Abstract
Membrane trafficking and cytoskeletal dynamics are important cellular processes that drive tip growth in root hairs. These processes interact with a multitude of signaling pathways that allow for the efficient transfer of information to specify the direction in which tip growth occurs. Here, we show that AGD1, a class I ADP ribosylation factor GTPase-activating protein, is important for maintaining straight growth in Arabidopsis (Arabidopsis thaliana) root hairs, since mutations in the AGD1 gene resulted in wavy root hair growth. Live cell imaging of growing agd1 root hairs revealed bundles of endoplasmic microtubules and actin filaments extending into the extreme tip. The wavy phenotype and pattern of cytoskeletal distribution in root hairs of agd1 partially resembled that of mutants in an armadillo repeat-containing kinesin (ARK1). Root hairs of double agd1 ark1 mutants were more severely deformed compared with single mutants. Organelle trafficking as revealed by a fluorescent Golgi marker was slightly inhibited, and Golgi stacks frequently protruded into the extreme root hair apex of agd1 mutants. Transient expression of green fluorescent protein-AGD1 in tobacco (Nicotiana tabacum) epidermal cells labeled punctate bodies that partially colocalized with the endocytic marker FM4-64, while ARK1-yellow fluorescent protein associated with microtubules. Brefeldin A rescued the phenotype of agd1, indicating that the altered activity of an AGD1-dependent ADP ribosylation factor contributes to the defective growth, organelle trafficking, and cytoskeletal organization of agd1 root hairs. We propose that AGD1, a regulator of membrane trafficking, and ARK1, a microtubule motor, are components of converging signaling pathways that affect cytoskeletal organization to specify growth orientation in Arabidopsis root hairs.
Cells that undergo tip growth provide an excellent experimental system to uncover fundamental mechanisms underlying polarity establishment in plants. The most widely studied tip-growing cells in plants are root hairs and pollen tubes. In these cell types, expansion is restricted primarily to the extreme apex of the cell, leading to the formation of long tubular structures. Years of research have established that tip growth is modulated by a number of cellular processes, including a dynamic cytoskeleton, tip-focused cytoplasmic calcium gradients and oscillations, and vesicle/membrane trafficking events. These processes in turn are under tight regulation by an elaborate network of signaling molecules, such as reactive oxygen species (ROS), monomeric guanine nucleotide-binding proteins (small GTPases), mitogen-activated protein kinases, and phosphoinositides (Cole and Fowler, 2006; Samaj et al., 2006; Campanoni and Blatt, 2007; Kost, 2008).
An intriguing question in regard to studies on root hairs and pollen tubes is how the directionality of tip growth is maintained. Whereas a change in growth direction in pollen tubes occurs partly in response to chemotropic factors from the stigma (Kim et al., 2003), root hairs maintain an endogenous polarity that guides the tip to grow straight and away from the primary root body (Bibikova et al., 1997). In both cases, calcium appears to play an important role in maintaining tip growth, since artificially relocalizing the tip-focused cytoplasmic calcium gradient using caged calcium ionophores resulted in a change in tip growth direction that often followed the new calcium gradient (Malho and Trewavas, 1996; Bibikova et al., 1997).
In addition to cytoplasmic calcium, the cytoskeleton, which consists of microtubules, actin filaments (F-actin), and several accessory proteins (Blancaflor et al., 2006; Hussey et al., 2006), plays an essential role in regulating tip growth direction. The important role of the cytoskeleton in this process is supported by the observation that exposure to microtubule inhibitors or down-regulation of tubulin, the major protein subunit of microtubules, resulted in the formation of root hairs with multiple tips and wavy growth (Bibikova et al., 1999; Bao et al., 2001). Recently, root hairs of Arabidopsis (Arabidopsis thaliana) knockouts to an armadillo repeat-containing kinesin displayed wavy and branched root hairs, further establishing the link between microtubules and the maintenance of root hair growth direction (Jones et al., 2006; Yang et al., 2007; Sakai et al., 2008). The actin cytoskeleton also is essential for maintaining tip-focused growth. Actin-disrupting drugs inhibited or delocalized root hair growth (Bibikova et al., 1999; Ketelaar et al., 2003), consistent with reports showing that knockouts to the vegetative ACT2 gene in Arabidopsis have distorted root hair morphology (Ringli et al., 2002). Moreover, altering the expression of genes encoding proteins that affect actin turnover, such as profilin, actin-depolymering factor, formin, actin-interacting protein, and cyclase-associated protein, disrupted normal root hair development (Ramachandran et al., 2000; Dong et al., 2001; Yi et al., 2005; Deeks et al., 2007; Ketelaar et al., 2007).
In coordination with the cytoskeleton and calcium gradients, vesicle trafficking at the tip contributes largely to membrane turnover and deposition of cell wall precursors necessary to sustain the rapid growth of pollen tubes and root hairs. Several components of the vesicle-trafficking machinery have been implicated in tip growth, including the Rab, Rho, and ADP-ribosylation factor (ARF) subfamilies of small GTPases (Cole and Fowler, 2006; Kost, 2008). Small GTPases act as molecular switches that cycle between an active (GTP-bound) and an inactive (GDP-bound) conformation. In their active form, small GTPases interact with specific effector proteins that modulate cytoskeletal organization, vesicle budding and docking, and protein trafficking. Whereas conversion of the inactive to the active GTPase is facilitated by guanine nucleotide exchange factors (GEFs), inactivation occurs through the action of GTPase-activating proteins (GAPs), which stimulate the hydrolysis of GTP to GDP (Bos et al., 2007).
The ROPs (for Rho of plants) and Rabs have been the most widely studied subfamilies of small GTPases with regard to the control of tip growth in plants. ROPs and Rabs have been shown to localize to the tips of root hairs and pollen tubes (Molendijk et al., 2001; Jones et al., 2002; Preuss et al., 2004), and consistent with its proposed function in polarized growth, tip localization of Rab4Ab was only observed in growing root hairs but not in mature root hairs that had ceased growing or root hair mutants with defective tip growth (Preuss et al., 2004). Furthermore, constitutively active or dominant negative ROPs altered tip growth, resulting in the formation of bulbous tips in pollen tubes (Kost et al., 1999; Li et al., 1999) and root hairs with wavy growth, multiple tips, and partially swollen tips (Molendijk et al., 2001; Jones et al., 2002; Bloch et al., 2005). Disrupting ROP GTPase function by the expression of constitutively active or dominant negative ROP has been shown to affect apical F-actin dynamics, calcium signaling, and ROS production. In pollen tubes, ROPs appear to modulate F-actin dynamics and calcium oscillations via effector proteins with CRIB (for Cdc42/Rac-interactive binding) motifs (Gu et al., 2005). Moreover, Arabidopsis mutants in a ROP guanine dissociation inhibitor, a protein that prevents ROP activation by GEFs, have multiple tip-growing sites in their root hairs. This particular ROP guanine dissociation inhibitor was demonstrated to function in root hair growth by restricting ROS production to a single growing point (Carol et al., 2005).
Small GTPases also have been shown to coordinate tip growth via their interaction with lipid signaling pathways. In Arabidopsis root hairs, for example, RabA4b specifically interacts with a phosphatidylinositol 4-OH kinase at the tip, and this interaction has been proposed to be relevant for maintaining the cytoplasmic calcium gradient that drives polar root hair growth (Preuss et al., 2006). In this regard, inhibiting the production of lipid signaling molecules such as phosphatidic acid disrupted tip growth, disorganized the actin cytoskeleton, and dissipated the tip-focused cytoplasmic calcium gradient (Potocký et al., 2003; Monteiro et al., 2005; Motes et al., 2005). Also, a mutation in an Arabidopsis phosphatidylinositol transfer protein compromised polarized growth in root hairs, which coincided with a disorganization of the microtubule cytoskeleton (Vincent et al., 2005).
The ROP and Rab GTPases are themselves under tight regulation by other small GTPases, such as those belonging to the ARF subfamily. In Arabidopsis, the fungal macrolide brefeldin A (BFA), an inhibitor of ARF-GEFs, disrupted the polar localization of ROPs in root hairs (Molendijk et al., 2001). Similarly, expression of the GTP-locked form of ARF1 induced the formation of bulged root hairs, which occurred concomitantly with altered ROP localization (Xu and Scheres, 2005). Recently, an Arabidopsis mutant in a class II ARF-GAP displayed deformed root hairs and reduced growth of pollen tubes, further implicating ARFs in tip growth maintenance (Song et al., 2006).
It is clear that a number of interconnected signaling pathways and cellular processes are required for the polarized delivery of vesicles and new cell wall material important for driving tip growth in plants. It is likely, however, that additional molecular components of the tip growth machinery remain to be identified. In an effort to uncover new players involved in maintaining root hair growth directionality, we conducted a forward genetic screen of T-DNA activation-tagged Arabidopsis seedlings to isolate root hair mutants with altered growth direction (Weigel et al., 2000). Here, we characterize two loss-of-function Arabidopsis mutants with wavy root hair growth. We show that one mutant is disrupted in a gene that encodes a class I ARF-GAP (AGD1; Vernoud et al., 2003) while the other mutant is a new allele of an armadillo repeat-containing kinesin previously designated MRH2 or ARMADILLO REPEAT KINESIN1 (ARK1; Jones et al., 2006; Yang et al., 2007; Sakai et al., 2008). AGD1 localized to punctate intracellular compartments that partially associated with the endocytic marker FM4-64, consistent with its function as a mediator of vesicle trafficking. ARK1, on the other hand, localized to microtubules, consistent with its function as a microtubule motor protein. Low concentrations of BFA that did not inhibit root hair growth reversed the wavy root hair phenotype of agd1 mutants, supporting the notion that AGD1 modulates root hair growth direction, organelle trafficking, and cytoskeletal organization through the activity of an AGD1-dependent ARF substrate. The more severe root hair phenotype of double agd1 ark1 mutants compared with single mutants indicates that AGD1 and ARK1 are components of independent but possibly intersecting signaling pathways that specify growth orientation in Arabidopsis root hairs by affecting cytoskeletal dynamics at the tip.
RESULTS
Identification and Characterization of Arabidopsis Mutants with Altered Root Hair Growth Direction
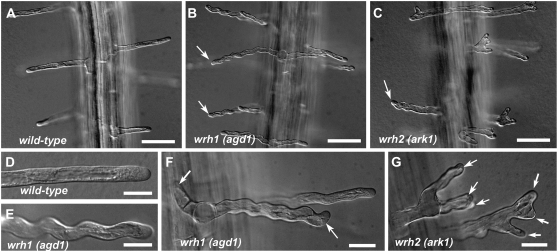
When germinated in semisolid medium, wild-type Arabidopsis root hairs typically grow straight and away from the primary root axis (Fig. 1A; Supplemental Movie S1). This highly predictable growth of Arabidopsis root hairs allows for rapid genetic screening of mutants with altered root hair morphology (Parker et al., 2000). We isolated two mutants from a T-DNA activation-tagged population in the Columbia (Col-0) background that exhibited wavy root hair growth (Fig. 1, B and C). When the mutants were backcrossed to the wild type, the F1 progeny did not exhibit the wavy root hair phenotype. In the F2 generation, the root hair phenotype for both mutants segregated 3:1 (wild-type to wavy), indicating that both mutants were recessive in a single Mendelian locus (data not shown). We initially designated these two mutants wrh1 and wrh2 (for wavy root hair) but renamed them agd1-1 and ark1-4, respectively, upon subsequent molecular identification of the disrupted genes (see below).
Figure 1.
Root hair morphology of wild-type, wrh1(agd1), and wrh2(ark1) mutant plants. Whereas wild-type root hairs grow straight (A), some root hairs of wrh1(agd1) and wrh2(ark1) display wavy growth (B and C; arrows). High-magnification images of a straight-growing wild-type root hair (D) and a wrh1(agd1) root hair with a distinct wavy growth pattern (E) are also shown. Roots of wrh1(agd1) would sometimes have two root hairs emerging from a common initiation point (F; arrows). Root hairs of wrh2(ark1) occasionally had root hairs with multiple tips (G; arrows). Bars = 50 μm (A–C) and 20 μm (D–G).
After back-crossing two times to the wild type to remove possible extraneous mutations, we conducted a more detailed microscopic analysis of the root hairs of wrh1(agd1) and wrh2(ark1). In contrast to wild-type root hairs, which typically maintained straight growth (Fig. 1D; Supplemental Movie S1), root hairs of wrh1(agd1) redirected their growth at almost regular intervals, leading to the wavy phenotype (Fig. 1E; Supplemental Movie S2). Although the phenotype of wrh1(agd1) generally appeared wavy, other time-lapse movies revealed that some root hairs grew in a spiral manner (data not shown). Also, roots of wrh1(agd1) occasionally exhibited two hairs originating from a single initiation site (Fig. 1F). Whereas some root hairs of wrh2(ark1) displayed similar wavy growth as wrh1(agd1), wrh2(ark1) had a greater tendency to form root hairs with multiple tips and branches (Fig. 1G). Time-lapse imaging of growing root hairs of wrh2(ark1) revealed occasional cessation of tip growth, which was followed by gradual broadening of the tip. The broader tip apex eventually gave rise to two or more tips that either grew simultaneously or displayed alternating pulses of growth (Supplemental Movie S3). We measured the growth rate of root hairs from 3- to 4-d-old wild-type and wrh mutant plants located at 500 to 700 μm from the primary root tip, since this is the region where root hairs are actively elongating. The average growth rate of wild-type root hairs was 1.15 ± 0.25 μm min−1, while the growth rates of wrh1 and wrh2 were significantly less, with values at 0.77 ± 0.29 and 0.74 ± 0.23 μm min−1, respectively (values are means ± sd of 50–60 root hairs; Student's t test; P < 0.001).
Molecular Identification of the Disrupted Genes in the Root Hair Mutants
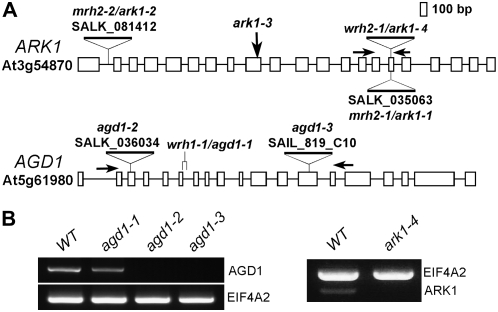
Examination of root hairs of the F1 progeny resulting from a cross between wrh1 and wrh2 showed wild-type phenotypes, indicating that WRH1 and WRH2 were not allelic. We then conducted thermal asymmetric interlaced (TAIL)-PCR to identify flanking T-DNA insertions in the wrh mutants (Liu et al., 1995). We found that wrh2 had a T-DNA insertion in the 16th exon of the gene At3g54870, which encodes an armadillo repeat-containing kinesin motor protein (Fig. 2A). Two independent groups had previously identified mutants in this gene (Jones et al., 2006; Yang et al., 2007; Sakai et al., 2008). These mutants were designated mrh2 but were recently renamed ark1 (Sakai et al., 2008). Since root hairs of the previously described ark1 mutants exhibited similar wavy and multiple tip phenotypes as our wrh2 mutant, it is likely that wrh2 is another allele of ARK1. This was confirmed by our independent identification of two homozygous SALK lines with T-DNA insertions in the ARK1 gene (SALK_081412 and SALK_035063; Alonso et al., 2003). We found that root hairs from these insertion lines exhibited similar phenotypes as wrh2 (data not shown). Therefore, we renamed wrh2 as ark1-4, since three other ark1 mutants that included the homozygous SALK lines that we identified here have already been described (Fig. 2A; Jones et al., 2006; Sakai et al., 2008).
Figure 2.
Mutant alleles of agd1 and ark1. A, Schematic diagrams of the genome organization of ARK1 and AGD1. Boxes indicate exons, and lines indicate introns. The deletion site (agd1-1) and T-DNA insertion sites for each of the mutants are indicated. ark1-3 is a point mutation described by Sakai et al. (2008). B, Semiquantitative RT-PCR of AGD1 and ARK1 in the wild type (WT), agd1-1, agd1-2, agd1-3, and ark1-4. Total RNA was prepared from root tissue, and arrows in A indicate the positions of the primers used for RT-PCR. EIF4A2 primers were used as a control.
Since our attempts to identify the disrupted gene in wrh1 using TAIL-PCR were unsuccessful, we tested for the presence of the T-DNA insert in this line using BASTA (glufosinate ammonium) and found that wrh1 was not resistant. Therefore, we performed map-based cloning to identify the WRH1 gene. We found that wrh1 had a deletion of 46 bp spanning the splice donor site of exon 6 of the At5g61980 gene (Fig. 2A). Comparison of wild-type and mutant WRH1 cDNA revealed a 31-bp deletion resulting from aberrant splicing of intron 6 that caused a frame-shift mutation that introduced a TAA stop codon after seven irrelevant amino acid residues (Supplemental Fig. S1).
The At5g61980 gene encodes a class I ARF-GAP, which encompasses 19 exons and encodes a predicted 850-amino acid protein (Fig. 2A). An analysis of the Arabidopsis genome identified 15 ARF-GAP domain (AGD) proteins belonging to four classes. In addition to a GAP domain, class I ARF-GAP proteins, which consist of four members, contain Bin1-amphiphysin-Rvs167p/Rvs161p (BAR) and pleckstrin homology (PH) domains and two ankyrin repeats. At5g61980 was designated AGD1 for ARF-GAP domain-containing protein 1 (Vernoud et al., 2003). Previously, a closely related class I ARF-GAP (AGD3) was shown to be involved in auxin efflux and vein patterning in Arabidopsis (Koizumi et al., 2005; Sieburth et al., 2006). Functional analyses of single mutants in the other class I ARF-GAPs, including mutations in the AGD1 gene, however, did not reveal any apparent phenotype (Sieburth et al., 2006). Therefore, to verify whether disruption of the AGD1 gene is indeed the cause of the wavy root hair phenotype of wrh1, we searched the publicly available SALK collection for T-DNA insertions in the At5g61980 gene (Alonso et al., 2003). We genotyped the segregating progeny by PCR and identified homozygous plants for one SALK line (SALK_036034), which was similar to the agd1 mutant reported by Sieburth et al. (2006), and one SAIL line (SAIL_819_C10), which had an insertion in the 13th exon of AGD1 (Fig. 2A). Root hairs of these two T-DNA insertion mutants exhibited similar wavy phenotypes as wrh1 (data not shown), suggesting that these were alleles of wrh1. Therefore, we renamed wrh1 as agd1-1 while SALK_036034 and SAIL_819_C10 were designated as agd1-2 and agd1-3, respectively (Fig. 2A). All of the subsequent cellular analyses reported here were conducted with agd1-1 mutants. Reverse transcription (RT)-PCR analysis showed that agd1-2, agd1-3, and ark1-4 had no detectable transcript using primers downstream of the T-DNA insertion. As noted, agd1-1, being a frame-shift mutant, still produced a transcript, but this transcript is likely nonfunctional (Fig. 2B; Supplemental Fig. S1).
Sequencing of three independently cloned, PCR-amplified wild-type AGD1 cDNA fragments showed a discrepancy with the National Center for Biotechnology Information-annotated At5g61980 gene. From our sequencing results, we found that the cDNA encodes a predicted 828-amino acid protein, which differs from National Center for Biotechnology Information database prediction due to the use of a GC splice acceptor site in intron 9 between the BAR and PH domains rather than the predicted GT splice site (Supplemental Fig. S1). This result is consistent with a previous report that 1% of Arabidopsis introns have GC at their 5′ ends (Brown et al., 1996).
Cytoskeletal Organization Is Disrupted in Growing Root Hairs of agd1-1 and ark1-4
The wavy root hair phenotype of agd1-1 and ark1-4 was reminiscent of root hairs treated with microtubule inhibitors, suggesting that microtubule organization could be affected in these mutants (Bibikova et al., 1999). Indeed, Sakai et al. (2008) and Yang et al. (2007) recently reported that microtubule organization was altered in root hairs of other ark1 alleles. Therefore, we asked whether root hairs of agd1-1 also had disrupted cytoskeletal organization, since both agd1 and ark1 often exhibited similar wavy root hair phenotypes. We crossed agd1-1 and ark1-4 with Arabidopsis plants expressing a GFP construct that labels microtubules (GFP-MBD; Marc et al., 1998) and F-actin (GFP-ABD2-GFP; Wang et al., 2008) and analyzed cytoskeletal organization in growing root hairs using confocal microscopy (Figs. 3–5).
Figure 3.
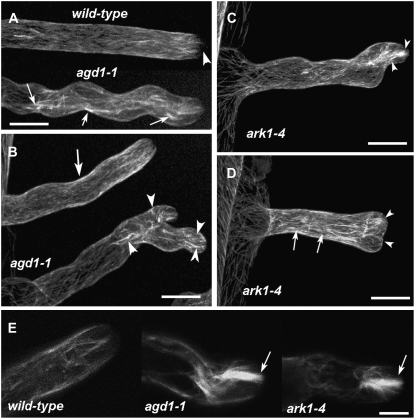
Microtubule organization in living wild-type, agd1-1, and ark1-4 roots as revealed by GFP-MBD. A, Microtubules in a straight-growing wild-type root hair are mostly longitudinal in orientation. The apical 5 μm of the tip appears devoid of distinct microtubules (arrowhead). Thick bundles of microtubules are observed throughout the length of a wavy root hair in agd1-1 mutants (arrows). B, In a root hair of agd1-1 that shows mild waving, the pattern of microtubules resembles that of the wild type (arrow). In a root hair with two tips, thick bundles of microtubules are abundant along the tips and regions of root hair branching (arrowheads). C and D, A root hair of ark1-4 with wavy growth (C) and a root hair that has developed a broad tip (D). Like agd1-1, bundles of microtubules are abundant along the length of the root hair (arrows in D) and at the extreme tip (arrowheads in C and D) of ark1-4. E, Single optical sections of root hair tips showing thick bundles of endoplasmic microtubules in agd1-1 and ark1-4 (arrows) but not in the wild type. The images in A to D are projections of 30 optical sections taken at 0.5-μm intervals. Bars = 20 μm (A–D) and 10 μm (E).
Figure 4.
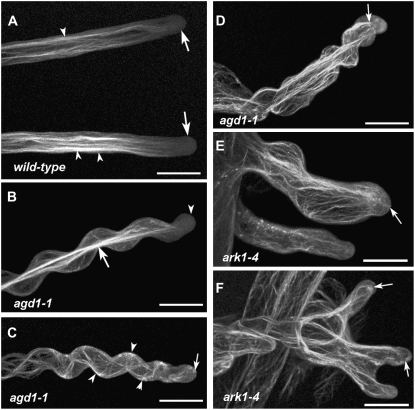
F-actin organization in living wild-type, agd1-1, and ark1-4 roots as revealed by the GFP-ABD2-GFP reporter. A, Straight-growing wild-type root hairs show longitudinal actin cables (arrowheads) and diffuse fluorescence at the apical 10 to 20 μm (arrows). B, A wavy root hair of agd1-1 shows diffuse labeling at the tip (arrowhead) and thick longitudinal bundles along the center of the cell (arrow). C, A root hair of agd1-1 showing extreme spiral growth. In this root hair, distinct F-actin bundles follow the outline of the cell (arrowheads) and extend to the very tip of the root hair (arrow). D, A root hair of agd1-1 showing extreme wavy growth. In this cell, longitudinal F-actin bundles extend very close to the tip (arrow). E, A wavy and swollen root hair of ark1-4 showing F-actin bundles at the extreme tip (arrow). F, A root hair of ark1-4 with three tips also exhibits distinct F-actin bundles that entirely occupy the tip regions of the cell (arrows). Images are projections of 30 to 40 optical sections taken at 0.5-μm intervals. Bars = 20 μm.
Figure 5.
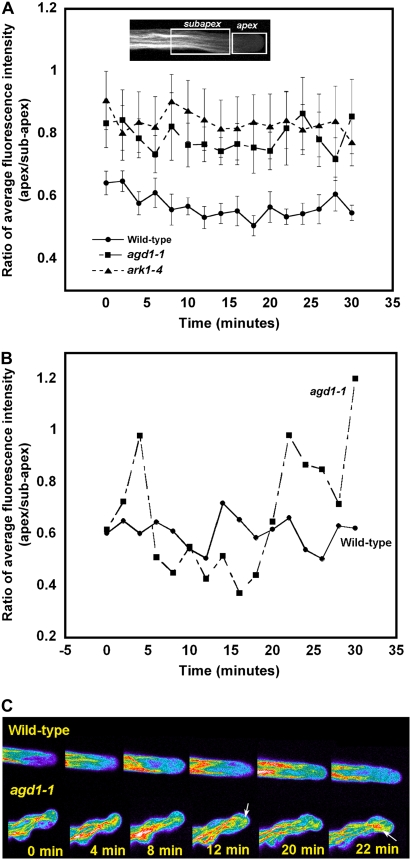
Quantification of F-actin organization in growing root hairs of agd1-1 and ark1-4. A, The average fluorescence intensity from the apex and base of 20 root hair movie sequences was acquired using Image J, as indicated by the white boxes in the inset. The ratio of apical and basal fluorescence was plotted at 2-min intervals for 30 min. The higher apex-to-base fluorescence ratio in agd1-1 and ark1-4 mutants is indicative of fluorescent F-actin bundles that occasionally extend into the extreme root hair apex. Values are means ± sd from 20 root hairs. B, Apical-to-basal fluorescence ratio from single wild-type and agd1-1 root hairs. Note the greater fluctuations in apical-to-basal fluorescence ratio in the root hair of agd1-1 compared with the wild type. C, Heat map of growing root hairs of the wild type and agd1-1 expressing GFP-ABD2-GFP. Intense patches of fluorescence, as indicated by red-white color (arrows), extend into the very tip of agd1-1 root hairs but not in wild-type root hairs. Blue-purple color indicates regions of lowest fluorescence.
We observed that the extent of cytoskeletal disruption in the agd1-1 and ark1-4 mutants was somewhat dependent on the severity of the root hair phenotype. A projection of several optical sections from a wild-type root hair expressing GFP-MBD showed predominantly longitudinal microtubules, consistent with a previous report (Van Bruaene et al., 2004). On the other hand, root hairs of agd1-1 that displayed regular wavy growth accumulated thick microtubule bundles along their length (Fig. 3A). In addition, root hairs of agd1-1 that formed branches had thick bundles of microtubules close to the point of branching. Root hairs of agd1-1 that displayed a milder wavy phenotype, however, contained predominantly longitudinal microtubules and a low incidence of thick microtubule bundles (Fig. 3B). Similar observations were made in root hairs of the ark1-4 mutant. Thick microtubule bundles were observed at the tips of root hairs that had changed their growth direction and those that developed a broad tip (Fig. 3, C and D). Differences in microtubule organization between the wild type and agd1-1 or ark1-4 were most apparent in single optical sections and time-lapse confocal movies of endoplasmic microtubules of growing root hairs. Whereas bundles of endoplasmic microtubules in wild-type root hairs were generally less dense, root hairs of both agd1-1 and ark1-4 mutants displayed thick bundles of endoplasmic microtubules at the tip (Fig. 3E; Supplemental Movies S4–S6).
Since the actin cytoskeleton is also relevant for sustaining tip growth, we next examined whether the organization of F-actin is affected in root hairs of agd1-1 and ark1-4 mutants. When decorated with the GFP-ABD2-GFP reporter described by Wang et al. (2008), F-actin bundles in growing wild-type root hairs are typically oriented parallel to the length of the root hair. These thick bundles of longitudinal F-actin did not extend into the apical 10 to 15 μm of growing root hairs. Within the apical region, labeling with the GFP-ABD2-GFP reporter was typically diffuse (Wang et al., 2008; Fig. 4A; Supplemental Movie S7). In agd1-1 root hairs, F-actin organization was clearly disorganized. The extent of actin disruption also was partly dependent on the severity of the root hair phenotype. For example, in growing root hairs with regular waves, the apical 10 to 15 μm of the tip was devoid of distinct F-actin bundles, similar to wild-type root hairs. Highly fluorescent longitudinal actin cables, however, appeared to congregate at the center of the root hair cell (Fig. 4B). In root hairs with more compressed waves and strong spiral growth, F-actin bundles typically followed the contour of the cell, and distinct F-actin cables were observed to protrude into the extreme tip (Fig. 4, C and D; Supplemental Movie S8). Similar disruptions in F-actin organization were detected in ark1-4 mutants, such that distinct F-actin bundles often extended into the extreme apex of swollen or branched root hairs (Fig. 4, E and F).
To quantify differences in F-actin organization in wild-type and mutant root hairs, we took the ratio of the average fluorescence intensity of the apical 20 μm of growing root hairs and a 30-μm area basal to the tip region (Fig. 5A, inset). Compared with the wild type, the average fluorescence ratio in agd1-1 and ark1-4 mutants was higher at all time points measured (Fig. 5A). Furthermore, measurements from an extremely wavy root hair of agd1-1 revealed strong oscillations in fluorescence ratio compared with a wild-type root hair (Fig. 5, B and C). Our tip-to-basal fluorescence ratio measurements were consistent with the observation that distinct actin bundles occasionally extended into the extreme root hair tips of agd1-1 and ark1-4 mutants.
Other evidence indicative of altered actin dynamics in agd1-1 and ark1-4 mutants was obtained using spinning disc confocal microscopy. Previously, we were able to augment our ability to image dynamic F-actin in Arabidopsis seedlings by a simple modification to the ABD2-GFP construct (Wang et al., 2008). We asked whether spinning disc confocal microscopy, which significantly minimizes sample bleaching, combined with the enhanced fluorescence brought about by our improved GFP F-actin reporter, would allow us to observe the finer details of actin dynamics at the tip of growing wild-type and mutant root hairs. We obtained time-lapse sequences of wild-type and mutant root hairs at 300-ms intervals. Despite the short time interval between image collections, we did not observe bleaching of GFP-labeled F-actin in the root hairs. In fact, the combination of spinning disc confocal microscopy and the enhanced fluorescence of our improved GFP-ABD2-GFP construct allowed us to resolve highly dynamic fine F-actin at the tip of growing wild-type root hairs, which was typically more difficult to resolve with conventional point scanning confocal microscopy (Supplemental Movie S9). In agd1-1 root hairs that exhibited extremely wavy growth, actin bundles at the tip, although still highly dynamic, were generally thicker than the fine F-actin arrays characteristic of wild-type root hairs (Supplemental Movie S10). Similar observations were made in wavy and branched root hair tips of ark1-4 mutants (data not shown).
Organelle Motility in agd1 Root Hairs
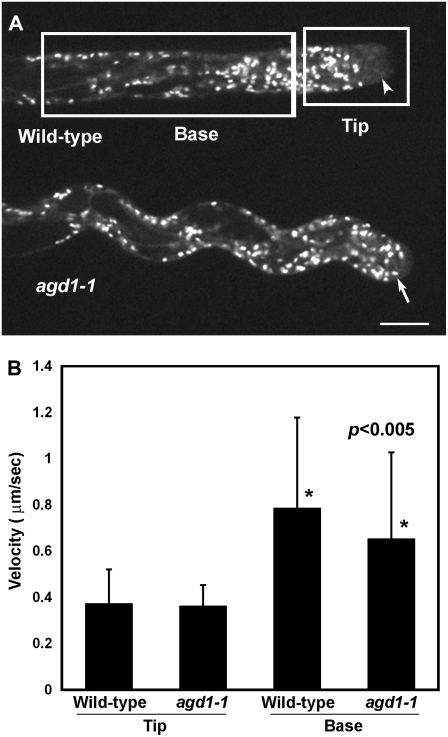
As mutants in the ARK1 gene have already been described (Yang et al., 2007; Sakai et al., 2008), we focused our efforts on further characterizing the cellular phenotypes of agd1 root hairs. Since root hairs of agd1-1 had obvious defects in cytoskeletal dynamics, we asked whether other cellular processes that are dependent on normal cytoskeletal function were affected. One important process regulated by the cytoskeleton is organelle transport. The movement of Golgi stacks in root hairs, for example, is regulated by class XI myosin motors that facilitate their transport along F-actin (Peremyslov et al., 2008). To evaluate whether the AGD1 mutation affected organelle trafficking in root hairs, we crossed agd1-1 mutants with plants expressing a yellow fluorescent protein (YFP) fused to the rat sialyl-transferase transmembrane domain (ST-YFP), which is used to fluorescently mark Golgi stacks in plants (Saint-Jore et al., 2002). There were no obvious gross morphological differences in the appearance of Golgi stacks between growing wild-type and agd1-1 root hairs (Fig. 6A). Furthermore, spinning disc confocal microscopy of growing wild-type and agd1-1 root hairs expressing ST-YFP revealed that Golgi stacks were highly dynamic along the length of the root hair (Supplemental Movies S11 and S12). Using Volocity classification software (Peremyslov et al., 2008), we tracked individual Golgi stacks from the apical 20 μm and the basal region of the root hair, as indicated by the white boxes in Figure 6A, to obtain Golgi velocity measurements. In both the wild type and agd1-1, the average velocity of Golgi stacks differed significantly between the apical and basal regions of the root hair, with mean velocity in the tip region being about 2-fold less than that of Golgi stacks at the basal region (Fig. 6B). There were no significant differences in the velocity of Golgi stacks at the tip region between the wild type and agd1-1. In the basal region, however, the average velocity of Golgi stacks in agd1-1 was slightly reduced compared with that in the wild type, and this difference was statistically significant (Fig. 6B; Student's t test; P < 0.005). More importantly, time-lapse spinning disc confocal microscopy revealed that Golgi stacks in root hairs of agd1-1 occasionally extended into the root hair tip, whereas Golgi stacks in wild-type root hairs maintained approximately a 10-μm distance from the extreme apex (Fig. 6A; compare Supplemental Movies S11 and S12).
Figure 6.
Golgi motility in living root hairs of the wild type and the agd1-1 mutant. A, Representative images of wild-type and agd1-1 root hairs expressing the Golgi marker ST-YFP. Whereas Golgi stacks are excluded from a 10-μm region at the tip of growing wild-type root hairs (arrowhead), Golgi stacks extend closer to the apical region in agd1-1 root hairs (arrow). Bar = 10 μm. B, Mean velocity of individual Golgi stacks from the apical 20 μm (tip) and basal region of wild-type and agd1-1 root hairs. Velocity measurements were taken from the areas delineated by the white boxes in A. Values are means + sd of about 200 individual Golgi stacks from root hairs of five to 10 independent seedlings (Student's t test; P < 0.005).
BFA Rescues the Wavy Root Hair Phenotype and Cytoskeletal Defects of agd1
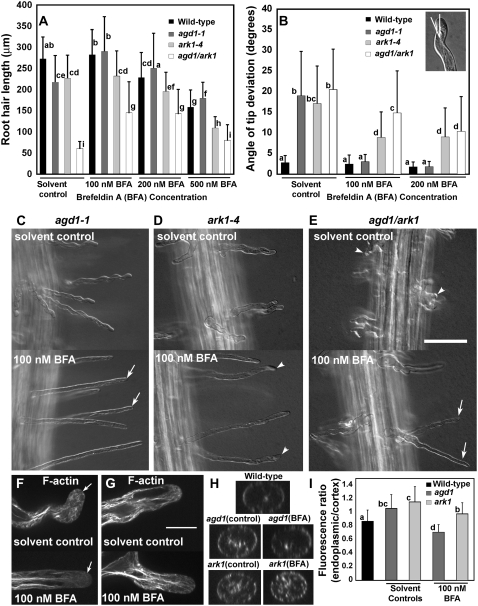
Given that AGD1 encodes a class I ARF-GAP, we hypothesized that root hair growth in agd1 might be differentially affected by inhibitors of vesicle trafficking. One compound that has been used extensively for such studies is the fungal macrolide BFA. BFA is known to inhibit the activity of ARF-GEFs in mammalian cells, and there is evidence that similar targets for BFA exist in plants (Nebenfuhr et al., 2002). We found that 100 nm BFA did not inhibit root hair growth of wild-type seedlings, but BFA concentrations above 200 nm did. Strikingly, we found that average root hair length of agd1-1 increased significantly when germinated in 100 nm BFA (Fig. 7A). More importantly, the increase in average root hair length of agd1-1 mutants upon exposure to 100 nm BFA coincided with root hairs reverting to the straight growth pattern characteristic of wild-type root hairs (Fig. 7, B and C). We quantified the effect of BFA on root hair waviness by measuring the angle of tip deviation from the main axis of the root hair (Fig. 7B, inset). The average angle of tip deviation in root hairs of agd1-1 decreased from 19° to 3° when grown on 100 to 200 nm BFA, which was similar to that of untreated and BFA-treated wild-type root hairs (Fig. 7B). Furthermore, confocal microscopy showed that the abundant bundles of endoplasmic microtubules and F-actin in the tips of agd1-1 root hairs were reduced by incubation in BFA (Fig. 7F; Supplemental Movie S13). When BFA was washed out, agd1-1 root hairs reverted back to the wavy growth pattern (data not shown).
Figure 7.
Effects of BFA on root hair morphology and cytoskeletal organization of agd1-1 and ark1-4 mutants. A, Average root hair length of seedlings germinated and maintained in BFA for 4 d. Note that incubation in 100 nm BFA results in a statistically significant increase in mean root hair length of agd1-1 single and agd1 ark1 double mutants but not on that of the ark1-4 mutant and the wild type. B, Average angle of root hair tip deviation as measured by white lines in the inset. Note that the angle of tip deviation of agd1-1 but not ark1-4 or agd1 ark1 was restored to wild-type values. Values are means of 100 to 200 root hairs from 15 to 20 independent seedlings for each genotype. Error bars indicate sd. Means with different letters are significantly different (P < 0.005; Tukey's test). C to E, Representative images of root hairs from 4-d-old agd1-1, ark1-4, and agd1 ark1 seedlings germinated in Murashige and Skoog medium with or without 100 nm BFA. Root hairs of agd1-1 are completely straightened when grown on BFA (arrows in C). Root hairs of ark1-4 are only partially reversed by incubation in BFA, as is evident from the persistence of kinks and bends in the cell (arrowheads in D). Root hairs of agd1 ark1 are more deformed than those in single agd1 and ark1 mutants (arrowheads in E). Incubation of agd1 ark1 in BFA results in an increase in average root hair length (A), but deformed root hairs are still apparent (arrows in E). F and G, Representative single optical confocal sections showing F-actin in root hair tips of agd1-1 (F) and ark1-4 (G). Note that incubation in BFA diminishes the bundling of F-actin in the apex of agd1 root hairs (arrow in F) but not in ark1 (G). H, Computer-generated cross sections 5 μm from the root hair apex. Note the more abundant endoplasmic microtubules in agd1 and ark1 in root hair tips of seedlings grown in the control solution and in ark1 mutants exposed to BFA. I, Ratio of fluorescence of endoplasmic to cortical microtubules. BFA treatment reduces the fluorescence ratio of agd1 more strongly than ark1. Data are means from 25 to 30 root hairs + sd. Means with different letters are significantly different (P < 0.005; Tukey's test). Bar = 50 μm (for C–E) and 20 μm (F and G).
To quantify the effect of BFA on cytoskeletal organization, we focused on studying further the bundling of endoplasmic microtubules at the root hair tip, since means to evaluate this population of microtubules were established recently (Sakai et al., 2008). We projected Z-stacks obtained with the spinning disc microscope using Volocity classification software to generate cross sections at a region 5 μm from the root hair tip (Fig. 7H). Mean fluorescence intensity ratios of endoplasmic and cortical microtubules from these computer-generated cross sections were obtained following the methods of Sakai et al. (2008). The ratio of endoplasmic to cortical microtubule fluorescence in agd1-1 root hairs was higher than that of wild-type root hairs (Fig. 7I), consistent with visual observations of abundant endoplasmic microtubules at the root hair tip of agd1-1 (Figs. 3E and 7H; Supplemental Movie S5). When grown on 100 nm BFA, the mean ratio of endoplasmic to cortical microtubule fluorescence in agd1-1 root hairs decreased to a value slightly less than that of wild-type root hairs (Fig. 7I). These measurements were consistent with visual observation that endoplasmic microtubule organization in untreated agd1-1 root hairs reverted to that of wild-type root hairs after BFA treatment (Fig. 7H; Supplemental Movie S13). BFA at 100 to 200 nm did not affect the overall organization of Golgi bodies in wild-type root hairs (data not shown). However, many agd1-1 root hairs treated with BFA displayed wild-type Golgi distribution consistent with their reversal to straight growth (Supplemental Movie S14).
We then asked whether BFA had similar effects on the root hair phenotype of the ark1-4 mutant. In contrast to agd1-1, the root hair length of ark1-4 did not increase in response to BFA (Fig. 7A). Although the wavy phenotype of ark1-4 was somewhat dampened by BFA, as is evident from a reduction in the frequency of waves along the root hair, ark1-4 root hairs, unlike agd1-1 root hairs, did not completely revert to the wild-type phenotype. When incubated in BFA, many root hairs of ark1-4 still contained branches and occasional kinks at the tip (Fig. 7D). The partial rescue of ark1-4 root hairs by BFA was manifested as a reduction in the angle of root hair tip deviation from 17° to 9° (Fig. 7B). Furthermore, in contrast to agd1-1, abundant F-actin and endoplasmic microtubule bundles in root hair tips of ark1-4 persisted despite BFA treatment (Fig. 7, G–I).
Like single mutants, when agd1 ark1 double mutants were examined they exhibited no other obvious plant phenotype (data not shown). However, we found that root hair growth of agd1 ark1 double mutants was more strongly inhibited compared with single mutants. The average root hair length of agd1 ark1 was only about 20% that of controls and 25% that of single mutants (Fig. 7A). Furthermore, root hairs of agd1 ark1 double mutants were more severely deformed, as is evident from the formation of root hairs with short branches that ceased elongating soon after initiation (Fig. 7E). The tips of these short root hairs also had the tendency to bend, resulting in a tip angle of deviation comparable to that of single agd1 and ark1 mutants (Fig. 7B). When germinated in 100 to 200 nm BFA, average root hair length of agd1 ark1 double mutants increased by about 50% (Fig. 7, A and B), but root hairs were still characterized by swollen bases, kinks, and waves (Fig. 7, B and E). The tip angle of deviation of agd1 ark1 double mutants, particularly after incubation in 200 nm BFA, was similar to that of ark1-4 single mutants (Fig. 7B).
AGD1 Partly Localizes to the Endocytic Marker FM4-64, and ARK1 Associates with Microtubules
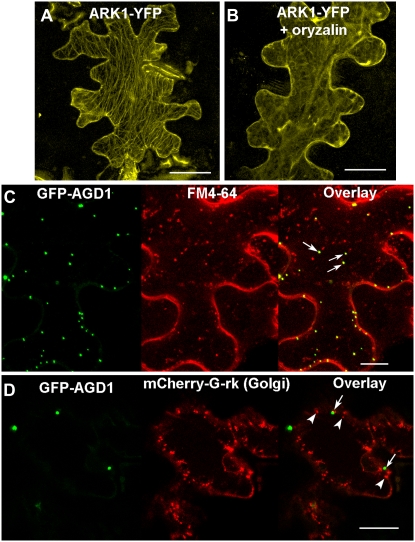
Cosedimentation assays have shown that the N-terminal motor domain of ARK1 binds strongly to polymerized tubulin and the C-terminal armadillo repeat binds weakly to polymerized actin (Yang et al., 2007). The in vivo localization of ARK1, however, has not yet been reported. Therefore, we created translation fusions of YFP to the C terminus of full-length ARK1 under the control of the 35S cauliflower mosaic virus (35S) promoter (35S∷ARK1-YFP). This construct was biolistically bombarded into tobacco (Nicotiana tabacum) leaf epidermal cells, and transformed cells were imaged using confocal microscopy. The 35S∷ARK1-YFP construct localized to filamentous structures that were reminiscent of microtubules (Fig. 8A). To verify that the 35S∷ARK1-YFP-labeled filaments were microtubules, leaf cells were incubated in either oryzalin or latrunculin B, which disrupt microtubules or F-actin, respectively. Incubation in oryzalin but not latrunculin B caused the dissipation of the filamentous structures in ARK1-YFP-expressing cells, indicating that ARK1 indeed localized to microtubules (Fig. 8B; data not shown).
Figure 8.
Transient localization of ARK1 and AGD1 in tobacco epidermal cells. A, ARK1-YFP decorates filamentous structures in the cell. B, Incubating cells in the microtubule-depolymerizing drug oryzalin disrupts the filamentous labeling pattern of ARK1-YFP. C, Tobacco leaves inoculated with 35S∷GFP-AGD1 and counterstained with FM4-64. GFP-AGD1 localizes to punctate bodies of varying size that partly overlap with internalized FM4-64 vesicles (arrows). D, Tobacco leaf epidermal cells transiently expressing GFP-AGD1 and mCherry-G-rk. GFP-AGD1 bodies (arrows) do not colocalize with Golgi stacks (arrowheads). Bars = 50 μm (A and B) and 20 μm (C and D).
We also created GFP translational fusions to the full-length AGD1 protein under the control of the 35S promoter. The 35S∷GFP-AGD1 fusion transiently expressed in tobacco epidermal cells decorated punctate structures of varying sizes that resembled Golgi stacks (Fig. 8C). To determine whether these fluorescent structures represented Golgi stacks, we cobombarded the GFP-AGD1 with mCherry-G-rk, which localizes to the Golgi based on a fusion with the transmembrane domain of α-1,2-mannosidase I (Nelson et al., 2007). We also incubated leaves that were agroinfiltrated with GFP-AGD1 with the lipophilic styryl dye FM-4-64, which labels diverse components of the endocytic pathway (Müller et al., 2008). We found that the punctate GFP-AGD1 structures partially overlapped with FM4-64-labeled vesicles but not with Golgi stacks (Fig. 8, C and D).
DISCUSSION
A forward genetics approach enabled us to identify two Arabidopsis mutants with wavy root hair growth but with no other obvious phenotype. The phenotype of these mutants was reminiscent of root hairs treated with microtubule-disrupting drugs (Bibikova et al., 1999), prompting us to hypothesize that the genetic lesions in these mutants might encode proteins that are important for cytoskeletal regulation. Indeed, TAIL-PCR revealed that one of our mutants had a T-DNA insertion in ARK1, which encodes a kinesin microtubule motor protein. At the time we were characterizing our mutants, two groups had independently isolated mutants in the ARK1 gene, which showed similar defects in root hair morphology as our mutants (Yang et al., 2007; Sakai et al., 2008). Although in vitro biochemical studies demonstrated that the N terminal motor domain of ARK1 binds to microtubules (Yang et al., 2007), our transient localization of ARK1-YFP to oryzalin-sensitive filaments provides supporting evidence that ARK1 is a microtubule-associated protein in vivo (Fig. 8, A and B).
Map-based cloning of our other wavy root hair mutant revealed a disruption in AGD1, a gene encoding an ARF-GAP (Vernoud et al., 2003). ARF-GAPs are negative regulators of ARFs, small GTP-binding proteins that mediate the formation of membrane trafficking intermediates in the cell. In the classical model of ARF function, the active ARF-bound GTP recruits coat proteins to membranes to form protein-coated vesicles. Hydrolysis of GTP by ARF-GAP inactivates ARF, triggering the dissociation of coat proteins, which then allows vesicles to fuse with acceptor membranes (Nie and Randazzo, 2007). In Arabidopsis, ARF-GAPs belong to a 15-member family that is further subdivided into four classes (Vernoud et al., 2003). AGD1 encodes a class I ARF-GAP, which is closely related to the multidomain-containing mammalian AZAP-type ARF-GAPs (Vernoud et al., 2003; Inoue and Randazzo, 2007). Among the Arabidopsis class I ARF-GAPs, only AGD3 has been demonstrated to function in plant development by affecting auxin-dependent vascular differentiation (Koizumi et al., 2005; Sieburth et al., 2006). Also, the overexpression of a rice (Oryza sativa) ARF-GAP (OsAGAP) hampered root development by interfering with the normal trafficking of auxin influx carriers (Zhuang et al., 2006). Here, we show that an additional function for class I ARF-GAPs in plants is in the maintenance of root hair growth directionality. The identification of two independent SALK T-DNA insertion alleles for AGD1 that exhibited wavy root hair phenotypes indicates that altered AGD1 expression is the cause of the root hair defect in our mutant and that it is not due to secondary mutations elsewhere in the genome.
The function of AGD1 in tip growth is likely specific to root hairs, since we did not observe any defects in pollen germination or pollen tube growth in agd1 mutants (data not shown). In a study of expression patterns of the four class I ARF-GAPs using real-time RT-PCR, AGD1 was not detected in hypocotyls and cotyledons and had very low expression in roots, siliques, and leaves (Sieburth et al., 2006). The low expression of AGD1 is also apparent from an in silico analysis of AGD1 from the Genevestigator database, which collates data from Arabidopsis Affymetrix Gene Chip experiments (Zimmermann et al., 2004; data not shown). As speculated by Sieburth et al. (2006), the low abundance of AGD1 likely reflects its function in only a few cell types, and the phenotype we observed in agd1 mutants strongly indicates that AGD1 might operate specifically in root hairs. It was shown recently that AGD10, a class II ARF-GAP, also is important for tip growth in Arabidopsis root hairs. The root hair phenotype of agd10, however, was different from that of agd1 in that agd10 had very short bulbous root hairs and also exhibited defects in pollen germination (Song et al., 2006). This indicates that some of the mechanisms by which AGD10 regulates tip growth in plants might be distinct from those of AGD1. AGD10 is more closely related to the mammalian ARF-GAP1, which has a simpler domain organization than the AZAP class of ARF-GAPs to which AGD1 belongs. AGD10 and AGD7, another class II ARF-GAP, only contain a GAP domain and localize to Golgi stacks (Vernoud et al., 2003; Song et al., 2006; Min et al., 2007), in agreement with mammalian ARF-GAP1 localization (Liu et al., 2005). AGD1 and AGD3, on the other hand, partially localized to FM4-64-labeled endocytic vesicles and the trans-Golgi network (Fig. 8C; Koizumi et al., 2005). The partial localization of AGD1 to FM4-64-labeled bodies that we observed here is consistent with the endosomal localization of the closely related animal AZAPs (Jackson et al., 2000; Nie et al., 2002). The differences in localization, domain organization, and possibly ARF substrate preference between AGD1 and AGD10 could likely explain the different nature of tip growth defects caused by their respective mutants. Whereas AGD10 activates the GTPase-hydrolyzing activity of plant ARF1 (Song et al., 2006), the precise plant ARF proteins that are substrates of the class I ARF-GAPs remain to be determined.
AGD3, which has similar domain organization to AGD1, has been shown to activate the GTPase-hydrolyzing activity of yeast ARF1p (Koizumi et al., 2005). Although we have yet to assay for AGD1 GAP activity, the possibility that the root hair phenotype of agd1 results from modified endogenous ARF function is evident from our results with BFA. Low concentrations of BFA not only caused a statistically significant increase in root hair length but also caused wavy root hairs of agd1 to grow straight and revert to wild-type cytoskeletal organization (Fig. 7). A likely explanation for this observation is that loss of AGD1 function leads to an increase in the active GTP-bound form of an AGD1-dependent ARF in the cell. Since BFA is known to inhibit endogenous ARF-GEF activity, which activates ARF by facilitating the exchange reaction of GDP for GTP (Bos et al., 2007), BFA treatment could conceivably cause the buildup of GDP-bound (inactive) ARF in the cell. Therefore, despite the absence of AGD1 activity, agd1 root hairs treated with BFA reverted to straight growth, because the opposing biochemical reaction triggered by a yet to be identified BFA-sensitive ARF-GEF in the cell was inhibited. In this regard, it is worth noting that AGD3 knockouts, which display discontinuous venation patterns (Koizumi et al., 2005; Sieburth et al., 2006), have roughly opposite phenotypes to mutants of the ARF-GEF, GNOM, which display extensive venation (Geldner et al., 2003). If the effect of BFA on agd1 root hairs is due to inhibited ARF-GEF activity counteracting the loss of AGD1, one would predict that overexpression of a constitutively active AGD1-dependent ARF will lead to similar wavy root hair phenotypes as agd1. Arabidopsis has a number of ARF and ARF-like proteins, some of which are expressed in roots (Genevestigator; Zimmermann et al., 2004). Future studies will attempt to identify which among these ARF or ARF-like proteins are substrates of AGD1.
Since the overexpression of dominant negative ROP2 induced the formation of wavy root hairs similar to that of agd1 mutants (Jones et al., 2002) and modified ARF1 activity disrupted normal root hair development by inhibiting the polar localization of ROP2 (Xu and Scheres, 2005), the root hair phenotype of agd1 could partly be explained by altered ROP signaling. Also, RabA4b, whose root tip localization is actin dependent (Preuss et al., 2004), might be misdirected in agd1. Interestingly, ROP2-induced root hair defects were accompanied by disrupted apical F-actin organization similar to that observed in agd1 mutants (Figs. 4 and 5; Jones et al., 2002). Collectively, these observations reinforce the concept of cross talk between the different small GTPases in defining the highly polarized growth of root hairs by impinging on cytoskeletal function (Xu and Scheres, 2005; Yang et al., 2007). Such cross talk with other signaling molecules is likely facilitated by the other protein domains of AGD1. For instance, the PH domain of class I ARF-GAPs, as demonstrated in the closely related AGD3, binds to phosphatidylinositol-4,5-bisphosphate (Koizumi et al., 2005). In mammalian systems, GAP activity of the AZAP-type ARF-GAPs is stimulated by phosphoinositide binding to the PH domain (Jackson et al., 2000). In this regard, overexpression or down-regulation of a type B phosphatidylinositol-4-phosphate 5-kinase 3, which regulates endogenous phosphatidylinositol-4,5-bisphosphate levels, leads to deformed root hairs (Kusano et al., 2008; Stenzel et al., 2008). Further support for the interdependence of lipid signaling and small GTPases in root hair growth comes from the observation that RabA4b recruits phosphatidyl 4-OH kinase to the growing root hair tip and that knockouts of this gene induced the formation of wavy root hairs similar to agd1 (Preuss et al., 2006). More recently, the root hair defective4 (rhd4) mutant, which has short, branched, and bulged root hairs, was shown to be disrupted in a gene encoding a phosphoinositide phosphatase that regulates the levels of phosphatidylinositol-4-phosphate (Thole et al., 2008). One possible effect of such changes in steady-state levels of phosphoinositides is a modification in the activity of AGD1. For technical reasons, we have been unsuccessful in our initial attempts to express recombinant AGD1 in a heterologous system to assay for lipid binding. Identification of the type and specificity of phosphoinositide binding to AGD1 will shed light on how AGD1 interacts with lipid signaling to specify polarized root hair growth. We are currently crossing our agd1 mutants with fluorescent markers for RabA4b and ROPs to determine whether targeting of these tip growth effectors is affected in the wavy root hairs of agd1.
In animal cells, AZAP-type ARF-GAPs, by virtue of their effects on membrane trafficking, induce a remodeling of the actin cytoskeleton (Nie et al., 2002; Randazzo and Hirsch, 2004; Randazzo et al., 2007). Modification in membrane structure by AZAP has been shown to be mediated in part by the BAR domain at the N terminus of the protein (Nie et al., 2006) and has been proposed to facilitate the binding of other classes of small GTPases (Habermann, 2004). Here, live cell imaging allowed us to gain additional insight into the underlying cytoskeletal defects that led to the root hair phenotype of agd1. Extensive bundles of F-actin periodically extended into the extreme apex of growing agd1 root hairs (Figs. 4 and 5; Supplemental Movies S8 and S10). This pattern of cytoskeletal reorganization was in contrast to that of growing wild-type root hairs, which typically display a region at the tip with diffuse or fine arrays of F-actin (Wang et al., 2008; Supplemental Movies S7 and S9). The abnormally high levels of F-actin bundles have a number of consequences that could explain the root hair phenotype of agd1. One possibility is the disruption of organelle trafficking in the mutant root hair. Indeed, agd1 displayed Golgi stacks extending into the very tip of growing root hairs and slightly reduced Golgi stack velocity at the root hair base (Fig. 6). Recently, knockouts to a class XI myosin, an actin-based molecular motor, resulted in short root hairs and reduced Golgi motility (Peremyslov et al., 2008). The impact of the AGD1 mutation on Golgi trafficking (i.e. the protrusion of Golgi stacks into the extreme root hair apex) was similar to that observed in depolarized pollen tubes expressing a catalytically inactive phospholipase C (Dowd et al., 2006). The movement of organelles to regions of growing root hairs where they are typically excluded could affect targeted delivery and localized recycling of vesicles, leading to differential tip growth (Ovecka et al., 2005). Alternatively, disrupted F-actin dynamics could interfere with the tip recruitment of lipid signaling components that bind to actin, such as phosphatidylinositol phosphate kinase 1 (Davis et al., 2007) or trafficking of RHD2, the ROS-generating NADPH oxidase, which was recently shown to be dependent on actin for proper root hair tip localization (Takeda et al., 2008). As alterations in the metabolism of phosphoinositide lipid mediators have been shown to affect root hair growth (Bohme et al., 2004; Vincent et al., 2005; Kusano et al., 2008; Stenzel et al., 2008; Thole et al., 2008), the disruption of apical F-actin dynamics at the root hair tip, regardless of whether it is a direct result of the loss of AGD1 function, continues to point to an elaborate system of feedback regulation between lipids, small GTPases, and the cytoskeleton in the maintenance of tip growth (Nibau et al., 2006; Kost, 2008).
Another consequence of the loss of AGD1 function was a modification in microtubule organization, as is evident from thick bundles of endoplasmic microtubules at the root hair tip of agd1. Intriguingly, the pattern of microtubule distribution in agd1 was similar to that of ark1 root hairs, confirming previous observations in other ark1 alleles (Fig. 3; Supplemental Movie S6; Sakai et al., 2008). In Arabidopsis root hairs, endoplasmic microtubules have been proposed to function as microtubule nucleation complexes (Van Bruaene et al., 2004). Thus, ARK1 and AGD1 could conceivably be part of a signaling pathway that regulates microtubule nucleation, and their altered activity could result in abnormally high levels of polymerized tubulin at the root hair tip, leading to unstable growth. The extensive bundles of endoplasmic microtubules of agd1 mutants could also explain the abnormal Golgi movement at the root hair tip. In this regard, it was shown that Golgi stacks can interact with both microtubules and actin and that certain kinesins facilitate the dispersal of Golgi stacks in plant cells (Lu et al., 2005). Since ark1 root hairs exhibited bundled arrays of F-actin at the tip similar to agd1, both AGD1 and ARK1 may have common downstream targets that regulate cytoskeletal dynamics. The more severe root hair phenotype of agd1 ark1 double mutants, however, indicates that ARK1 and AGD1 are components of separate but possibly convergent signaling pathways that impinge on cytoskeletal function to specify root hair growth orientation. The complete reversal of the wavy root hair phenotype of agd1 by low doses of BFA suggests that AGD1 affects tip growth and the cytoskeleton via a BFA-sensitive pathway involving unknown ARFs. On the other hand, since BFA did not completely reverse the root hair and cytoskeletal phenotypes of ark1, the pathway in which ARK1 modulates tip growth and cytoskeletal organization occurs in part via a BFA-independent process. It is possible that AGD1, by virtue of its impact on membrane and actin remodeling, indirectly modulates ARK1 trafficking, since ARK1 was shown to bind F-actin in vitro (Yang et al., 2007). Such an effect on ARK1 trafficking in agd1 could in turn affect microtubule polymerization, explaining the very similar nature of endoplasmic microtubule bundling at the root hair tips of both agd1 and ark1 mutants. Additional studies using functional ARK1 or AGD1 fluorescent protein fusions will be needed to determine the manner in which ARK1 and AGD1 interact to specify root hair orientation. In the future, it will be important to compare the cytoskeletal organization in root hairs treated with cytoskeleton-disrupting drugs with that of agd1 and ark1 root hairs to determine whether the cytoskeletal defects in the mutant root hairs are the direct result of the loss of AGD1 or ARK1 function.
In summary, we present new evidence showing that AGD1, a class I ARF-GAP protein, is essential for maintaining straight root hair growth in Arabidopsis. One function of AGD1 is in the maintenance of normal cytoskeletal turnover in the root hair, and this process is facilitated by a BFA-dependent pathway involving ARF and ARF-GEF proteins. We propose that the multidomain structure of AGD1 plays similar roles in plants to their animal AZAP-type ARF-GAP counterparts, by mediating cross talk and feedback regulation between phosphoinositides, small GTPases, and the cytoskeleton in polarized root hair growth.
MATERIALS AND METHODS
Isolation of Root Hair Mutants with Altered Growth Direction and Molecular Identification of the Disrupted Genes
An Arabidopsis (Arabidopsis thaliana) Col-0 T-DNA mutant seed stock, CS31100, from the Arabidopsis Biological Resource Center was used to screen root hairs with altered growth direction. Upon confirmation of the root hair phenotype in the progeny, mutants were backcrossed to Col-0 at least two times. The gene of the T-DNA insertion in wrh2(ark1) was identified by TAIL-PCR essentially as described by Liu et al. (1995).
For map-based cloning, homozygous wrh1(agd1) was crossed to the Landsberg erecta ecotype. wrh1(agd1) mutant plants from the resulting F2 seedlings were phenotyped based on the occurrence of wavy root hairs. Publicly available simple sequence length polymorphism and cleaved amplified polymorphic sequences markers were used to map the wrh1(agd1) loci (Lukowitz et al., 2000). We mapped the WRH1 locus to a 95-kb region on chromosome 5. Candidate gene sequencing revealed a 46-bp deletion in the annotated gene At5g61980 (Supplemental Fig. S1). There were no other mutations in this region. Additional T-DNA knockout lines, namely, agd1-2 (SALK_036034), agd1-3 (SAIL_819_C10), ark1-1 (SALK_081412), and ark1-2 (SALK_035063), were obtained from the Arabidopsis Biological Resource Center.
Complementary DNA Isolation and Semiquantitative RT-PCR Analysis
Total RNA was extracted from roots of 2-week-old wild-type Arabidopsis (Col-0) and mutant agd1 seedlings using the RNeasy plant mini-kit (Qiagen). RT was carried out using 500 ng of total RNA with the Omniscript RT kit (Qiagen) following the manufacturer's protocol to obtain a pool of cDNAs. The full-length wild-type AGD1 and mutant agd1 cDNA were amplified using gene-specific primers (see below) and sequenced completely.
For semiquantitative RT-PCR, root cDNA was prepared from the wild type and mutants as described above. Primers used for AGD1 were AGD1-RT-f (5′-AAGGTTGCAGAAAATACAC-3′) and AGD1-RT-r (5′-TGATCCTGTGCATTCTCTGC-3′), to generate a 1,135-bp product. For ARK1, the primers used were ARK1-RT-f (5′-AGGATCAGCAGAATCTGGAGCTC-3′) and ARK1-RT-r (5′-TCAGCGGCTAGATTAGCAAGGAC-3′), to generate a 195-bp product. Arabidopsis translation initiation factor primers EIF4A2 forward (5′-GAATCTTCTTAGGGGTATCTATGC-3′) and EIF4A2 reverse (5′-CTATGACATATTCCAGCTTCTCCC-3′) were used as a control.
Microscopic Analysis of Root Hair Growth
Seeds of mutants were germinated on 48- × 64-mm coverslips layered with 0.5% phyta-agar supplemented with half-strength Murashige and Skoog medium as described by Bibikova et al. (1999). After 4 d, coverslips containing the seedlings were transferred directly onto the stage of an inverted Nikon TE300 compound microscope equipped with differential interference contrast optics. Movies and still images of growing root hairs were acquired using a Hamamatsu C2400-75i camera running on Metamorph 6.3 image-acquisition software (Molecular Devices).
Imaging of Cytoskeletal Organization and Golgi Stack Motility
agd1-1 and ark1-4 mutants were crossed with wild-type Arabidopsis plants harboring GFP constructs that label microtubules and F-actin (Marc et al., 1998; Wang et al., 2008). Seeds of homozygous lines expressing the cytoskeletal reporters were germinated in the coverslip system described above. Root hairs of 3- to 4-d-old seedlings were imaged using a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems) or a Perkin-Elmer UltraView ERS spinning disc confocal microscope (Perkin-Elmer Life and Analytical Sciences) equipped with 63× water-immersion objectives. GFP was excited using the 488-nm line of the argon laser, and emission was detected at 510 nm. For quantification of F-actin organization, the average fluorescence from the apical 20-μm and basal 30-μm regions immediately adjacent to the tip was obtained using the rectangular selection marquee of Image J software (Fig. 5A). Measurements were collected from individual movie frames of growing root hairs that were each separated by a 2-min interval.
For studies of Golgi trafficking, agd1-1 was crossed with plants expressing ST-YFP (Saint-Jore et al., 2002). Time-lapse sequences of growing mutant and wild-type root hairs were obtained with a spinning disc confocal microscope at 1-s intervals and 60-s duration. Individual Golgi stacks were tracked using Volocity 3.70 classification software (Improvision). Velocity measurements were obtained from about 200 individual Golgi stacks from growing root hairs of five to 10 independent seedlings. Statistical analyses of Golgi velocity were conducted using SPSS 15.0 (SPSS).
BFA Assays
Wild-type and mutant seedlings were germinated in the layered agar-coverslip system described above, but this time the medium was supplemented with 100 to 500 nm BFA or with the equivalent volume of solvent control solution. After approximately 4 to 5 d of germination, digital images of the root hairs were taken using a Nikon SMZ1500 stereomicroscope equipped with a DXM1200 camera (Nikon). The length and angle of tip deviation of the root hairs from the digital images were measured using Image J 1.36b software (Wayne Rasband, National Institutes of Health). For the quantification of endoplasmic microtubules, wild-type and mutant seedlings expressing GFP-MBD were germinated on BFA for 4 d. Z-stacks from 25 to 30 growing root hairs from eight to 10 independent seedlings were acquired using the Perkin-Elmer spinning disc confocal microscope, and computer-reconstructed transverse sections of the root hair tip were generated using Volocity classification software. Laser power and camera gain settings were kept constant for each treatment. Average fluorescence from the cortical region and the endoplasmic region were measured using Image J software as defined by Sakai et al. (2008). One-way ANOVA was used to test statistical significance, and Tukey's honestly significant difference test was used for multiple comparison of means. Statistical analyses were conducted using SPSS software.
Generation of Expression Constructs
The plant expression vector pCAMBIA (CAMBIA) was used for expression assays. All of the constructs were driven by the cauliflower mosaic virus 35S promoter.
The full-length AGD1 cDNA was amplified using the wild-type cDNA pool as described above and primers AGD1-F-HindIII (5′-CTAAGCTTCATTTCGCCAAGCTCGATGATTCTC-3′) and AGD1-R-XhoI (5′-GACTCGAGTCATCTTTTGGAGTCTGTTAATAAAGC-3′). The resulting product was fused to the C terminus of GFP to generate the 35S∷GFP-AGD1. For full-length ARK1, primers ARK1-F-SalI (5′-ATGTCGACATGAGTTCGTCAAATTCCTCCTCC-3′) and ARK1-R-BamHI (5′-CTGGATCCTCGCTTGAGAAGTAAGGGTTTG-3′) were used to amplify the cDNA, and this product was fused to the N terminus of YFP to generate the 35S∷ARK1-YFP.
Transient Localization of AGD1 and ARK1 by Particle Bombardment and Direct Agrobacterium Infiltration
A total of 0.5 μg of plasmid DNA containing the 35S∷ARK1-YFP fusion construct was mixed with 25 μL of an aqueous suspension containing 1.6-μm gold particles. The gold-DNA suspension was mixed with moderate vortexing in the presence of CaCl2 and spermidine. After brief centrifugation, the plasmid-coated gold particles were washed and resuspended in ethanol. The gold was spread onto plastic carrier discs for biolistic bombardment of tobacco (Nicotiana tabacum) epidermal cells using a Bio-Rad 1000/HE particle delivery system. For cobombardment of GFP-AGD1 and mCherry-G-rk (Nelson et al., 2007), both plasmids were mixed in the same gold suspension. After 24 h, epidermal cells expressing the constructs were selected using a fluorescence stereomicroscope, and images were acquired using a Leica confocal microscope.
For localization of 35S∷GFP-AGD1 via leaf infiltration, the constructs were transformed in Agrobacterium tumefaciens strain C58C1. Overnight 5-mL cultures of each construct were used to inoculate 40 mL of Luria-Bertani culture plus antibiotics, 10 mm MES, and 20 μm acetosyringone. The overnight bacterial culture was centrifuged, and the pellet was resuspended in medium consisting of 10 mm MES, 10 mm MgCl2, and 150 μm acetosyringone to an optical density at 600 nm of approximately 0.3. After 2 h of incubation at room temperature, the bacterial solution was infiltrated into the abaxial side of expanded 3-week-old Nicotiana leaves using a 5-mL syringe. The leaves remained attached to the plants for 48 h. The infiltrated leaves were removed and counterstained with FM4-64 prior to imaging with a confocal microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_115344 (ARK1) and NM_125591 (AGD1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and amino acid sequence of AGD1.
Supplemental Movie S1. Time-lapse imaging of wild-type root hairs. Movie shows images taken every 1 min. Total elapsed time is 60 min.
Supplemental Movie S2. Time-lapse imaging of agd1-1 root hairs. Movie shows images taken every 1 min. Total elapsed time is 60 min.
Supplemental Movie S3. Time-lapse imaging of ark1-4 root hairs. Movie shows images taken every 1 min. Total elapsed time is 150 min.
Supplemental Movie S4. Confocal time-lapse imaging of a wild-type root hair expressing a microtubule marker (GFP-MBD). Movie shows images taken every 2 min. Total elapsed time is 60 min.
Supplemental Movie S5. Confocal time-lapse imaging of an agd1-1 root hair expressing GFP-MBD. Movie shows images taken every 2 min. Total elapsed time is 82 min.
Supplemental Movie S6. Confocal time-lapse imaging of an ark1-4 root hair expressing GFP-MBD. Movie shows images taken every 2 min. Total elapsed time is 60 min.
Supplemental Movie S7. Confocal time-lapse imaging of a wild-type root hair expressing GFP-ABD2-GFP. Movie shows images taken every 2 min. Total elapsed time is 84 min.
Supplemental Movie S8. Confocal time-lapse imaging of an agd1-1 root hair expressing GFP-ABD2-GFP. Movie shows images taken every 2 min. Total elapsed time is 106 min.
Supplemental Movie S9. Spinning disc confocal microscopy of a wild-type root hair expressing GFP-ABD2-GFP to label F-actin. Movie shows images taken every 300 ms. Total elapsed time is 180 s.
Supplemental Movie S10. Spinning disc confocal microscopy of an agd1-1 root hair expressing GFP-ABD2-GFP. Movie shows images taken every 300 ms. Total elapsed time is 180 s.
Supplemental Movie S11. Spinning disc confocal microscopy of a wild-type root hair expressing ST-YFP to label Golgi stacks. Movie shows images taken every 1 s. Total elapsed time is 60 s.
Supplemental Movie S12. Spinning disc confocal microscopy of an agd1-1 root hair expressing ST-YFP to label Golgi stacks. Movie shows images taken every 1 s. Total elapsed time is 60 s.
Supplemental Movie S13. Confocal time-lapse imaging of an agd1-1 root hair expressing GFP-MBD and germinated in 100 nM BFA. Movie shows images taken every 2 min. Total elapsed time is 60 min.
Supplemental Movie S14. Spinning disc confocal microscopy of BFA-treated agd1-1 root hair expressing ST-YFP to label Golgi stacks. Movie shows images taken every 1 s. Total elapsed time is 60 s.
Supplementary Material
Acknowledgments
We thank Dr. Jeremy Murray for critical reading of the manuscript and Dr. Ian Moore (Oxford University) for seeds of ST-YFP.
This work was supported by the Samuel Roberts Noble Foundation and by the Oklahoma Center for the Advancement of Science and Technology (grant no. PSB08–003 to E.B.B.). The confocal microscopes used in this study were acquired through Multi-User Equipment and Major Research Instrumentation grants from the National Science Foundation (grant nos. DBI–0400580 and DBI–0722635 to E.B.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Elison B. Blancaflor (eblancaflor@noble.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bao Y, Kost B, Chua NH (2001) Reduced expression of α-tubulin genes in Arabidopsis thaliana specifically affects root growth and morphology, root hair development and root gravitropism. Plant J 28 145–157 [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S (1999) Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J 17 657–665 [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Zhigilei A, Gilroy S (1997) Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta 203 495–505 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Wang YS, Motes CM (2006) Organization and function of the actin cytoskeleton in developing root cells. Int Rev Cytol 252 153–198 [DOI] [PubMed] [Google Scholar]
- Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, Sadot E, Yalovsky S (2005) Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol Biol Cell 16 1913–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohme K, Li Y, Charlot F, Grierson C, Marrocco K, Okada K, Laloue M, Nogué F (2004) The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J 40 686–698 [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129 865–877 [DOI] [PubMed] [Google Scholar]
- Brown JW, Smith P, Simpson CG (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 312 531–535 [DOI] [PubMed] [Google Scholar]
- Campanoni P, Blatt MR (2007) Membrane trafficking and polar growth in root hairs and pollen tubes. J Exp Bot 58 65–74 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438 1013–1016 [DOI] [PubMed] [Google Scholar]
- Cole RA, Fowler JE (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9 579–588 [DOI] [PubMed] [Google Scholar]
- Davis AJ, Im YJ, Dubin JS, Tomer KB, Boss WF (2007) Arabidopsis phosphatidylinositol phosphate kinase 1 binds F-actin and recruits phosphatidylinositol 4-kinase beta1 to the actin cytoskeleton. J Biol Chem 282 14121–14131 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Rodrigues C, Dimmock S, Ketelaar T, Maciver SK, Malhó R, Hussey PJ (2007) Arabidopsis CAP1: a key regulator of actin organisation and development. J Cell Sci 120 2609–2618 [DOI] [PubMed] [Google Scholar]
- Dong CH, Xia GX, Hong Y, Ramachandran S, Kost B, Chua NH (2001) ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd PE, Coursol S, Skirpan AL, Kao TH, Gilroy S (2006) Petunia phospholipase c1 is involved in pollen tube growth. Plant Cell 18 1438–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230 [DOI] [PubMed] [Google Scholar]
- Gu Y, Fu Y, Dowd P, Li S, Vernoud V, Gilroy S, Yang Z (2005) A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J Cell Biol 169 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann B (2004) The BAR-domain family of proteins: a case of bending and binding. EMBO Rep 5 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ (2006) Control of actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57 109–125 [DOI] [PubMed] [Google Scholar]
- Inoue H, Randazzo PA (2007) Arf GAPs and their interacting proteins. Traffic 8 1465–1475 [DOI] [PubMed] [Google Scholar]
- Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, Randazzo PA (2000) ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol 151 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identify of six genes with roles in root-hair development in Arabidopsis. Plant J 45 83–100 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Allwood EG, Hussey PJ (2007) Actin organization and root hair development are disrupted by ethanol-induced overexpression of Arabidopsis actin interacting protein 1 (AIP1). New Phytol 174 57–62 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, de Ruijter NC, Emons AM (2003) Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 15 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mollet JC, Dong J, Zhang K, Park SY, Lord EM (2003) Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc Natl Acad Sci USA 100 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Naramoto S, Sawa S, Yahara N, Ueda T, Nakano A, Sugiyama M, Fukuda H (2005) VAN3 ARF-GAP mediated vesicle transport is involved in leaf vascular network formation. Development 132 1699–1711 [DOI] [PubMed] [Google Scholar]
- Kost B (2008) Spatial control of Rho(Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol 18 119–127 [DOI] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH (1999) Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 145 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JEM, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T (2008) The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Duden R, Phair RD, Lippincott-Schwartz J (2005) ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J Cell Biol 168 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Lu L, Lee YRJ, Pan R, Maloof JN, Liu B (2005) An internal motor kinesin in associated with the Golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Mol Biol Cell 16 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malho R, Trewavas AJ (1996) Localized apical increases of cytosolic free calcium control pollen tube orientation. Plant Cell 8 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao TH, McCubbin AG, Cyr RJ (1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk A, Bischoff F, Rajendrakumar SV, Friml J, Braun M, Gilroy S, Palme K (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro D, Liu Q, Lisboa S, Scherer GE, Quader H, Malhó R (2005) Phosphoinositides and phosphatidic acid regulate pollen tube growth and reorientation through modulation of [Ca2+]c and membrane secretion. J Exp Bot 56 1665–1674 [DOI] [PubMed] [Google Scholar]
- Motes CM, Pechter P, Yoo CM, Wang YS, Chapman KD, Blancaflor EB (2005) Differential effects of two phospholipase D inhibitors, 1-butanol and N-acylethanolamine, on in vivo cytoskeletal organization and Arabidopsis seedling growth. Protoplasma 226 109–123 [DOI] [PubMed] [Google Scholar]
- Min MK, Kim SJ, Miao Y, Shin J, Jiang L, Hwang I (2007) Overexpression of Arabidopsis AGD7 causes relocation of Golgi-localized proteins to the endoplasmic reticulum and inhibits protein trafficking in plant cells. Plant Physiol 143 1601–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Mettbach U, Menzel D, Šamaj J (2008) Molecular dissection of endosomal compartments in plants. Plant Physiol 145 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr A, Ritzenthaler C, Robinson DG (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY (2006) RAC/ROP GTPases: “hubs” for signal integration and diversification in plants. Trends Plant Sci 11 309–315 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hirsch DS, Luo R, Jian X, Stauffer S, Cremesti A, Andrade J, Lebowitz J, Marino M, Ahvazi B, et al (2006) A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr Biol 16 130–139 [DOI] [PubMed] [Google Scholar]
- Nie Z, Randazzo PA (2007) Arf GAPs and membrane traffic. J Cell Sci 119 1203–1211 [DOI] [PubMed] [Google Scholar]
- Nie Z, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Taketi J, Randazzo PA (2002) AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biochem (Tokyo) 277 48965–48975 [DOI] [PubMed] [Google Scholar]
- Ovecka M, Lang I, Baluska F, Ismail A, Illes P, Lichtscheidl IK (2005) Endocytosis and vesicle trafficking during tip growth of root hairs. Protoplasma 226 39–54 [DOI] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis thaliana. Plant Physiol 146 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocký M, Eliás M, Profotová B, Novotná Z, Valentová O, Zárský V (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217 122–130 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E (2006) A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol 172 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E (2004) The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HEM, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA, Hirsch DS (2004) Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodeling. Cell Signal 16 401–413 [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Inoue H, Bharti S (2007) ArfGAPs as regulators of the actin cytoskeleton. Biol Cell 99 583–600 [DOI] [PubMed] [Google Scholar]
- Ringli C, Baumberger N, Diet A, Frey B, Keller B (2002) ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol 129 1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J 29 661–678 [DOI] [PubMed] [Google Scholar]
- Sakai T, Honing H, Nishioka M, Uehara Y, Takahashi M, Fujisawa N, Saji K, Seki M, Shinozaki K, Jones MA, et al (2008) Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J 53 157–171 [DOI] [PubMed] [Google Scholar]
- Samaj J, Muller J, Beck M, Bohm N, Menzel D (2006) Vesicular trafficking, cytoskeleton and signaling in root hairs and pollen tubes. Trends Plant Sci 11 594–600 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E, Van Norman JM (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Yang CY, Liu J, Yang WC (2006) RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiol 141 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Ischebeck T, König S, Holubowska A, Sporysz M, Hause B, Heilmann I (2008) The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20 124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L (2008) Local positive feedback regulation determines cell shape in root hair cells. Science 319 1241–1244 [DOI] [PubMed] [Google Scholar]
- Thole JM, Vermeer JEM, Zhang Y, Gadella TWJ, Nielsen E (2008) ROOT HAIR DEFECTIVE4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bruaene N, Joss G, Van Oostveldt P (2004) Reorganization and in vivo dynamics of microtubules during Arabidopsis root hair development. Plant Physiol 136 3905–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA (2005) A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol 168 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Yoo CM, Blancaflor EB (2008) Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C and N termini of the fimbrin actin binding domain 2. New Phytol 177 525–536 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP-ribosylation factor-1 function in epidermal cell polarity. Plant Cell 17 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gao P, Zhang H, Huang S, Zheng ZL (2007) A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLOS One 10 e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Guo C, Chen D, Zhao B, Yang B, Ren H (2005) Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol 138 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Jiang J, Li J, Ma Q, Xu Y, Xue Y, Xu Z, Chong K (2006) Over-expression of OsAGAP, an ARF-GAP, interferes with auxin influx, vesicle trafficking and root development. Plant J 48 581–591 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.