Abstract
Strategies employed for the production of genetically modified (GM) crops are premised on (1) the avoidance of gene transfer in the field; (2) the use of genes derived from edible organisms such as plants; (3) preventing the appearance of herbicide-resistant weeds; and (4) maintaining transgenes without obstructing plant cell propagation. To this end, we developed a novel vector system for chloroplast transformation with acetolactate synthase (ALS). ALS catalyzes the first step in the biosynthesis of the branched amino acids, and its enzymatic activity is inhibited by certain classes of herbicides. We generated a series of Arabidopsis (Arabidopsis thaliana) mutated ALS (mALS) genes and introduced constructs with mALS and the aminoglycoside 3′-adenyltransferase gene (aadA) into the tobacco (Nicotiana tabacum) chloroplast genome by particle bombardment. Transplastomic plants were selected using their resistance to spectinomycin. The effects of herbicides on transplastomic mALS activity were examined by a colorimetric assay using the leaves of transplastomic plants. We found that transplastomic G121A, A122V, and P197S plants were specifically tolerant to pyrimidinylcarboxylate, imidazolinon, and sulfonylurea/pyrimidinylcarboxylate herbicides, respectively. Transplastomic plants possessing mALSs were able to grow in the presence of various herbicides, thus affirming the relationship between mALSs and the associated resistance to herbicides. Our results show that mALS genes integrated into the chloroplast genome are useful sustainable markers that function to exclude plants other than those that are GM while maintaining transplastomic crops. This investigation suggests that the resistance management of weeds in the field amid growing GM crops is possible using (1) a series of mALSs that confer specific resistance to herbicides and (2) a strategy that employs herbicide rotation.
Plastid transformation was first reported in studies using the unicellular alga Chlamydomonas reinhardtii (Boynton et al., 1988) and was followed by investigations involving tobacco (Nicotiana tabacum; Svab et al., 1990). Genetic engineering approaches that utilize chloroplasts possess a number of benefits in comparison with nuclear transformation, including (1) a high level of transgene expression (DeCosa et al., 2001); (2) the delivery of multiple genes in a single transformation event (Daniell and Dhingra, 2002); (3) the absence of gene silencing (Lee et al., 2003); (4) the absence of position effects due to site-specific transgene integration (Daniell et al., 2004); and (5) the absence of pleiotropic effects since transgene products are localized within the chloroplast (Daniell et al., 2001). These benefits were exploited by chloroplast transformation trials utilizing a variety of plants, including Arabidopsis (Arabidopsis thaliana; Sikdar et al., 1998), potato (Solanum tuberosum; Sidorov et al., 1999), rice (Oryza sativa; Khan and Maliga, 1999), tomato (Solanum lycopersicum; Ruf et al., 2001), oilseed rape (Brassica napus; Hou et al., 2003), carrot (Daucus carota; Kumar et al., 2004a), cotton (Gossypium hirsutum; Kumar et al., 2004b), soybean (Glycine max; Dufourmantel et al., 2004), lettuce (Lactuca sativa; Lelivelt et al., 2005), and cabbage (Brassica capitata; Liu et al., 2007). However, transplastomic plants were not stable in Arabidopsis, oilseed rape, or rice. Recently, Lee et al. (2006) made progress in rice. Lesquerella fendleri (oilseed Brassicacea) belongs to the mustard family (Brassicaceae), as does Arabidopsis and oilseed rape. Skarjinskaia et al. (2003) demonstrated that transplastomic Lesquerella plants were fertile and produced seed. Methods and selection conditions for the chloroplast transformation of these species were summarized by Verma and Daniell (2007). The basic plastid transformation vector consists of flanking sequences and chloroplast-specific expression cassettes. Flanking sequences are species specific and function as regions for homologous recombination. Chloroplast-specific expression cassettes have selectable markers and genes of interest for additional functions in plants (Verma and Daniell, 2007).
Transformation technology utilizing nuclear genomes was developed in an effort to eliminate antibiotic marker genes (Yoder and Goldsbrough, 1994) and is referred to as Nuclear genome-Clean Gene Transformation Technology (N-CGTT). This approach has recently been applied to genetic engineering studies utilizing chloroplasts and is referred to as Chloroplast genome-Clean Gene Transformation Technology (C-CGTT) in which antibiotic marker genes, such as the aminoglycoside 3′-adenyltransferase gene (aadA) introduced into the chloroplast genome, are eliminated (Iamtham and Day, 2000; Ye et al., 2003). Use of these methodologies prevents the transfer of antibiotic genes to surrounding weeds and microorganisms in the soil and to bacteria in the gut following oral intake. Integration of foreign genes into the plastid genome enhances gene containment because plastids are inherited maternally in many crop plants, thereby avoiding the pollen-mediated spread of transgenes (Maliga, 1993; Daniell et al., 1998; Scott and Wilkinson, 1999). Homologous recombination in plastids allows for accurate gene targeting into a well-characterized genome and the elimination of bacterial vector sequences (Svab et al., 1990). High levels of gene expression have been observed for a variety of foreign genes when located within plastids (McBride et al., 1995; Staub et al., 2000; Kanamoto et al., 2006).
Notwithstanding the advantages associated with C-CGTT, use of this technology remains problematic in terms of potential effects in the field. Genetically modified (GM) and non-GM plants must be distinguished correctly and easily. Although use of the PCR method is the most convenient for the determination of contamination in bulk samples, it is unsuitable for examining single seeds or plants in terms of efficiency and resources. Although the use of herbicides was proposed as a suitable method to resolve this problem, the generation of herbicide-resistant plants must be considered. Studies indicate that herbicides inhibiting ALS (acetohydroxyacid synthase [AHAS]; EC 2.2.1.6) have accelerated the generation rate of tolerant weeds and crops (Preston and Powles, 2002; Shimizu et al., 2002; Tranel and Wright, 2002; Tranel et al., 2007). Rotating the supply of some herbicides was proposed as a countermeasure against the occurrence of herbicide-resistant weeds (Gressel, 1984).
The solution to this problem was investigated in this investigation through the use of an herbicide-tolerant gene of plant origin. ALS catalyzes the first step in the biosynthesis of the branched amino acids Val, Leu, and Ile, and is composed of catalytic and regulatory subunits. Plant ALSs possess catalytic subunits similar to the equivalent bacterial and yeast (Saccharomyces cerevisiae) enzymes, except for the N-terminal signal peptide sequences required for translocation of the protein to the chloroplast (Ott et al., 1996). Genes that express the plant regulatory subunit have been cloned and characterized (Hershey et al., 1999). The deduced amino acid sequences are as much as twice the length of bacterial sequences and comprise two domains thought to be involved in feedback inhibitor mechanisms. It is well known that mutations at several amino acid residues in ALS are responsible for tolerance to herbicides shown in Figure 1. Herbicide-tolerant plants have been reported for rice, tobacco, and Arabidopsis (Chang and Duggleby, 1998; Shimizu et al., 2002; Tan et al., 2005; Kawai et al., 2007; Okuzaki et al., 2007). Several mutated species of Arabidopsis ALS have been expressed in Escherichia coli and sensitivity to inhibitors has been examined (Kawai et al., 2008). These results showed that, in the ALS polypeptide, P197S and S653I changes can produce sulfonylurea (SU) resistance, whereas W574L and P197H/R198S (double mutations of P197H and R198S) changes can produce imidazolinon (IM) resistance, and W574L/S653I (double mutations of W574L and S653I) changes can produce resistance to SU, IM, and pyrimidinylcarboxylate (PC). However, little is known about plant growth tolerance to herbicides in specimens with mALSs. Nuclear genetic engineering studies have shown that mutation of W548L/S627I and G95A in rice ALS confers tolerance to the aforementioned herbicides, including PC herbicides (Kawai et al., 2007; Okuzaki et al., 2007). The introduction of mALSs into the chloroplast genome had not been attempted because ALS was thought to be unsuitable as a selectable marker as in the case of 5-enol-pyruvyl shikimate-3-P synthase (EPSPS; Ye et al., 2003).
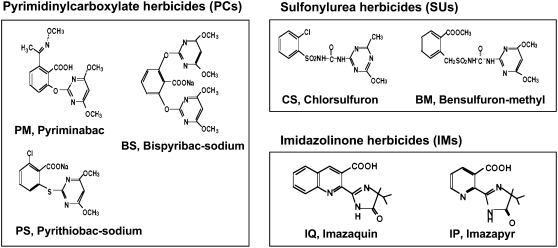
Figure 1.
Herbicides inhibiting ALS activity. These herbicides can be classified into the three classes PC, SU, and IM. BS, PS, and PM belong to the PC class of herbicides, CS and BM to the SU class of herbicides, and IQ and IP to the IM class of herbicides.
We report here the introduction of mALS genes into the chloroplast genome and investigate the sensitivity of transformants with respect to ALS-inhibiting herbicides. The mALS genes proved useful as sustainable markers that functioned to exclude nontransformed plants while maintaining the transplastomic plants. These markers showed selectable tolerance to various herbicides. Our findings suggest that rotation of the herbicide supply to transgenic plants harboring different sustainable markers can be an effective strategy in countering the occurrence of herbicide-resistant weeds.
RESULTS
Generation and Analysis of Transplastomic Plants
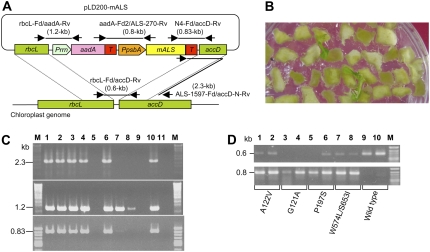
Chloroplast transformation vectors, pLD200-mALS possessing the aadA and mALS (transit peptide truncated) genes inserted between tobacco rbcL (gene for the large subunit of Rubisco) and accD (gene for acetyl-CoA carboxylase) sequences for homologous recombination, were introduced by particle bombardment. The integration of mALS into the chloroplast genome of regenerated tobacco plants was confirmed by PCR using five primer sets as shown in Figure 2A. Tobacco chloroplast transformation was performed with 10 shots of bombardment on 10 different plates per vector pLD200-mALS harboring mALS comprising G121A, A122V, P197S, or W574L/S653I (double mutant). The resultant transplastomic candidates (10 per vector) were maintained on hormone-free Murashige and Skoog medium (Fig. 2B). PCR was performed with primer sets comprising rbcL-Fd (annealing to the homologous region of rbcL) and ALS-433-Rv to generate 0.83-kb products in lanes 1 to 4, 6, and 10 (Fig. 2C, bottom; results of P197S), and ALS-1597-Fd and accD-Rv (annealing to the homologous region of accD) to generate 1.2-kb products in lanes 1 to 4, 6 to 8, and 10 (Fig. 2C, middle; results of P197S), confirming the presence of transgenes in candidate lines. To discriminate between nuclear and chloroplast transgenic lines, an accD-N-Rv primer was designed that would anneal to the endogenous chloroplast genome and enable a 2.3-kb PCR product to be generated when paired with an internal ALS primer, ALS-1597-Fd (Fig. 2A). Transplastomic lines were identified by PCR with the primer set accD-N-Rv and ALS-1597-Fd, with 2.3-kb products visible in lanes 1 to 4, 6, and 10 (Fig. 2C, top; results of P197S), confirming site-specific integration of the transgenes into the tobacco chloroplast genome. The size of the PCR product amplified in lanes 7 and 8 was 1.2 kb, not 2.3 kb and 0.8 kb, indicating that the mALS region was not integrated into the chloroplast genome. The overall results indicate that plant samples shown in lanes 1 to 4, 6, and 10 were derived from genuine transplastomic lines containing full-length transgenes. The same PCR approach was employed for all candidates transformed with the other vectors and confirmed the production of several transplastomic lines.
Figure 2.
Chloroplast transformation with aadA and mutated ALS (mALS). A, Structure of the chloroplast transformation vector. Prrn-aadA and PpsbA-mALS represent transgenes introduced into the chloroplast genome. The region located between rbcL and accD may be integrated in the chloroplast genome by homologous recombination. The primer set rbcL-Fd and aadA-Rv was used to amplify rbcL-aadA in the transformation vector, generating a 1.2-kb product. The primer set N4-Fd and accD-Rv was used for the mALS-accD region, yielding a 0.83-kb product. The primer set ALS-1597-Fd (forward primer of mALS) and accD-N-Rv (reverse primer for endogenous accD) generated a 2.3-kb product. The primer set aadA-Fd2 and ALS-270-Rv was used for the aadA-mALS region, generating a 0.8-kb product. The primer set rbcL-Fd and accD-Rv was used to amplify the endogenous region between rbcL and accD of the chloroplast genome, generating a 0.6-kb fragment. B, Following bombardment, leaf slices were grown on RMOP containing 0.5 g L−1 SP. A SP plate following a 6-week growing period is shown. The regenerated plants represent potential transformants. C, Confirmation of transgenes integrated into the chloroplast genome by PCR. PCR analysis was performed using three primer sets. Lanes 1 to 10, DNA from regenerated plants; lane 11, nontransformed tobacco; M, size markers. The indicated sizes of 2.3 kb, 1.2 kb, and 0.83 kb are products derived from the use of primer sets ALS-1597-Fd and accD-N-Rv, rbcL-Fd and aadA-Rv, and N4-Fd and accD-Rv, respectively. D, The population representing the transformed chloroplast genome. PCR analysis of two different lines of each chloroplast transformant harboring mALS: A122V (lanes 1 and 2), G121A (lanes 3 and 4), P197S (lanes 5 and 6), W574L/S653I (lanes 7 and 8), and the wild type (lanes 9 and 10). PCR was performed in 25 cycles. The 0.6-kb product represents the endogenous chloroplast genome without a transgene insert, whereas the 0.8-kb product represents part of the transgenes introduced into the chloroplast genome.
To examine the amount of transplastomic chloroplast genome in each line, the population representing the native chloroplast genome was determined by PCR with rbcL-Fd and accD-Rv. Two different lines transformed with the same construct were subjected to PCR analysis. The wild-type lines markedly amplified the 0.6-kb product (Fig. 2D, top; lanes 9 and 10), whereas the 0.6-kb products were barely amplified in lanes 1 to 8 (Fig. 2D, top), indicating that the native regions amplified by rbcL-Fd and accD-Rv were split by the transgenes. The internal insert sequence was detected to a greater extent using primers aadA-Fd2 and ALS-270-Rv in all transgenic plants (Fig. 2D, bottom). However, the 0.8-kb products were not detected in wild-type plants. Therefore, it is concluded that the majority of the chloroplast genome in these transgenic plants was transplastomic. Subsequent studies used the transplastomic plants of lane 2 for A122V, lane 4 for G121A, lane 6 for P197S, and lane 8 for W574L/S653I.
ALS Activity in Leaves of Transplastomic Plants
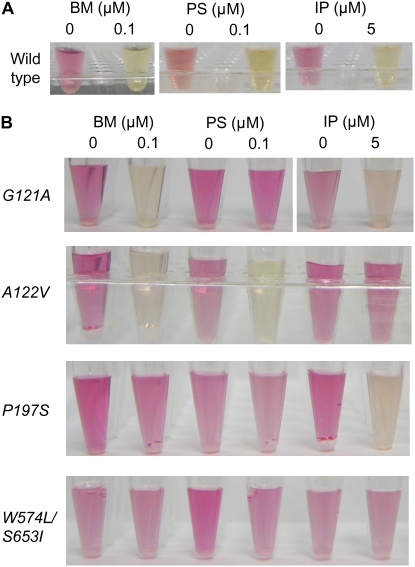
The activity of native ALS from wild-type tobacco in the absence of ALS-inhibiting herbicides was determined by a colorimetric assay and appeared as a red color in samples (Fig. 3A). The red color changed to a transparent or pale yellow color following the addition of SU herbicide (0.1 μm bensulfuron-methyl [BM]), PC herbicide (0.1 μm pyrithiobac-sodium [PS]), and IM herbicide (5 μm imazapyr [IP]; Fig. 3A), indicating that these herbicides inhibited ALS activity. This assay was employed for the evaluation of mALS activity in transplastomic plants (G121A, A122V, P197S, and W574L/S653I), as determined by the PCR methods described above. The ALS activity of G121A plants was strongly resistant to PS, weakly resistant to BM and sensitive to IP (Fig. 3B), whereas A122V plants were specifically resistant to IP (Fig. 3B), and P197S plants were strongly resistant to BM, middle resistant to PS, and sensitive to IP (Fig. 3B). The ALS activity of W574L/S653I plants was strongly resistant to PS, BM, and IP (Fig. 3B). These results show a mALS-mediated herbicide-specific resistance resulting from a transgene introduced by chloroplast transformation.
Figure 3.
Inhibition of ALS activity with herbicides in transplastomic plants with mALSs. ALS activity in transplastomic tobacco plants with mALSs was determined using a colorimetric assay. ALS activity of tobacco, wild-type (A), and mALS-transformed plants G121A, A122V, P197S, and W574L/S653I (B) was determined in the presence of 0.1 μm BM, 0.1 μm PS, or 5 μm IP. Experimental details are described in “Materials and Methods.”
Effects of Herbicides on Plant Growth
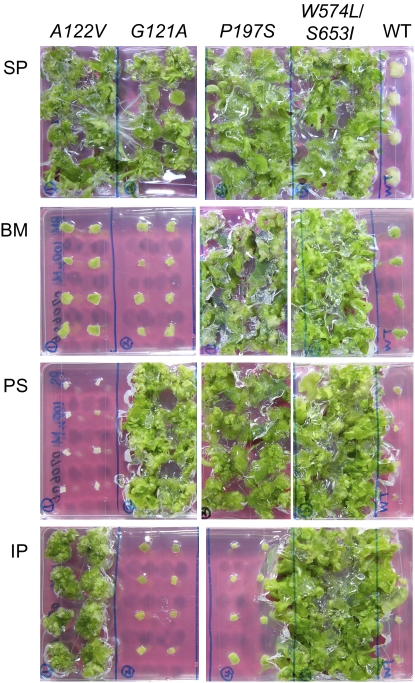
Herbicide resistance was examined using a combination of three different herbicides and a variety of mALSs. In an effort to examine the in vivo effect of each herbicide on transplastomic plants, the leaves of plants were transferred to regeneration medium containing 0.5 g L−1 spectinomycin (SP), 0.1 μm BM, 0.1 μm PS, or 1 μm IP, and cultured for 3 weeks. Wild-type tobacco was unable to grow on medium containing any of the herbicides. On the other hand, G121A plants regenerated on 0.1 μm PS medium (Fig. 4), and A122V plants regenerated on 1 μm IP medium (Fig. 4). P197S plants were tolerant to 0.1 μm BM and 0.1 μm PS (Fig. 4). W574L/S653I plants regenerated on medium containing 0.1 μm BM, 0.1 μm PS, or 1 μm IP (Fig. 4).
Figure 4.
Regeneration of transplastomic plants on medium containing ALS-inhibiting herbicides. Transgenic plants comprising A122V, G121V, P197S, or W574L/S653I mALSs were regenerated on RMOP medium supplemented with 0.5 g L−1 SP, 0.1 μm BM, 0.1 μm PS, or 1 μm IP.
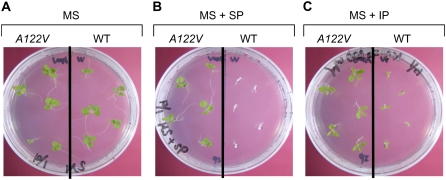
T1 seeds, the self-pollinated progeny of A122V plants, and wild-type seeds were planted on medium supplemented with 1 μm IP or 0.5 g L−1 SP. Both seed types were able to grow on Murashige and Skoog (Fig. 5A). Although wild-type plants were sensitive to IP and SP, all A122V seeds were uniformly resistant to SP and IP (Fig. 5, B and C). Green tissues of A122V plants on 1 μm IP medium grew in a manner similar to that of wild-type tissue grown on herbicide-free medium (Fig. 5C), although the root length of the former was shorter. The accumulation of mALS protein did not appear to be sufficient to impart resistance to the herbicide in root plastids because mALS expression was driven by the psbA promoter, which is known to be a strong promoter in green tissues.
Figure 5.
Influence of psbA promoter on the growth of transplastomic plants. T1 seeds transplastomic with PpsbA-A122V (on the left in all images, A122V) and wild-type (on the right in all images, WT) seeds were germinated on Murashige and Skoog medium alone (A, MS), or on medium containing 0.5 g L−1 SP (B, MS + SP) or 1 μm IP (C, MS + IP).
DISCUSSION
ALS Activity in Transplastomic Plants
Herbicide-specific resistance conferred by mALSs and directed by transplastomic genes in the chloroplast genome has been demonstrated by this investigation, even though mALS genes had previously only been delivered into the nuclear genomes of certain species (Kawai et al., 2007; Okuzaki et al., 2007). ALS activity is thought to be controlled by the regulatory subunit, which also plays a role in feedback regulation by Val, Ile, and Leu (Lee and Duggleby, 2001). Although the tobacco mosaic virus 35S promoter has frequently been used in nuclear transformation studies, our study used the psbA promoter because it is the strongest known promoter in chloroplasts (Hayashi et al., 2003). Moreover, given that 100 to 1,000 copies of the chloroplast genome exist in a cell, it would be interesting to determine whether highly expressed mALSs in chloroplasts can influence plant growth. This study has revealed that transplastomic plants with mALSs grow normally on Murashige and Skoog medium without significant differences when compared to wild-type plants, showing that hyperexpression of mALSs does not influence plant growth.
We investigated the involvement of the regulatory subunit in ALS activity. Regulatory subunits play roles in feedback regulation and full enzyme activity (Lee and Duggleby, 2001). The determination of ALS activity in leaves where regulatory subunits exist was done in the presence of 1,1-cyclopropanedicarboxylic acid, which blocked the acetolactate metabolism, resulting in no feedback regulation. The selectable tolerance of plants transplastomic with G121A, A122V, and W574L/S653I (Fig. 3) were similar to those obtained when using the same recombinant mALSs that only expressed the catalytic subunit in E. coli, with which endogenous E. coli regulatory subunits was not concluded to be associated (Kawai et al., 2008). Therefore, the regulatory subunits do not affect the sensitivity of these mALSs to herbicides in transplastomic plants. On the other hand, P197S mALS exhibited different behavior from the above mutations. The novel tolerance of P197S plants to PC and SU herbicides was demonstrated with regard to mALS activity in response to herbicides in leaves (Fig. 3), whereas P197S mALS expressed in E. coli was resistant to SU herbicides, but not to PC herbicides (Kawai et al., 2008). This result suggested that the regulatory subunit contributed to the acquisition of resistance of P197S activity to the PC herbicides.
In an effort to investigate the influence of feedback regulation, transplastomic plants were grown on medium containing an herbicide. We analyzed herbicide resistance in transplastomic plants harboring four different mALSs. W574L/S653I plants showed synergistic tolerance, similar to that observed when the corresponding mALS gene was introduced into the nuclear genome of rice (Kawai et al., 2007). The tolerance of P197S plants to PC and SU herbicides was demonstrated also in plant growth (Fig. 4). In addition, two other transplastomic plants (G121A and A122V) showed sensitivities to herbicides both in the activity in leaves (Fig. 3) and in plant growth (Fig. 4). Our results provide evidence suggesting that the sensitivity of mALSs to herbicides in plants is not affected by feedback regulation. The highly expressed mALS molecules may not be fully active due to the resultant stoichiometrically insufficient number of regulatory subunits (Lee and Duggleby, 2001). Therefore, the ALS activity of transplastomic plants was almost equivalent to that of wild-type plants in the absence of herbicides. It is concluded that transplastomic mALSs are useful as sustainable markers that can be employed to distinguish GM and non-GM plants when using appropriate herbicides in the field.
Benefit of mALSs as Sustainable Markers
Herbicide-resistant weeds have been reported in many countries (Tranel and Wright, 2002; Tranel et al., 2007) and include weeds resistant to ALS-inhibiting herbicides. New technology is required to assist in the management of weeds resistant to these herbicides. We propose a strategy involving herbicide rotation to overcome the aforementioned problem. To this end, we have developed transplastomic plants that possess tolerance to PC, IM, and SU/PC. Although it is known that some ALS mutations are associated with plant resistance to a single herbicide, there have been no reports detailing the relationship between a single ALS mutation and resistance to the three classes of herbicide PC, SU, and IM. In this article, transplastomic plants were generated and their resistance to herbicides was characterized. We determined three kinds of ALS mutations that conferred specific resistance to the three classes of herbicides used with the transplastomic plants and showed that G121A, A122V, and P197S plants were resistant to PC, IM, and SU/PC herbicides, respectively (Fig. 4). Use of these transplastomic markers in crop plants would enable a new strategy based on the rotation of three or more combinations of herbicides. The advanced technology described in this article allows for the efficient and strict management of weeds resistant to ALS-inhibiting herbicides.
Investigations concerning herbicide resistance have been made using chloroplast transformation. For example, the petunia EPSPS gene was introduced into the tobacco chloroplast genome and resulted in transplastomic plants resistant to glyphosate (Daniell et al., 1998). Similarly, the bar gene for phosphinothricin resistance was used to investigate the resulting plant phenotype (Lutz et al., 2001). Because this gene is derived from microorganisms and not plants, it is less suitable for use in C-CGTT-based approaches. However, EPSPS is worthy of consideration in strategies involving herbicide rotation schemes as described above because EPSPS is present in higher plants. The glyphosate and ALS-inhibiting herbicides are thought to be nontoxic to living organisms, except plants and microorganisms (Peterson and Shama, 2005). Plant-derived EPSPS might be useful as an additional tool for use in an herbicide rotation system for the management of herbicide-resistant weeds. The technology developed in this study may be employed in C-CGTT-based methodologies in association with aadA elimination following transformation.
MATERIALS AND METHODS
Construction of Plastid Transformation Vectors
Genes for the transit peptide-truncated ALS mutants A122V, P197S, and W574L/S653 placed in pBluescript (pBS; Stratagene; Kawai et al., 2008) were used as templates for PCR. ALS(G121A) was generated by replacement of the EcoRV-AvrII fragment of pBS-ALS(A122V) with a G121A-containing product amplified using G121A Fd and G121A Rv, followed by digestion with EcoRV and AvrII. pBS-ALS(G121A) was also used as a template for PCR. Fragments of ALS mutants were amplified using KOD-plus DNA polymerase for higher fidelity (Toyobo). Primers corresponding to the ALS coding region were ALS Fd and ALS Rv. The PCR reaction was performed by employing 30 cycles of denaturation for 30 s at 94°C, annealing for 20 s at 55°C, and extension for 2 min at 68°C. PCR-generated fragments were ligated to the SphI site of pLD6 (GenBank accession no. CS165374; Adachi et al., 2007) harboring the 16S rRNA gene promoter-aadA-psbA gene terminator, psbA gene promoter-SphI-TpsbA, and Prrn-aadA-TpsbA-PpsbA-SphI-TpsbA to drive mALS gene expression using the psbA promoter. Sequencing analysis was performed using a BigDye terminator cycle sequence kit (Applied Biosystems). Vectors generated were named pLD6-mALS, pLD6-A122V, and so on. The SalI-NotI region of pLD6-mALS was introduced into the SalI and NotI sites of pLD200 (GenBank accession no. BD174938; Adachi et al., 2007), which possesses a sequence between rbcL and accD derived from tobacco for homologous recombination. The absence of unexpected mutations was confirmed by sequence analysis. Finally, each chloroplast transformation vector comprised two transgene constructs, Prrn-aadA-TpsbA and PpsbA-mALS-TpsbA, referred to as pLD200-mALS. The nucleotide sequences of primers used in this study are listed in Table I.
Table I.
List of primer sequences
| Name | Nucleotide Sequence (5′ to 3′) |
|---|---|
| G121A Fd | GGCGCTGATATCCTGGTCGAAGCATTAGAACGTCAAGGCGTAGAAACCGTATTCGCTTACCCTGGAGCTGCATCAATGG |
| G121A Rv | CTCAATAATCCTAGGAATATCTTCAACATCCATCACAAG |
| ALS Fd | TCATGACATTTATCTCTCCCGATTCGCTCCAGATCAACC |
| ALS Rv | CCGGTTTGAGCTCTTAGTATTTAATCCGGCCATCTCC |
| rbcL Fd | AGCAGTGGACGTTTTGGATAA |
| aadA Rv | ATCTCGCCTTTCACGTAGTGG |
| N4-Fd | GAGAATCTTCCAGTGAAGG |
| pLD200-Rv | TATGTCGTGTCAAATTCGCATCG |
| ALS-1597-Fd | GTGGATATTGACGGAGATGGAAGC |
| accD-N-Rv | CAACATACCAAAACTAGCTGTCACTCC |
| aadA-Fd2 | GGACAAGAAGAAGATCGCTTGG |
| ALS-270-Rv | TCCCGATTCGCTCCAGAT |
Transformation and Transgene Confirmation
Chloroplast transformation and the preparation of genomic DNA to confirm the presence of transgenes were performed using the tobacco (Nicotiana tabacum) cultivar Xanthi according to previously described methods (Adachi et al., 2007). SP-resistant shoots were selected on RMOP medium containing 0.5 g L−1 SP rooted on Murashige and Skoog agar and self-pollinated to obtain transplastomic seeds. PCR analysis was performed using the following five primer sets: the rbcL-aadA region of pLD200-mALSs with rbcL-Fd and aadA-Rv, the ALS-accD region with N4-Fd and accD-Rv, the insert integrated into the chloroplast genome with ALS-1597-Fd (annealed with ALS in the vector) and accD-N-Rv (for accD in the endogenous chloroplast genome, but not included in the vector), the aadA-ALS region with aadA-Fd2 and ALS-270-Rv, and the rbcL and accD regions of the endogenous chloroplast genome with rbcL-Fd and accD-Rv. The nucleotide sequences of primers used in this study are listed in Table I.
Herbicides Inhibiting ALS Activity
BS, PS, and PM were used as representative PC herbicides, chlorsulfuron (CS) and BM were used as representative SU herbicides, and imazaquin (IQ) and IP were used as representative IM herbicides (Fig. 1). These chemical compounds were provided by KI Chemical Research Institute Co., Ltd.
ALS Activity Assay in Leaves
ALS activity was determined as follows. A leaf section (50 mg) was floated and incubated on 25% Murashige and Skoog medium containing 0.5 mm 1,1-cyclopropanedicarboxylic acid, an inhibitor of acetolactate metabolism, with or without ALS-inhibiting herbicides for 42 h under the same conditions used for plant growth (Adachi et al., 2007). The sample was then placed at −80°C for 1 h, transferred into 200 μL of 0.025% Triton X-100, and incubated at 60°C for 5 min followed by incubation at room temperature for 60 min to effect extraction of acetolactate synthesized by ALS. The extract (100 μL) was transferred to a 1.5-mL tube, to which was added 10 μL of 1 n H2SO4. The sample was then incubated at 60°C for 30 min to convert acetolactate to acetoin. An aliquot (50 μL) of 0.5% (w/v) creatine and 50 μL of 5% (w/v) 1-naphthol dissolved in 2.5 n NaOH were added and the resultant mixture was subsequently incubated at 37°C for 30 min. The amount of acetoin formed was determined by a colorimetric assay.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CS165374 (pLD6), BD174938 (pLD200), and BT020540 (ALS).
Acknowledgments
We are grateful to Kiyoshi Kawai for technical support concerning the ALS activity assay.
This work was supported by the Intellectual Cluster (Keihanna, 2002–2006), Center of Excellence (COE) Program in the 21st Century (2002–2006), and Global COE Program (2007), and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Monbukagakusho), and by the Goto Research Grant from University of Shizuoka (to H. Kobayashi). M.S. was a postdoctoral fellow supported by the COE Program in the 21st Century.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hirokazu Kobayashi (hirokoba@u-shizuoka-ken.ac.jp).
Open Access articles can be viewed online without a subscription.
References
- Adachi T, Takase H, Tomizawa K (2007) Introduction of a 50 kbp DNA fragment into the plastid genome. Biosci Biotechnol Biochem 71 2266–2273 [DOI] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, et al (1988) Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240 1534–1538 [DOI] [PubMed] [Google Scholar]
- Chang AK, Duggleby RG (1998) Herbicide-resistant forms of Arabidopsis thaliana acetohydroxyacid synthase: characterization of the catalytic properties and sensitivity to inhibitors of four defined mutants. Biochem J 333 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Carmona-Sanchez O, Burns B (2004) Chloroplast derived antibodies, biopharmaceuticals and edible vaccines. In R Fischer, S Schillberg, eds, Molecular Farming. Wiley-VCH Verlag Publishers, Berlin, pp 113–133
- Daniell H, Datta R, Varma S, Gray S, Lee SB (1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol 16 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO (2001) Expression of cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol 311 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee SB, Miller M, Daniell H (2001) Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourmantel N, Pelissier B, Garçon F, Peltier G, Ferullo JM, Tissot G (2004) Generation of fertile transplastomic soybean. Plant Mol Biol 55 479–489 [DOI] [PubMed] [Google Scholar]
- Gressel J (1984) Evolution of herbicide-resistant weeds. Ciba Found Symp 102 73–93 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Shiina T, Ishii N, Iwai K, Ishizaki Y, Morikawa K, Toyoshima Y (2003) A role of the -35 element in the initiation of transcription at psbA promoter in tobacco plastids. Plant Cell Physiol 44 334–341 [DOI] [PubMed] [Google Scholar]
- Hershey HP, Schwartz LJ, Gale JP, Abell LM (1999) Cloning and functional expression of the small subunit of acetolactate synthase from Nicotiana plumbaginifolia. Plant Mol Biol 40 795–806 [DOI] [PubMed] [Google Scholar]
- Hou BK, Zhou YH, Wan LH, Zhang ZL, Shen GF, Chen ZH, Hu ZM (2003) Chloroplast transformation in oilseed rape. Transgenic Res 12 111–114 [DOI] [PubMed] [Google Scholar]
- Iamtham S, Day A (2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol 18 1172–1176 [DOI] [PubMed] [Google Scholar]
- Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa K (2006) Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res 15 205–217 [DOI] [PubMed] [Google Scholar]
- Kawai K, Kaku K, Izawa N, Shimizu T, Fukuda A, Tanaka Y (2007) A novel mutant acetolactate synthase gene from rice cells, which confers resistance to ALS-inhibiting herbicides. J Pestic Sci 32 89–98 [Google Scholar]
- Kawai K, Kaku K, Izawa N, Shimizu M, Kobayashi H, Shimizu T (2008) Herbicide sensitivities of mutated enzymes expressed from artificially generated genes of acetolactate synthase. J Pestic Sci 33 128–137 [Google Scholar]
- Khan MS, Maliga P (1999) Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol 17 910–915 [DOI] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H (2004. a) Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots and leaves confers enhanced salt tolerance. Plant Physiol 136 2843–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H (2004. b) Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol Biol 56 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H (2003) Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed 11 1–13 [Google Scholar]
- Lee SM, Kang K, Chung H, Yoo SH, Xu XM, Lee SB, Cheong JJ, Daniell H, Kim M (2006) Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol Cells 21 401–410 [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Duggleby GR (2001) Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry 40 6836–6844 [DOI] [PubMed] [Google Scholar]
- Lelivelt CL, McCabe MS, Newell CA, Desnoo CB, van Dun KM, Birch-Machin I, Gray JC, Mills KH, Nugent JM (2005) Stable plastid transformation in lettuce (Lactuca sativa L.). Plant Mol Biol 58 763–774 [DOI] [PubMed] [Google Scholar]
- Liu CW, Lin CC, Chen JJ, Tseng MJ (2007) Stable chloroplast transformation in cabbage (Brassica oleracea L. var. capitata L.) by particle bombardment. Plant Cell Rep 26 1733–1744 [DOI] [PubMed] [Google Scholar]
- Lutz KA, Knapp JE, Maliga P (2001) Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol 125 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P (1993) Towards plastid transformation in flowering plants. Trends Biotechnol 11 101–107 [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P (1995) Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of insecticidal protein in tobacco. Biotechnology (N Y) 13 362–365 [DOI] [PubMed] [Google Scholar]
- Okuzaki A, Shimizu T, Kaku K, Kawai K, Toriyama K (2007) A novel mutated acetolactate synthase gene conferring specific resistance to pyrimidinyl carboxy herbicides in rice. Plant Mol Biol 64 219–224 [DOI] [PubMed] [Google Scholar]
- Ott KH, Kwagh JG, Stockton GW, Sidorov V, Kakefuda G (1996) Rational molecular design and genetic engineering of herbicide resistant crops by structure modeling and site-directed mutagenesis of acetohydroxyacid synthase. J Mol Biol 263 359–368 [DOI] [PubMed] [Google Scholar]
- Peterson RK, Shama LM (2005) A comparative risk assessment of genetically engineered, mutagenic, and conventional wheat production systems. Transgenic Res 14 859–875 [DOI] [PubMed] [Google Scholar]
- Preston C, Powles SB (2002) Evolution of herbicide resistance in weeds: initial frequency of target site-based resistance to acetolactate synthase-inhibiting herbicides in Lolium rigidum. Heredity 88 8–13 [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Ralph B (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19 870–875 [DOI] [PubMed] [Google Scholar]
- Scott SE, Wilkinson MJ (1999) Low probability of chloroplast movement from oilseed rape (Brassica napus) into wild Brassica rapa. Nat Biotechnol 17 390–392 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Nakayama I, Nagayama K, Miyazawa T, Nezu Y (2002) ALS inhibitors. In P Boeger, K Wakabayashi, K Hirai, eds, Herbicide Classes in Development. Springer-Verlag, Berlin, pp 1–41
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, Nehra NS (1999) Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19 209–216 [DOI] [PubMed] [Google Scholar]
- Sikdar SR, Serino G, Chaudhuri S, Maliga P (1998) Plastid transformation in Arabidopsis thaliana. Plant Cell Rep 18 20–24 [Google Scholar]
- Skarjinskaia M, Svab Z, Maliga P (2003) Plastid transformation in Lesquerella fendleri, an oilseed Brassicacea. Transgenic Res 12 115–122 [DOI] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PT, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, et al (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18 333–338 [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P (1990) Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA 87 8526–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Evans RR, Dahmer ML, Singh BK, Shaner DL (2005) Imidazolinone-tolerant crops: history, current status and future. Pest Manag Sci 61 246–257 [DOI] [PubMed] [Google Scholar]
- Tranel PJ, Wright TR (2002) Resistance of weeds to ALS-inhibiting herbicides: What have we learned? Weed Sci 50 700–712 [Google Scholar]
- Tranel PJ, Wright TR, Heap IM (2007) ALS mutations from herbicide-resistant weeds. WeedScience. http://www.weedscience.org/mutations/MutDisplay.aspx (December 30, 2007)
- Verma D, Daniell H (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye GN, Colburn SM, Xu CW, Hajdukiewicz PTJ, Staub JM (2003) Persistence of unselected transgenic DNA during a plastid transformation and segregation approach to herbicide resistance. Plant Physiol 133 402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JI, Goldsbrough AP (1994) Transformation systems for generating marker-free transgenic plants. Nat Biotechnol 12 263–267 [Google Scholar]