem. 1992;267:9478–9480. [PubMed] [Google Scholar]
- 32.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 33.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: The genes encoding blue, green and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION AVAILABLE Over 200 absorption maxima of various vertebrate red cone pigments used to generate the histograms shown in Figure 3, as well as a summary of the quantum mechanical calculations. This information is available free of charge via the Internet at http://pubs.acs.org.