Abstract
Potent and selective active-site-spanning inhibitors have been designed for cathepsin K, a cysteine protease unique to osteoclasts. They act by mechanisms that involve tight binding intermediates, potentially on a hydrolytic pathway. X-ray crystallographic, MS, NMR spectroscopic, and kinetic studies of the mechanisms of inhibition indicate that different intermediates or transition states are being represented that are dependent on the conditions of measurement and the specific groups flanking the carbonyl in the inhibitor. The species observed crystallographically are most consistent with tetrahedral intermediates that may be close approximations of those that occur during substrate hydrolysis. Initial kinetic studies suggest the possibility of irreversible and reversible active-site modification. Representative inhibitors have demonstrated antiresorptive activity both in vitro and in vivo and therefore are promising leads for therapeutic agents for the treatment of osteoporosis. Expansion of these inhibitor concepts can be envisioned for the many other cysteine proteases implicated for therapeutic intervention.
Cathepsin K, a cysteine protease of the papain superfamily, has been implicated in the process of bone resorption (1–3). Selective inhibitors of cathepsin K therefore could be promising therapeutic agents for the treatment of diseases characterized by excessive bone loss, such as osteoporosis.
Cysteine proteases have been broadly implicated as targets for therapeutic intervention (e.g., cancer, arthritis, viral and parasitic diseases) (4). Generally, the known inhibitors of cysteine proteases occupy only one-half of the enzyme active site and often contain an inherently reactive functional group (e.g., epoxide, chloromethyl ketone) (ref. 4, pp. 47–63; ref. 5). Evaluation of the x-ray structures of various inhibitors bound to papain suggests that inhibitor design spanning both sides of the active site may be a critical aspect of selectivity. Members of the papain superfamily are relatively invariant on the S′ side of the active site cysteine whereas most of the differences are seen on the S side of the active site [nomenclature of Schechter and Berger (6)]. When good binding can be achieved in the S′ direction by an inhibitor that binds in only one-half of the active site, selectivity seems unlikely despite any selectivity achieved by alternate binding in the S direction. The presence of binding elements on both sides of the active site assures an increased likelihood of binding in a single direction, and that S-site recognition will be utilized during inhibitor binding. Additionally, S′-site recognition appears to be an important aspect of inhibitor potency, as has been demonstrated by Abeles with azapeptide esters and amides (7). Also important to the successful design of protease inhibitors suitable for chronic therapeutic applications is the avoidance of inherently reactive functional groups that may lead to undesired antigenic and immunologic responses (8). This constraint has been applied to a successful therapeutic endpoint in the design of inhibitors of angiotensin converting enzyme (9), a metalloprotease, and inhibitors of HIV protease (10), an aspartyl protease, but not for serine or cysteine proteases. X-ray crystallographic studies carried out in our laboratories on papain complexed to peptide aldehyde inhibitors have revealed an unexpected mode of binding for such compounds.
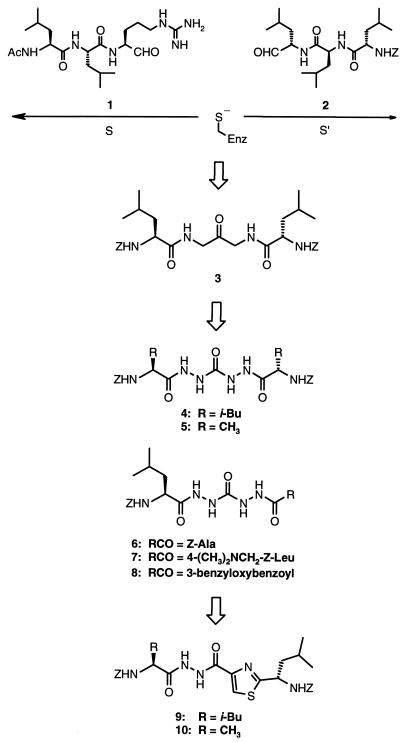
Although leupeptin (Ac-Leu-Leu-Arg-H) (compound 1) was observed to bind on the S side of the active site as had been previously reported (11), the closely related aldehyde 2 was observed to bind only in the S′ direction (Fig. 1). The overlay of these two crystal structures led to the successful design of a potent class of 1,3-diamino-2-propanones that span both sides of the active site (compound 3) (12). The present report describes the design and synthesis of the bis(aza) analogs of 3 as well as diacylhydrazines containing a thiazole amide bond isostere that are potent and selective inhibitors of cathepsin K and span both sides of its active site.
Figure 1.
The evolution of inhibitors 4–10.
MATERIALS AND METHODS
Inhibitor Synthesis.
Symmetric inhibitors 4 and 5 were prepared by treatment of carbohydrazide with 2 equivalents of a Z amino acid, 2 equivalents of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC⋅HCl), and 0.2 equivalents of 1-hydroxybenzotriazole (HOBT) in N,N-dimethylformamide (DMF). Tritiated 4 was prepared analogously by using [3H2]Z-leucine. The (13C,15N4)-labeled 4 was prepared by treatment of 15N2-hydrazine sulfate with 13C-labeled phosgene and potassium hydroxide in water/ether/toluene, followed by coupling with Z-leucine as above. Unsymmetric inhibitors 6–8 were prepared by sequential treatment of Z-leucine methyl ester with hydrazine hydrate in methanol, phosgene in toluene, hydrazine hydrate in methanol, and coupling with a carboxylic acid as above, except using 1 equivalent each of the carboxylic acid and EDC⋅HCl. The carboxylic acid used in the synthesis of 7 was prepared by sequential treatment of methyl 4-bromomethylbenzoate with dimethylamine in methanol, lithium aluminum hydride in ether, α-isocyanato-l-leucine methyl ester (13) in toluene, and lithium hydroxide (LiOH) in tetrahydrofuran (THF)/water. The 3-benzyloxybenzoic acid used in the synthesis of 8 was prepared by treatment of methyl 3-hydroxybenzoate with sodium hydride and benzyl bromide in DMF followed by treatment with LiOH in THF/water. Inhibitors 9 and 10 were prepared by treatment of Z-leucine with isobutyl chloroformate and N-methylmorpholine in THF at −40°C and subsequent addition of gaseous ammonia, followed by sequential treatment with Lawesson’s reagent in THF, ethyl bromopyruvate in acetone, trifluoroacetic anhydride in methylene chloride, then hydrazinolysis and coupling with a Z amino acid as above. [3H2]-9 was prepared as above by using [3H2]Z-leucine in the final step. All compounds were characterized by satisfactory proton NMR and MS data.
Protein Supply and Enzyme Inhibition Assays.
Cathepsin K was expressed in baculovirus-infected cells and activated to its mature form as described previously (2). Cathepsin B and cathepsin L were obtained from Calbiochem. Cathepsin S was expressed in baculovirus-infected cells and purified by a method similar to that reported for cathepsin K (2). The details will be described separately. The protein was activated as previously described (14) except that the activation was carried out for 45–55 min at 37°C instead of 3 hr. Enzyme inhibition was evaluated as previously described (15).
MS and NMR Analysis of Cathepsin K/Inhibitor Complexes.
For the MS analysis of products released on inhibition of cathepsin K by compound 4, cathepsin K (10 μM) in 100 mM sodium acetate (pH 5.5)/5 mM EDTA/5 mM cysteine was incubated with compound 4 (50 μM) for 0.5 hr. Reaction was halted by the addition of 1.5 volumes of dimethyl sulfoxide (DMSO). The reaction mixtures were injected (200 μL) on a Beckman Ultrasphere ODS 4.6 mm × 250 mm column at a flow rate of 1 ml/min. The gradient program used solvent A [99.9% water, 0.1% trifluoroacetic acid (TFA)] and solvent B [90% acetonitrile (CH3CN), 9.9% water, 0.1% TFA]. The gradient was programmed to start at 33% B (hold for 11 min) and increase to 55% B in 19 min (hold for 3 min), then increase to 100% B in 7 min (hold for 20 min). The column effluent was split approximately 125:1 with the minor portion going to the mass spectrometer and the remainder going to the UV detector (214 nm). Electrospray mass spectra were recorded on a PE-Sciex API-III triple quadrupole mass spectrometer (Thornhill, Canada). The mass spectrometer was scanned from m/z 83 to 93 (OR = 160 V, to produce marker ion m/z 91) and m/z 250 to 2,000 (OR = 55 V, to produce molecular ions with no fragmentation). For the detection of the cathepsin K adducts with inhibitors, compounds 4, 8, and 9 were incubated with 27 μM cathepsin K in 20 mM 2-(N-morpholino)ethanesulfonic acid (Mes)/10 mM sodium chloride (NaCl)/2 mM Cys, pH 6.0 at inhibitor concentrations and incubation times of 270 μM for 2 hr, 270 μM for 3 hr, and 135 μM for 1.25 hr, respectively. All inhibitors were in 20% DMSO. The reaction mixtures were injected (50–100 pmol) directly onto a 0.5-mm i.d. Reliasil C18 RP HPLC column at a flow rate of 20 μl/min connected on-line to the mass spectrometer. The gradient program used solvent A (97.9% water/2% CH3CN/0.1% TFA) and solvent B (90% CH3CN/9.9% water/0.1% TFA). The gradient was programmed to start at 5% B and increase to 95% B in 20 min (hold for 20 min). The column effluent was split approximately 5:1 with the minor portion going to the mass spectrometer and the remainder going to the UV detector (214 nm). The mass spectrometer was scanned from m/z 1,150 to 2,250 with OR = 80 V. Each m/z scan was 5.48 s by using 0.25 m/z steps. Alternatively, the samples were subjected to direct electrospray MS analysis. The sample for NMR analysis was dialyzed into 90% water/10%D2O, 50 mM acetate-d3, 250 mM NaCl, and 2 mM l-Cys, pH 4.0.
Crystallography.
Protein was prepared as described previously (16). Crystals of mature, activated cathepsin K complexed with inhibitor 4 grew to a size of ≈0.2 mm3 in about 6 days at 20°C. The concentration of inhibited cathepsin K used in the crystallization was approximately 8 mg/ml. Crystals were grown by using the vapor diffusion method with the reservoir containing 30% MPD, 0.1 M Mes, and 0.1 M Tris at pH 7. Crystals of the complex are orthorhombic, space group P212121, with cell constants of a = 38.4, b = 50.7, and c = 104.9. X-ray diffraction data were measured from a single crystal by using a Siemens two-dimensional, position-sensitive detector on a Siemens rotating anode generator operating at 5 kW. The structure was determined by rigid body refinement by using x-plor, and the starting model consisted of the protein atoms from the crystal structure of cathepsin K in complex with the cysteine protease inhibitor E64 (16). Fourier maps with coefficients |Fo-Fc| and |2Fo-Fc| were used to fit the atomic model of the inhibitor by using the molecular graphics program frodo. Conventional positional refinement was used to refine the structure during model building by using x-plor. Several cycles of map fitting and refinement were carried out to a final Rc of 0.208 at 2.2 Å resolution.
Crystals of mature, activated cathepsin K complexed with inhibitor 8 grew as above from a solution of 22.5% PEG 8000/0.075 M sodium acetate, pH 4.5, containing 0.15 M lithium sulfate. Crystals of the complex are tetragonal, space group P43212, with cell constants of a = 57.6 Å and c = 131.2 Å. The data were collected and the structure was determined at 2.4 Å resolution as described. The final Rc was 0.237.
Crystals of mature, activated cathepsin K complexed with inhibitor 9 grew as above for inhibitor 4 and are very similar to those of the complex with 4. Diffraction data were collected and the structure was determined at 2.4 Å resolution as described. The final Rc was 0.211.
Human Osteoclast Resorption Assay.
Human osteoclasts were disaggregated from fresh osteoclastoma tissue and used in the human osteoclast resorption assay as described (15). Osteoclastic resorption was measured by using a competitive ELISA according to the manufacturer’s protocol (Osteometer, Rodovre, Denmark). The assay measures carboxyl-terminal telopeptides of the α1 chain of human type I collagen that are released during the resorption process (17). The results were expressed as percent inhibition of resorption compared with supernatants derived from osteoclasts cultured in the absence of inhibitors.
In Vivo Studies in the TPTX (Thyroparathyroidectomized) Rat Model.
Adult male thyroparathyroidectomized rats maintained on a calcium-deficient diet for 24 hr were sorted into groups (n = 5/group) having the same mean (±SD) blood-ionized calcium level and then infused (i.v.) for 6 hr with either human parathyroid hormone [hPTH(1–34)] (0.3 nM/kg per hr) or hPTH(1–34) plus compound 7 (13 mg/kg per hr). Blood levels of ionized calcium were measured at 0, 2, 4, and 6 hr during the infusion, and the data were expressed as mean percent change from baseline.
RESULTS AND DISCUSSION
Compound 4 is a potent time-dependent inhibitor of cathepsin K exhibiting an apparent second-order rate constant (kobs/[I]) of 3.1 × 106 M−1 s−1. X-ray analysis of the enzyme-inhibitor complex at 2.2 Å resolution revealed a mode of binding consistent with addition of the catalytic cysteine thiol to the central urea carbonyl. The inhibitor 4 is bound across both S and S′ sides of the active site (Fig. 2 A and B). On the S side of the active site, the leucine side chain is bound tightly in the S2 pocket and the phenyl portion of the carbobenzyloxy (Z) group appears to participate in an edge-face π–π interaction with Tyr-67. On the S′ side of the active site, the Z group is oriented to participate in a π–π interaction with Trp-184, whereas the isobutyl group is involved in a hydrophobic interaction with one face of the active site composed of residues Gln-21, Cys-22, and Gly-23. Compound 6, an analog of 4 in which one of the leucine residues was replaced with alanine, was 4-fold less potent than 4 (Table 1). However, replacement of both leucine residues with alanine (compound 5) gave rise to a >1,000-fold decrease in potency, demonstrating the necessity for at least one leucine residue to achieve potent inhibition. An aqueous-soluble analog (compound 7) exhibited potency and selectivity equivalent to that of 4. The relative binding affinities of compounds 4–6 as measured by Ki, app (Table 1) indicate that the differences in potency are a result of binding interactions and not differences in the degree of reactivity of the central carbonyl.§§
Figure 2.
(A) Ribbon diagram showing the crystallographically determined structure of cathepsin K complexed with inhibitor 4 with N and C termini labeled. (B) Stereoview of the x-ray crystal structure of inhibitor 4 bound to cathepsin K. The inhibitor is shown in ball-and-stick, and selected active site residues of cathepsin K are shown as sticks. (C) Stereoview of the x-ray crystal structure of inhibitor 8 bound to cathepsin K. (D) Stereoview of the x-ray crystal structure of inhibitor 9 bound to cathepsin K.
Table 1.
Inhibition of cathepsins K, B, L, and S by compounds 4–7
| Ki, app, nM
|
kobs/[I], m−1s−1
|
||||
|---|---|---|---|---|---|
| Compound | Cat K | Cat K | Cat B | Cat L | Cat S |
| 4 | 0.7 | 3.1 × 106 | 1.3 × 103 | 5.8 × 104 | Ki = 11 nM |
| 5 | >1,000 | 2.2 × 103 | Ki > 5,000 nM | Ki > 5,000 nM | Ki > 1,000 nM |
| 6 | 1.0 | 6.9 × 105 | 3.4 × 103 | 3.8 × 103 | Ki = 44 nM |
| 7 | — | 5.3 × 106 | 1.4 × 103 | 1.2 × 105 | Ki = 24 nM |
We have been unable to detect any unique, intrinsic reactivity of the central carbonyl of compounds of this class that would account for reaction with the catalytic cysteine. No detectable addition products were observed by proton NMR analysis on exposure of 4 to p-methoxybenzyl mercaptan in d4-methanol under neutral, acidic (acetic acid), or basic (triethylamine) conditions. This further indicates that the high inhibitory potency is specifically associated with molecular interactions within the binding domain along with the unique nucleophilic nature of the catalytic cysteine of the enzyme.
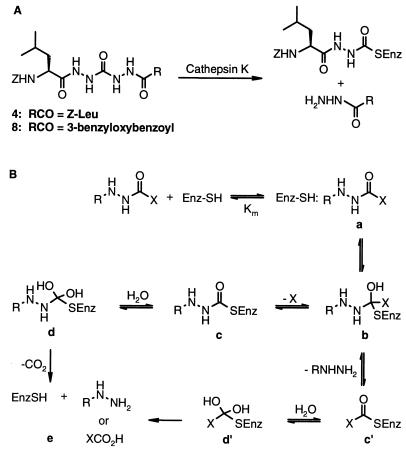
That 4 contains a potential leaving group attached to the carbonyl targeted by the cysteine nucleophile is consistent with its time-dependent inhibition kinetics, prompting us to investigate the mechanism of inhibition in greater detail. Initial studies of the mechanism of inhibition by 4 showed it to be essentially irreversible on dilution or dialysis at pH 5.5–9. In experiments using [3H]-4 having tritium in both leucyl side chains, inhibition of cathepsin K was accompanied by release of 0.92–0.96 equivalents of [3H]-Z-leucinylhydrazide (on analysis by HPLC after dilution with 1.5 volumes of DMSO), consistent with a single binding for inactivation. Whether the release of Z-leucinylhydrazide is part of the inactivation or is driven by the dialysis or HPLC steps remains an unresolved issue that is the subject of continuing study. The apparent irreversibility of this inhibitor, in contrast to the structurally similar, phenylalanine-based azapeptide esters and amides reported by Abeles (6) to be slow-turnover papain inhibitors, may also be because of the formation of a more stable cathepsin K/inhibitor complex relative to Abeles’ phenylalanine-based papain intermediate. However, the propensity of cathepsin K to undergo self-degradation in its uninhibited form has made it difficult to determine whether there is no release of free enzyme from an enzyme/inhibitor complex or whether a very slow release of active enzyme is occurring with subsequent enzyme self-degradation. Further studies are also in progress to clarify this point. To obtain direct evidence of a stable acyl enzyme species, cathepsin K was inactivated independently with 4 and the unsymmetric inhibitor 8, and the mixtures were analyzed by on-line LC-MS. In the case of inactivation with 4, the products observed were the Z-acyl-cathepsin K adduct (Mr = M + 305, where M is the molecular weight of free cathepsin K) and Z-leucinylhydrazide (Mr = 279) (Fig. 3A). The Z-leucinylhydrazinyl acyl-cathepsin K adduct was also observed by 13C NMR spectroscopic analysis of the complex of cathepsin K with 4, which was 13C-labeled at the central carbonyl and 15N-labeled at all four hydrazine nitrogens (Fig. 4). The downfield shift of the carbonyl resonance relative to the free ligand 4 (Fig. 4A) is consistent with the acyl enzyme adduct (Fig. 4B). Again, formation of this intermediate may be a result of either the inactivation step or the dialysis step used in the preparation of the NMR sample. We are unable to determine in this experiment whether the Z-leucinylhydrazine is still bound in the active site, as was observed crystallographically, without a covalent attachment to the labeled carbonyl. Such complexes, where the released S′ side fragment remains bound, have been observed with cyclic, proteinaceous, Kunitz inhibitors of trypsin (19), but have not been reported for small acyclic molecules. On inactivation with 8, the same Z-leucinylhydrazinyl acyl-cathepsin K adduct was observed by MS analysis as was seen with 4. The absence of a benzyloxybenzoylhydrazinyl acyl-cathepsin K adduct is indicative of a single mode of binding and is also consistent with the complex observed crystallographically (Fig. 2C), where 8 is observed as an apparent tetrahedral adduct spanning the S and S′ sites in a single orientation, with the nonpeptide portion bound on the S′ side of the active site. Although the tetrahedral form is the most satisfactory crystallographic solution, equilibrating, cleaved forms with both components still bound cannot be excluded.
Figure 3.
(A) Inhibition of cathepsin K by 4 and 8. (B) Proposed general mechanism of proteolysis by cathepsin K.
Figure 4.
13C NMR spectra of [13C-15N4]-4 and cathepsin K at 125.76 MHz. (A) Compound 4 (15 mM) in DMSO-d6 at 20°C, 3,000 scans, 1.5-s repetition period, 1H broadband decoupling. The triplet structure of the 13C-labeled carbonyl resonance peak is caused by the one-bond 13C-15N spin–spin coupling with two adjacent 15N-labeled amines. (B) Cathepsin K (0.45 mM) complexed with 4 in 90% water/10%D2O, 50 mM acetate-d3, 250 mM NaCl, and 2 mM l-Cys, pH 4.0, 5°C, 123,283 scans, 1.5-s repetition period, 1H broadband decoupling. The doublet structure is caused by the 13C-15N spin–spin coupling with only one adjacent 15N-labeled amine. (C) Same sample and experimental conditions as in B but with simultaneous 1H and 15N broadband decoupling, 240,000 scans, 1.5-s repetition period. The doublet seen in B coalesces into a singlet because of 15N decoupling.
All four of the bis(hydrazide) analogs 4–7 show good selectivity for inhibition of cathepsin K vs. human cathepsins B, L, and S. Interestingly 4, 6, and 7 did not exhibit time-dependent inhibition kinetics with cathepsin S, nor did 5 with cathepsins B, L, and S, albeit with relatively poor inhibitory potency. Alternate mechanisms of inhibition are possible for different enzymes, and more detailed evaluations are in progress. Nonetheless, it is clear that the mechanism of inhibition depends in subtle ways on the specific recognition by the enzyme being inhibited. One can therefore expect that selective inhibition of the various cysteine proteases may be achieved by exploiting the distinctive selectivity elements of each while using the same urea backbone. The importance of highly specific cysteine proteases in numerous intracellular processes suggests numerous opportunities for therapeutic application of this inhibitor concept (4).
A key aspect of the design of therapeutically useful enzyme inhibitors has been the replacement of amino acid elements by so-called peptidomimetic elements. Compound 8 represents a step in that direction. The previous use in our laboratories of heterocyclic amide bond isosteres in enzyme inhibitors (20, 21) suggested another route into peptidomimetics by replacement of one of the acylhydrazine groups in 4 with a thiazole ring, giving rise to diacylhydrazine 9. Acylhydrazones have been reported to be micromolar inhibitors of cruzain (22) and the cysteine protease from Plasmodium falciparum (23). However, these compounds differ substantially from compound 9 and may inhibit by other mechanisms. Compound 9 is a potent inhibitor of cathepsin K (Ki, app = 10 nM), but now exhibits initial inhibition kinetics (1–30 min) consistent with a rapidly reversible mechanism of inhibition. X-ray analysis of 9 complexed to cathepsin K (2.3 Å resolution) shows it also is best described as a tetrahedral adduct with the active site cysteine thiol (Fig. 2D). Compound 9 is also bound in only one orientation, with the thiazole ring on the S′ side of the active site. The alanine analog 10 was found to be 12-fold less potent than 9, consistent with an important interaction of the leucine side chain in the enzyme active site (Table 2).§§ Compounds 9 and 10 are also highly selective for inhibition of cathepsin K vs. human cathepsins B, L, and S. Incubation of [3H2]leucine-labeled 9 (100 μM) with cathepsin K at high concentration (3 μM) for 3.5 hr results in loss of 9, consistent with multiple, slow turnovers by enzyme. On-line LC-MS analysis of a mixture of cathepsin K and 9 showed a thiazolyl acyl-cathepsin K adduct (Mr = M + 330) along with apparently unreacted cathepsin K. The enzyme adduct was confirmed by direct nanospray MS analysis of 9 in the presence of cathepsin K without purification. Thus, 9 may be partially inhibiting via acylation of the enzyme, possibly acting as a slow, multiple turnover substrate. However, we are unable to exclude possible collapse of a tetrahedral adduct in the mass spectrometer.
Table 2.
Inhibition of cathepsins K, B, L, and S by compounds 9 and 10
| Compound | Ki, app, nM
|
|||
|---|---|---|---|---|
| Cat K | Cat B | Cat L | Cat S | |
| 9 | 10 | 5,200 | 700 | >1,000 |
| 10 | 120 | >1,000 | >1,000 | >1,000 |
Examination of crystallographic data and the results of the solution-phase experiments in the context of a general mechanism of proteolysis by cathepsin K (Fig. 3B) indicates that different stages of the mechanistic pathway are observed in different experiments. For both inhibitor classes, the species observed crystallographically appear to be representative of the tetrahedral intermediate (Fig. 3B, b). For compounds 4 and 8, the solution-phase experiments suggest that a fairly low energy state is represented by (c). These observations may differ from those reported by Abeles et al. (6) for structurally similar inhibitors of papain that undergo slow hydrolysis via (d) to regenerate active enzyme (e). The mass spectral data suggest that compound 9 can follow an analogous pathway, b–c′–d′–e. Differentiation of the postulated intermediates a, b, and c or a, b′, and c′ from states on the pathway between them is beyond the resolution of the methods that have been thus far applied.
In all three of the structures studied by crystallography, the electron density between the Cys-25 sulfur and the carbonyl carbon is most consistent with a covalent bond. Specific stabilization of the bound form of these intermediates relative to product release appears to have resulted in freezing at a state that is not normally seen crystallographically. In most cases, so-called transition state analogs that are seen as bound to the enzyme have no apparent pathway for collapse to products. They have nonetheless been extremely useful tools for evaluation of enzyme mechanisms. All three of the inhibitors studied by crystallography contain a potential leaving group (acylhydrazine), and the studies involving incubation of these inhibitors with cathepsin K suggest turnover as substrates. Tetrahedral intermediates and other states along such mechanistic pathways have been observed crystallographically but have required the use of low temperature (24) or time-resolved (25) methods. Thus, we feel that these complexes reported herein may be the closest approximations of tetrahedral intermediates that occur during substrate proteolysis that have been observed under static conditions at room temperature. At the present resolution of 2.2 Å, we cannot rule out the possibility that C—N bond cleavage has occurred while retaining the released hydrazide. It may also be an equilibrating mixture of cleaved and tetrahedral forms. The issues raised here are quite similar to those considered for polypeptide Kunitz inhibitors (19), the precise mechanism of which remains similarly unresolved after decades of study. We hope that an X-ray analysis carried to beyond 1.8 Å will be helpful in understanding the precise processes of inhibition for our small molecules. Clearly, these processes are complicated and will involve complex analytical methods. Nonetheless, 9 and 4 are representative of potent classes of inhibitors of cathepsin K and have been valuable tools for cell-based and in vivo studies.
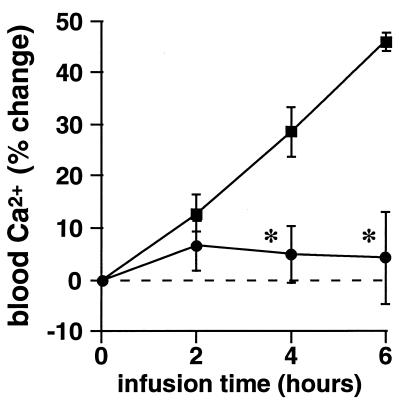
Compounds 4, 7, and 9 were evaluated in a human osteoclast resorption assay in vitro (15). Of the three, compounds 4 and 7 were the most potent, with IC50 values of 0.34 and 0.12 μM, respectively, and 9 was less potent, with an IC50 of 2 μM. Compound 7 was evaluated in the TPTX rat, an acute model of bone resorption in vivo (26). At a dose of 13 mg/kg per hr administered as a 6-hr i.v. coinfusion with hPTH(1–34), 7 caused a highly significant inhibition of the osteoclast-mediated calcemic response to hPTH(1–34), maintaining baseline blood levels of ionized calcium (P > 0.6 vs. baseline at 6 hr) at all time points (Fig. 5).
Figure 5.
Inhibition of bone resorption in the TPTX rat model by 7. hPTH(1–34) (▪) caused a significant calcemic response at 4 and 6 hr of infusion because of stimulation of osteoclastic bone resorption. Coinfusion with compound 7 (•) prevented the response to hPTH(1–34). ∗, P < 0.01 vs. hPTH(1–34) control. (Bars = SEM.)
In summary, we have designed two new classes of potent and selective inhibitors of the cysteine protease cathepsin K that have potent antiresorptive activity both in vitro and in vivo, and are therefore promising leads for therapeutic agents for the treatment of osteoporosis. The unique binding modes of the inhibitors have been detailed by x-ray crystallography, and the species observed crystallographically may be close approximations of tetrahedral intermediates that occur during substrate proteolysis. Hence, these structures may be useful in further elucidating the mechanisms of cysteine protease processing and inhibition.
Acknowledgments
We thank Drs. George Poste and Brian Metcalf for their helpful insights and significant support of this work. We also thank Dr. Thomas Meek and Dr. David Tew for helpful discussions, Lawrence Szewczuk for enzyme assays, Don Bennett for oligonucleotide synthesis, and Stephanie Van Horn for sequencing of the cathepsin S PCR fragment. We are also grateful to Dr. Allan Shatzman for his support of this work.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: Z, carbobenzyloxy.
Data deposition: The refined coordinates for the complexes of compounds 4, 8, and 9 with cathepsin K have been deposited under the file names 1AYU, 1AYW, and 1AYV, respectively, in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973.
The coincident 13C NMR chemical shifts of the central carbonyl carbons of 4 and 5 (157 ppm) are also supportive of this conclusion (17), as are the coincident 13C NMR chemical shifts of the central carbonyl carbons of 9 and 10 (159 ppm).
References
- 1.Drake F H, Dodds R A, James I E, Connor J R, Debouck C, Richardson S, Lee-Rykaczewski E, Coleman L, Rieman D, Barthlow R, Hastings G, Gowen M. J Biol Chem. 1996;271:12511–12516. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 2.Bossard M J, Tomaszek T A, Thompson S K, Amegadzie B Y, Hanning C R, Jones C, Kurdyla J T, McNulty D E, Drake F H, Gowen M, Levy M A. J Biol Chem. 1996;271:12517–12524. doi: 10.1074/jbc.271.21.12517. [DOI] [PubMed] [Google Scholar]
- 3.McQueney M S, Amegadzie B Y, D’Alessio K D, Hanning C R, McLaughlin M M, McNulty D, Carr S A, Ijames C, Kurdyla J, Jones C S. J Biol Chem. 1997;272:13955–13960. doi: 10.1074/jbc.272.21.13955. [DOI] [PubMed] [Google Scholar]
- 4.McKerrow, J. H. & James, M. N. G., eds. (1996) Perspect. Drug Dis. Des. 6, 1–125.
- 5.Demuth H-U. J Enzyme Inhibition. 1990;3:249–278. doi: 10.3109/14756369009030375. [DOI] [PubMed] [Google Scholar]
- 6.Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 7.Baggio R, Shi Y-Q, Wu Y-Q, Abeles R H. Biochemistry. 1996;35:3351–3353. doi: 10.1021/bi952879e. [DOI] [PubMed] [Google Scholar]
- 8.Amos H E, Park B K. In: Immunotoxicology and Immunopharmacology. Dean J H, Luster M I, Munson A E, Amos H E, editors. New York: Raven; 1985. pp. 207–228. [Google Scholar]
- 9.Ondetti M A, Rubin B, Cushman D W. Science. 1977;196:441–444. doi: 10.1126/science.191908. [DOI] [PubMed] [Google Scholar]
- 10.Roberts N A, Martin J A, Kinchington D, Broakhurst A V, Craig J C, Duncan I B, Galpin S A, Handa B K, Kay V, Krohn A, Lambert R W, Merrett J H, Mills J S, Parkes K E B, Redshaw S, Ritchie A J, Taylor D L, Thomas G J, Machin P J. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder E, Phillips C, Garman E, Harlos K, Crawford C. FEBS Lett. 1993;315:38–42. doi: 10.1016/0014-5793(93)81128-m. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita D S, Smith W W, Zhao B, Janson C A, Tomaszek T A, et al. J Am Chem Soc. 1997;119:11351–11352. [Google Scholar]
- 13.Nowick J S, Holmes D L, Noranha G, Smith E M, Nguyen T M, Huang S-L. J Org Chem. 1996;61:3929–3934. doi: 10.1021/jo960038n. [DOI] [PubMed] [Google Scholar]
- 14.Bromme D, Bonneau P R, Lachance P, Wiederanders B, Kirschke H, Peters C, Thomas D Y, Storer A C, Vernet T. J Biol Chem. 1993;268:4832–4838. [PubMed] [Google Scholar]
- 15.Votta B, Levy M A, Badger A, Bradbeer J, Dodds R A, James I E, Thompson S, Bossard M J, Carr T, Connor J R, Tomaszek T A, Szewczuk L, Drake F H, Veber D F, Gowen M. J Bone Miner Res. 1997;12:1396–1406. doi: 10.1359/jbmr.1997.12.9.1396. [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Janson C A, Amegadzie B Y, D’Alessio K, Griffin C, Hanning C R, Jones C, Kurdyla J, McQueney M, Qiu X, Smith W W, Abdel-Meguid S S. Nat Struct Biol. 1997;4:109–111. doi: 10.1038/nsb0297-109. [DOI] [PubMed] [Google Scholar]
- 17.Foged N T, Delaisse J-M, Hou P, Sato T, Winding B, Bonde M. J Bone Miner Res. 1996;11:226–237. doi: 10.1002/jbmr.5650110212. [DOI] [PubMed] [Google Scholar]
- 18.Suga T, Izumi S, Hirata T. Chem Lett. 1986;12:2053–2056. [Google Scholar]
- 19.Laskowski M. Adv Exp Med Biol. 1986;199:1–17. doi: 10.1007/978-1-4757-0022-0_1. [DOI] [PubMed] [Google Scholar]
- 20.Thompson S K, Murthy K H M, Zhao B, Winborne E, Green D W, Fisher S M, DesJarlais R L, Tomaszek T A, Meek T D, Gleason J G, Abdel-Meguid S S. J Med Chem. 1994;37:3100–3107. doi: 10.1021/jm00045a015. [DOI] [PubMed] [Google Scholar]
- 21.Thompson S K, Eppley A M, Frazee J S, Darcy M G, Lum R T, Tomaszek T A, Ivanoff L A, Morris J F, Sternberg E J, Lambert D M, Fernandez A V, Petteway S R, Meek T D, Metcalf B W, Gleason J G. Biomed Chem Lett. 1994;4:2441–2446. [Google Scholar]
- 22.Li R, Gong B, Selzer P M, Li Z, Davidson E, Kursban G, Miller R E, Nuzum E D, McKerrow J H, Fletterick R J, Gillmor S A, Craik C S, Kuntz I D, Cohen F E, Kenyon G L. Biomed Chem. 1996;4:1421–1427. doi: 10.1016/0968-0896(96)00136-8. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Chen X, Davidson E, Zwang O, Mendis C, Ring C S, Roush W R, Fegley G, Li R, Rosenthal P J, Lee G K, Kenyon G, Kuntz I D, Cohen F E. Chem Biol. 1994;1:31–37. doi: 10.1016/1074-5521(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 24.Fink A L, Petsko G A. Adv Enzymol Relat Areas Mol Biol. 1981;52:177–246. doi: 10.1002/9780470122976.ch3. [DOI] [PubMed] [Google Scholar]
- 25.Petsko G A. Philos Trans R Soc London, Ser A. 1992;340:323–334. [Google Scholar]
- 26.Thompson D D, Seedor J G, Fisher J E, Rosenblatt M, Rodan G A. Proc Natl Acad Sci USA. 1988;85:5673–5677. doi: 10.1073/pnas.85.15.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]