Abstract
Despite its water-soluble chlorophyll-binding protein (WSCP) function, the putative trypsin inhibitor (TI) activity of the Brassica napus drought 22 kD (BnD22) protein and its physiological function in young leaves during leaf nitrogen (N) remobilization promoted by stressful conditions remains an enigma. Therefore, our objectives were to determine (1) if BnD22 is related to the 19-kD TI previously detected in B. napus young leaves, and (2) if the levels of BnD22 transcripts, BnD22 protein, and TI activity in young leaves are associated with plant responses to stress conditions (N starvation and methyl jasmonate [MeJA] treatments) that are able to modulate leaf senescence. Compared to control, N starvation delayed initiation of senescence and induced 19-kD TI activity in the young leaves. After 3 d with MeJA, the 19-kD TI activity was 7-fold higher than the control. Using two-dimensional electrophoresis gel, TI activity, and electrospray ionization liquid chromatography tandem mass spectrometry analysis, it was demonstrated that two 19-kD proteins with isoelectric points 5.0 and 5.1 harboring TI activity correspond to BnD22 perfectly. BnD22 gene expression, TI activities, and BnD22 protein presented similar patterns. Using polyclonal anti-WSCP antibodies of Brassica oleracea, six polypeptides separated by two-dimensional electrophoresis were detected in young leaves treated with MeJA. Electrospray ionization liquid chromatography tandem mass spectrometry analysis of six polypeptides confirms their homologies with WSCP. Results suggest that BnD22 possesses dual functions (WSCP and TI) that lead to the protection of younger tissues from adverse conditions by maintaining metabolism (protein integrity and photosynthesis). By sustaining sink growth of stressed plants, BnD22 may contribute to a better utilization of recycling N from sources, a physiological trait that improves N-use efficiency.
To maintain or increase crop yields while benefiting the environment through reductions in nitrogen (N) fertilizer inputs, it will be necessary to improve both N-use efficiency (Good et al., 2004) and especially the recycling of endogenous N from vegetative organs to the growing tissues. When the N demand for growing vegetative or reproductive tissues exceeds the mineral N availability, the extent of endogenous N mobilization, particularly from senescing leaves, may lead to higher N-use efficiency (Aerts and Chapin, 2000; Yasumura et al., 2007). Proteolysis is one of the most important processes of N remobilization (Hörtensteiner and Feller, 2002; Zimmermann and Zentgraf, 2005). Moreover, the effective recycling of the N compounds from source leaves to sink growing tissues requires a fine coordination between sink demand and the processes of proteolysis. This is particularly important when plants are confronted by stresses that could lead to strong modification of the source/sink relationships. Therefore, understanding the mechanisms and associated regulation of (1) leaf proteolysis that presides over efficient protein-N remobilization and (2) maintenance of sink strength is a prerequisite for improving N recycling and subsequently N-use efficiency.
Among the factors able to modulate N-use efficiency, the inhibitors of proteases may have a pivotal role in both mechanisms, i.e. rate of protein-N remobilization in source leaves and maintenance of metabolism in young leaves. Indeed, a few studies (Sugawara et al., 2002; Shatters et al., 2004; Sin and Chye, 2004) have suggested the involvement of protease inhibitors in the control of proteolysis during the senescence process in which the Cys and Ser proteases are the proteolytic systems that have been the most frequently implicated (Buchanan-Wollaston and Ainsworth, 1997; Jiang et al., 1999). Ser proteinase inhibitors, including potato (Solanum tuberosum) inhibitors I and II (Ryan, 1990), Bowman-Birk inhibitors, and trypsin inhibitors (TIs) of Künitz type (Richardson, 1991), have been the most commonly studied. Many studies revealed that these protease inhibitors are more abundant in young leaves than in old or mature leaves (Reviron et al., 1992; Cipollini and Bergelson, 2000; Etienne et al., 2007). Inhibitors of Künitz-type Ser proteases (IkiL-1 and IkiL-2), identified in leaves of Citrus paradise, were down-regulated in mature leaves, whereas they were up-regulated in young leaves (Shatters et al., 2004). Gene expression of leaf protease inhibitors was stimulated by phytohormones that induced senescence or plant responses to abiotic and biotic stresses, such as abscisic acid (Peňa-Cortés et al., 1991; Horn et al., 2005), jasmonic acid (Farmer and Ryan, 1992; Sanchez-Hernandez et al., 2004), and ethylene (O'Donnell et al., 1996). Numerous studies demonstrated that environmental constraints that stimulated senescence (Yoshida, 2003), such as drought (Downing et al., 1992; Kang et al., 2002), N starvation (Etienne et al., 2007), and wounding (Shatters et al., 2004), can also modulate the expression of protease inhibitors in young leaves. Although it was clearly established that protease inhibitors acted as defense proteins in leaves against pathogen attacks (Ryan, 1990; Horn et al., 2005; Mosolov and Valueva, 2005), the physiological function of protease inhibitors induced in young leaves in response to modification of source/sink relationships induced by abiotic stresses remains unclear.
Oilseed rape (Brassica napus) is a particularly interesting plant for studying the involvement of protease inhibitors in leaves and their contribution to N-use efficiency. Indeed, this important crop plant requires high amounts of mineral N but is characterized by low N-use efficiency as only 50% of fertilizer N is recovered by the crop at harvest date (Schjoerring et al., 1995), leading to a large restitution of this element to the soil (Dejoux et al., 2000). Moreover, a high concentration of N in leaves shed before flowering (exceeding 2% of dry weight; Rossato et al., 2001; Malagoli et al., 2005) indicates that oilseed rape is not able to efficiently mobilize the N from leaves to growing tissues during senescence. During the vegetative stages of oilseed rape, this lack of mobilization of endogenous N in leaf is not due to a limitation in the amino acid transport from leaves to phloem (Tilsner et al., 2005). Therefore, the weak N remobilization could be related to an incomplete hydrolysis of foliar proteins, leading to low recycling of endogenous foliar N from source leaves and/or to a limitation of sink demand of young leaves. When oilseed rape is submitted to resource limitation such as nitrate starvation (Etienne et al., 2007) or water deficit (Reviron et al., 1992), senescence is induced and propagated from old to mature leaves while senescence is delayed in young leaves. These differences in behavior between leaves suggest a fine and high regulation of metabolism at a whole-plant level and have important consequences on N-use efficiency especially in the case of asynchronism between the N remobilization from source leaves and N demand in sink leaves. Recently, Etienne et al. (2007) have shown that the delay of senescence in N-deprived young leaves of oilseed rape was concomitant with an accumulation of TI activity detected by zymogram at 19 kD. Additionally, the pioneer works of Reviron et al. (1992) have reported that BnD22, a protein with high homology with TI, was accumulated in young leaves of drought-stressed oilseed rape characterized by a delay in senescence. Despite its homology with the Künitz protease inhibitor family, purified BnD22 protein did not significantly inhibit the endogenous leaf proteases or other Ser proteases tested, such as trypsin and chymotrypsin (Ilami et al., 1997; Nishio and Satoh, 1997). However, the high abundance of BnD22 in young leaves adapted to drought was consistent with the hypothesis that BnD22 may contribute to the protection of the young leaves by delaying their senescence. These studies suggested a role of protease inhibitors in the maintenance of growth in young leaves submitted to abiotic stresses. The accumulation of BnD22 transcripts in young leaves submitted to drought (Downing et al., 1992) and N starvation (Etienne et al., 2007) reinforces this assumption. Moreover, BnD22 is also known to be a water-soluble chlorophyll-binding protein (WSCP) of class II (specific class including Brassicacea; Nishio and Satoh, 1997). This WSCP is able to bind chlorophyll molecules, suggesting a role as a chlorophyll carrier (Satoh et al., 2001; Reinbothe et al., 2004) and an involvement in the regulation of both the development and senescence of chloroplasts. Recently, Horigome et al. (2007) have suggested this WSCP may contribute to the photoprotection of chlorophyll molecules.
Compared to its WSCP function, the putative protease inhibitor activity of BnD22 remains an enigma. Therefore, assuming that a protease inhibitor may have a central role in the maintenance of metabolism in young leaves, leading to the modulation of N-use efficiency, our objectives are (1) to determine if BnD22 is related to the 19-kD TI previously detected in oilseed rape young leaves, and (2) to verify if the levels of the BnD22 transcript and protein as well as TI activity in young leaves are associated with the plant tolerance response to stressful conditions such as N starvation and methyl jasmonate (MeJA). Proteomic approaches using high-resolution two-dimensional electrophoresis (2-DE) and identification of the protein by electrospray ionization liquid chromatography tandem mass spectrometry (ESI-LC MS/MS) revealed that (1) a TI activity could be detected after 2-DE gel separation, (2) TI activity was harbored by BnD22, and (3) BnD22 cross-reacted with the antibodies that recognized a WSCP of Brassica oleracea (Nishio and Satoh, 1997). The dual WSCP/TI functions of BnD22 in young leaves of oilseed rape submitted to stress conditions are discussed.
RESULTS
Effects of N Starvation and MeJA on Oilseed Rape Leaves
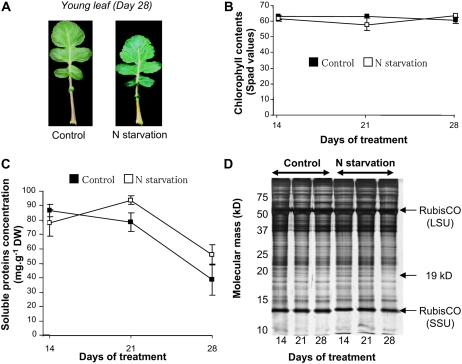
In mature leaves, N starvation led to precocious senescence symptoms as indicated by visible yellowing (data not shown) and a large decrease in chlorophyll content (Table I). The young leaves of N-deprived oilseed rape had no visible senescence symptoms (Fig. 1A). The chlorophyll content in laminae of young leaves remained high throughout the experiment and was not affected by N starvation (Fig. 1B). The decline in the soluble protein concentration in laminae of mature leaves occurred earlier in N-deprived plants than control plants (data not shown). In contrast, the soluble protein concentration in laminae of young leaves decreased between day 21 and day 28 in both treatments (Fig. 1C). To further investigate whether nitrate availability led to a modification of protein patterns in young leaves, SDS-PAGE was performed (Fig. 1D). However, protein at 19 kD that was present at day 14 in both treatments disappeared after 21 d in control plants and 28 d in N-deprived plants.
Table I.
Changes in chlorophyll content in mature leaves submitted to N starvation or MeJA treatments
Mean values of chlorophyll are expressed as percentage of control for each date of treatment ±se.
| Treatment | Days of Treatment
|
||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 3 | 7 | 14 | 21 | 28 | |
| MeJA | 100 | 95.5 (±7.2) | 75.3 (±12.4) | 22.0 (±14.1) | |||
| N starvation | 84.7 (±3.9) | 25.5 (±3.3) | 15.9 (±8.0) | ||||
Figure 1.
A, Photography of young leaves of oilseed rape supplied (control) or not (N starvation) with nitrate during 28 d. B to D, Changes in chlorophyll content (B), soluble protein amount in laminae (C), and SDS-PAGE profiles of soluble proteins (D) in the laminae of young leaf of oilseed rape supplied (control; ▪) or not (N starvation; □) with nitrate during 28 d. Vertical bars indicate ±se of the mean (n = 3) and fit within the plot if not visible. SDS-PAGE was obtained by loading a constant amount of 3 μg of soluble proteins per lane and using silver nitrate staining. Molecular mass markers (kD) are listed on the left of the gel. A protein at 19 kD with specific changes during N treatment is indicated on the right of the gel. LSU, Large subunit of Rubisco (55 kD); SSU, small subunit of Rubisco (14 kD).
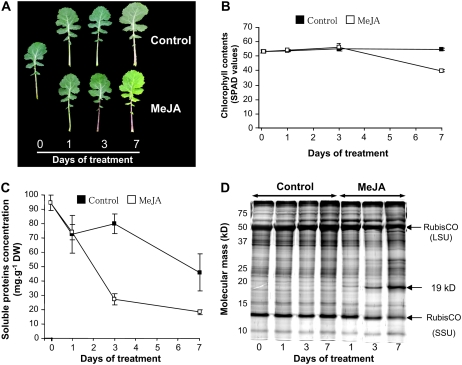
In response to MeJA treatment, leaf senescence was accelerated in mature leaves in comparison to control plants (Table I). Young leaves of MeJA-treated plants showed yellowing at day 7 when compared to control oilseed rape (Fig. 2A). There was also a decline in chlorophyll content in young leaves of MeJA-treated plants between day 3 and day 7 (Fig. 2B) and the concentration of soluble proteins at day 3 (−70% as compared to control; Fig. 2C). SDS-PAGE profiles revealed a decrease in both subunits of Rubisco after 3 d of MeJA treatment (Fig. 2D). In comparison to the controls, a protein of 19 kD was specifically expressed in young leaves of plants treated with MeJA since day 1 and was strongly accumulated at days 3 and 7 to reach a maximum of 9% of total soluble proteins (Fig. 2D).
Figure 2.
A, Photography of young leaves of oilseed rape in control or plant treated with MeJA pulverization during 7 d. B to D, Changes in chlorophyll content (B), soluble protein amounts in laminae (C), and SDS-PAGE profiles of soluble proteins (D) in the laminae of young leaves of oilseed rape treated (control; ▪) or not (MeJA; □) with MeJA during 7 d. Vertical bars indicate ±se of the mean (n = 3) and fit within the plot if not visible. SDS-PAGE was obtained by loading a constant amount of 3 μg of protein per lane and using silver nitrate staining. Molecular mass markers (kD) are listed on the left of the gel. A protein at 19 kD specifically induced by MeJA treatment is indicated on the right of the gel. LSU, Large subunit of Rubisco (55 kD); SSU, small subunit of Rubisco (14 kD).
Changes in TI Activities
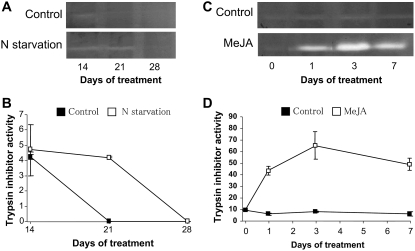
In laminae of young leaves of control plants (Fig. 3, A and C), 19-kD TI activity was weakly present until day 14 and disappeared at day 21 (Fig. 3B). Nitrate starvation induced TI activity in the laminae of young leaf after 14 and 21 d (Fig. 3, A and B). The laminae of young leaves from plants treated with MeJA had significantly higher TI levels than control plants on all dates of treatment (Fig. 3C). For instance, after 3 d of MeJA treatment, TI activity levels were 7-fold higher than in control plants (Fig. 3D). This elevated TI level was maintained until day 7.
Figure 3.
A and B, Changes in the 19-kD TI activities in laminae of young leaves of oilseed rape supplied (control; ▪) or not (N starvation; □) with nitrate between 14 and 28 d. C and D, Changes in the 19-kD TI activities in laminae of young leaves of oilseed rape treated (□) or not (▪) with MeJA during 0, 1, 3, and 7 d of treatment. Detection of a 19-kD protein possessing a TI activity was obtained after SDS-PAGE of soluble proteins (125 μg of proteins loaded per lane; A and C). TI activity corresponds to specific activity and is expressed as integrated intensity per milligram of soluble proteins. Vertical bars indicate ±se of the mean (n = 3) and fit within the plot if not visible.
Detection of TI Activity after 2-DE and Protein Identification
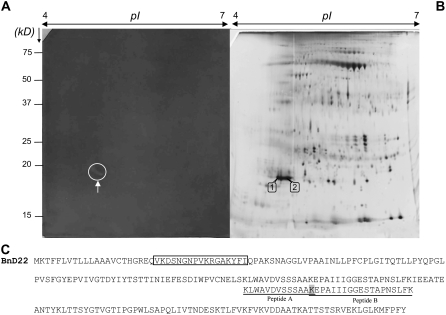
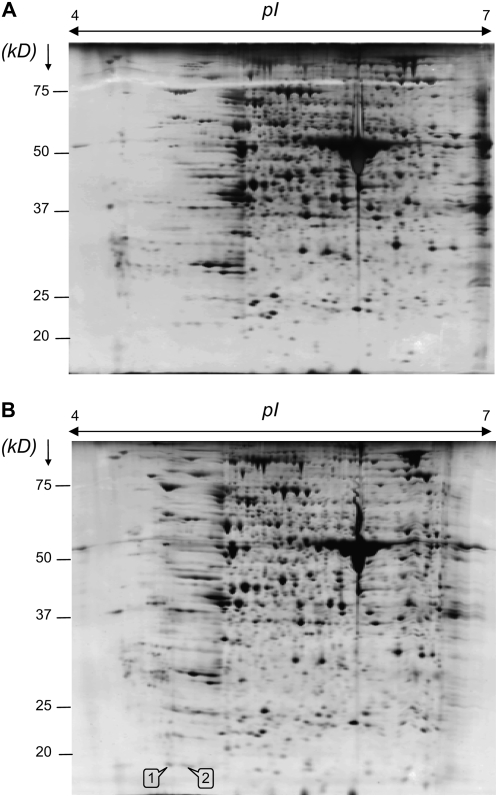
The TI activity in young leaves treated during 7 d with MeJA was also detected after separation of soluble proteins by 2-DE (Fig. 4A). A 2-DE gel stained with silver nitrate (Fig. 4B) indicated that TI activity corresponded to two spots with molecular mass of 19 kD and pI of 5.0 and 5.1, respectively. These two spots were analyzed using ESI-LC MS/MS, and the identified peptides A and B (Fig. 4C) revealed 100% homology to the amino acid sequence of BnD22 (accession no. X65637; Fig. 4C).
Figure 4.
A and B, 2-DE analysis of soluble proteins (340 μg) isolated from laminae of young leaves of plants treated with MeJA at 7 d after TI detection (A) or silver staining (B). Arrows indicate TI activity and two spots (spot nos. 1 and 2) corresponding with the TI activity are numbered. Identified spots were excised and digested with trypsin and analyzed by ESI-LC MS/MS. C, Alignment of 218-amino acid sequence of BnD22 (accession no. X65637) and sequenced peptides A and B by ESI-LC MS/MS from each spot. The shaded amino acid is shared by peptides A and B. Amino acid sequence of the motif of Künitz-type protease inhibitor is surrounded (Nishio and Satoh, 1997).
Identification of BnD22 after Separation of Total Proteins from Laminae by 2-DE
The identification of BnD22 in laminae of young leaves of N-deprived and MeJA-treated plants was confirmed by 2-DE analysis of total proteins (Figs. 5 and 6). After silver staining of total proteins from control and N-deprived plants during 21 d, approximately 950 individual protein spots could be separated by 2-DE across a pI range of 4 to 7 (Fig. 5, A and B). Among the proteins presenting a different pattern between the control and N-deprived plants, two proteins (spot nos. 1 and 2) were also observed at 19 kD and pI 5.0 and 5.1 in laminae of young leaves submitted to N starvation (Fig. 5B). Partial sequences from these two spots, identified by ESI-LC MS/MS, were identical to the peptides A and B previously identified by ESI-LC MS/MS after separation of soluble proteins by 2-DE (Fig. 4C). Consequently, database searches revealed that both polypeptides in Figure 5 also presented 100% homology to BnD22 (Fig. 4C).
Figure 5.
Silver-stained 2-DE gels of total proteins from laminae of young leaves of oilseed rape supplied (A) or not (B) with nitrate at 21 d; 125 μg of total protein was loaded on an IEF strip (for details see “Materials and Methods”). Two spots corresponding with BnD22 are numbered as 1 and 2. Molecular mass markers (kD) are listed on the left of the gel.
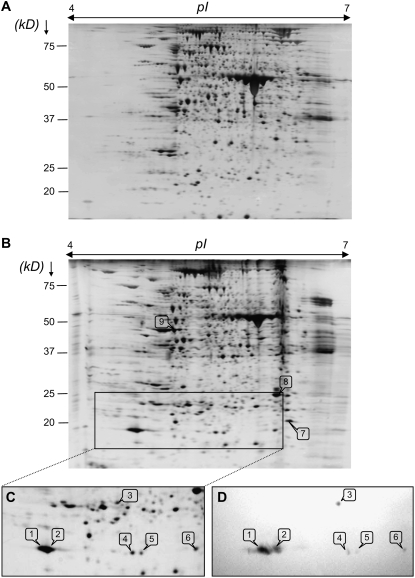
Figure 6.
A and B, Silver-stained 2-DE of total proteins from laminae of young leaves of control plants (A) and plants treated with MeJA (B) during 7 d. A total of 125 μg of total protein was loaded on an IEF strip (for details see “Materials and Methods”). Molecular mass markers (kD) are listed on the left of the gel. C, 2-DE immunoblot from the lamina of a plant treated with MeJA using anti-WSCP antibodies. Total proteins were transferred to a PVDF membrane before immunodetection using anti-WSCP antibodies. Only the region of the 2-DE gel displaying the protein after silver staining is shown in D. This region is surrounded by a box in B. Polypeptides induced by MeJA and/or recognized by the antibodies are numbered from 1 to 9, excised, digested, and identified by ESI-LC MS/MS (see Tables II and III).
Figure 6 shows representative images of 2-DE gels obtained from total proteins of laminae of young leaves from control or MeJA-treated plants after 7 d of experimentation. The total number of protein spots on 2-DE gels as well as the total protein content (data not shown) declined when plants were treated with MeJA (975 spots in control; Fig. 6A) versus 850 spots in MeJA treatment (Fig. 6B). Two spots with a molecular mass of 19 kD and pI of 5.0 and 5.1 were largely accumulated in the MeJA treatment (Fig. 6B). These two proteins (spot nos. 1 and 2) were analyzed using ESI-LC MS/MS (Fig. 4C). The obtained partial sequences were strictly identical to peptides A and B and consequently showed 100% homology to BnD22 as described above after 2-DE of soluble proteins (Fig. 4C).
Immunodetection of BnD22 and WSCP in Young Leaves of Oilseed Rape Using Anti-WSCP Antibodies from B. oleracea and Identification of Cross-Reacted Proteins by ESI-LC MS/MS
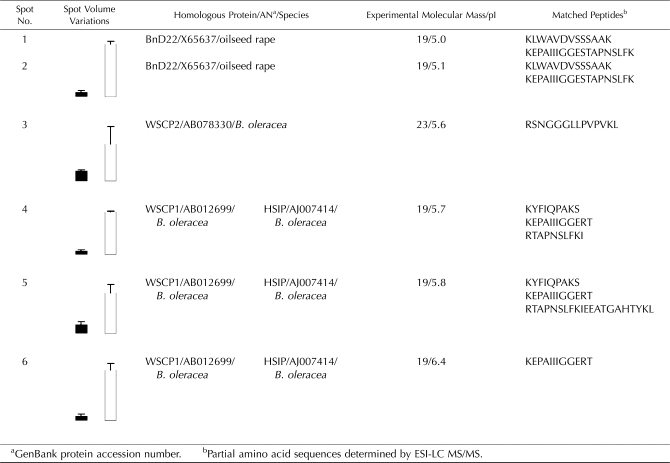
BnD22 is also considered to be a class II WSCP (Nishio and Satoh, 1997; Satoh et al., 1998). To evaluate the different forms of BnD22 and WSCP, 2-DE immunoblot analysis was performed using the anti-WSCP antibodies from B. oleracea (Fig. 6D). Six spots between 19 and 23 kD were recognized by antibodies against WSCP in plants treated with MeJA (Fig. 6, C and D). A pair of polypeptides of 19 kD and pI of 5.0 and 5.1 corresponding with BnD22 (spot nos. 1 and 2) were particularly noticeable. Antibodies against WSCP recognized four other spots with lower intensity signals: one spot at 23 kD with a pI of 5.6 (spot no. 3), and three spots at 19 kD and pI 5.7 (spot no. 4), 5.8 (spot no. 5), and 6.4 (spot no. 6), respectively. To identify spots corresponding with signals, a western blot was compared with a silver-stained 2-DE gel.
Identification by ESI-LC MS/MS revealed that polypeptides of 19 kD and pI 5.0 and 5.1 (spot nos. 1 and 2) were identical to BnD22 (Table II). Spot number 3 at 23 kD and pI 5.6 matched with WSCP2 of B. oleracea (accession no. AB078330; Table II). Using the Progenesis SameSpots software, image gel analysis revealed that BnD22 was induced 12-fold by MeJA, whereas WSCP2 was increased 4-fold (Table II). Spot numbers 4, 5, and 6 were induced 10-, 4-, and 6-fold, respectively (Table II).
Table II.
Identification of spots recognized by anti-WSCP antibodies after western blot of a 2-DE gel of total proteins of leaves treated by MeJA
Spots are presented in Figure 6C. Data were obtained by ESI-LC MS/MS analysis. The normalized volume of each spot identified is indicated for control (black) and MeJA treatment (white). Vertical bars indicate ±se of the mean (n = 3). Experimental pI and molecular mass for the ESI-LC MS/MS-identified peptides are also indicated. The assigned protein of the best matched is given with the organism in which it has been identified and its GenBank protein accession number.
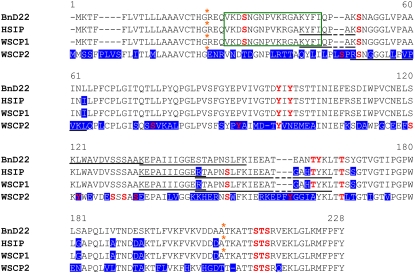
After BLASTp analysis (National Center for Biotechnology Information database), spot numbers 4, 5, and 6 at 19 kD and pI 5.7, 5.8, and 6.4, respectively, were identified as heat stress-induced protein (HSIP; accession no. AJ007414) or WSCP1 (accession no. AB012699) of B. oleracea. Multiple alignments of amino acid sequences (Fig. 7) allowed determination of the degree of homology between BnD22 and the three other proteins recognized by antibodies against WSCP (Fig. 6). Thus, HSIP and WSCP1 were strictly identical and shared 96% homology to BnD22 (Fig. 7). Consequently, these data suggest that WSCP1 and HSIP were the same protein and were different to BnD22. WSCP2 identified in spot number 3 at 23 kD and pI 5.6 showed only 54% homology to BnD22 and 55% homology to WSCP1.
Figure 7.
Sequence alignment of 228 amino acids of BnD22 (spot nos. 1 and 2) with HSIP, WSCP1 (spot nos. 4, 5, and 6), and WSCP2 (spot no. 3) that correspond to six spots detected by polyclonal anti-WSCP antibodies (Table II; Fig. 6). The peptides identified by ESI-LC MS/MS are underlined. Amino acid residues that are not identical to those to BnD22 are shaded in blue. Amino acid sequence of the motif of Künitz-type protease inhibitor is framed in green (Nishio and Satoh, 1997). Asterisks in orange indicate predicted sites of N- and C-signal peptide cleavages. Predicted sites of amino acid phosphorylation are in red and bold.
Figure 7 indicated that BnD22 and WSCP1 presented a similar N-signal peptide (cleaved between amino acid nos. 19 and 20) and C-signal peptide (cleaved between amino acid nos. 196 and 197), while WSCP2 harbored only an N-signal peptide that is cleaved between amino acid numbers 24 and 25. Interestingly, WSCP2 did not possess the specific motif of the Künitz-type protease inhibitor (Fig. 7), while this motif was present in BnD22 and WSCP1. Moreover, searches of predicted sites for phosphorylation of amino acids revealed 10 sites for BnD22 and WSCP1 and 16 sites for WSCP2 (Fig. 7).
Identification of Other Proteins Highly Induced by MeJA
Although proteins recognized by anti-WSCP antibodies were induced by MeJA, other proteins were also up-regulated by this hormone (Fig. 6). Among the induced proteins, three polypeptides were highly accumulated (at least 5-fold) and were identified by ESI-LC MS/MS (Table III). The selected proteins correspond to a putative TI at 21 kD (spot no. 7 induced 11-fold; accession no. U18995), a WSCP2 at 25 kD (spot no. 8 induced 11-fold; accession no. AB078330), and a Jacalin-related lectin family protein at 49 kD (spot no. 9 induced 5-fold; accession no. NM_112518). This last result is in accordance with those reported in recent works using transcriptional approaches (Jiang et al., 2006; Sarosh and Meijer, 2007). Indeed, in oilseed rape, Sarosh and Meijer (2007) have shown that MeJA induced the gene expression of Jacalin-related lectins that have a role in plant defense pathway.
Table III.
Identification of spots highly induced by MeJA after 7 d of treatment
Spots are presented in Figure 6. The normalized volume of each spot identified was indicated for control (black) and MeJA treatment (white). Vertical bars indicate ±se of the mean (n = 3). Experimental pI and molecular mass for the ESI-LC MS/MS-identified peptides are also indicated. The assigned protein of the best matched is given with the organism in which it has been identified and its GenBank protein accession number.
BnD22 Gene Expression
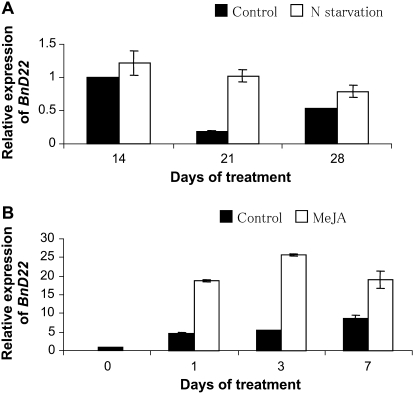
To determine whether TI activity was correlated with BnD22 gene expression, the level of BnD22 transcripts was studied by quantitative (Q)-PCR in the laminae of young leaves in the control, N-deprived, or MeJA-treated oilseed rape. Compared to day 14, the BnD22 transcript level in laminae of control plants decreased 5-fold at day 21 and 2-fold at day 28 (Fig. 8A). Under N-deprived conditions, BnD22 expression increased in young laminae throughout the experiment and was always higher than the control at all time points.
Figure 8.
A, Q-PCR relative BnD22 gene expression in laminae of young leaves of oilseed rape supplied (control; ▪) or not (N starvation; □) with nitrate during 28 d. B, Q-PCR relative BnD22 gene expression in the laminae of young leaves of oilseed rape treated (□) or not (▪) with MeJA during 7 d. Level of BnD22 gene expression at day 0 for MeJA experiment or day 14 for N-starvation experiment was equated to 1. Vertical bar indicates ±se of the mean (n = 3).
Figure 8B presented the accumulation of BnD22 transcripts in young laminae from control and MeJA-treated plants. In control plants, the transcript level of BnD22 increased during the experiment. Compared to the control plants, the expression of BnD22 in plants treated by MeJA significantly increased at all time points. For instance, between days 0 and 3, the expression of the BnD22 gene increased 27-fold in MeJA-treated plants versus only 8-fold higher in control plants.
DISCUSSION
The goals of this study were to verify if BnD22 (1) was homologous to the 19-kD protease inhibitor previously detected by Etienne et al. (2007) in young leaves of oilseed rape, and (2) could be involved in N-use efficiency through the maintenance of physiological functions of sink tissues in plants submitted to stressful conditions. N starvation and MeJA treatment were used to induce leaf senescence (Rossato et al., 2002; Gombert et al., 2006) and TI activity (Cipollini and Sipe, 2001; Etienne et al., 2007).
Detection of TI activity after SDS-PAGE revealed that 19-kD TI activity was induced in young leaves of N-deprived and MeJA-treated plants compared to the controls. In young leaves treated with MeJA, TI activity was 7-fold higher than in controls after 3 d of treatment (Fig. 3D). To identify the proteins harboring TI activity, soluble protein extracts of young leaves treated with MeJA were analyzed by 2-DE gels and through detection of TI activity (Fig. 4). Two polypeptides that were common in both gels were identified precisely at a molecular mass of 19 kD and a pI of 5.0 and 5.1, respectively. ESI-LC MS/MS analysis (Table II) revealed that both polypeptides were homologous to BnD22, a protein previously identified in the youngest leaves of oilseed rape subjected to drought or salinity stress (Reviron et al., 1992). Thus, our study revealed that (1) a TI activity could be detected after 2-DE gel separation and (2) BnD22 has an in vitro protease inhibitor activity. This protease inhibitor activity was consistent with its homology to the Künitz-type proteinase inhibitor family that contains a specific signature motif (Ilami et al., 1997). In fact, this protein of 22 kD contains N- and C-terminal signal peptides (Fig. 7) as reported by Ilami et al. (1997). After the cleavage of the N-signal peptide (between amino acid nos. 19 and 20) and the C-signal peptide (between amino acid nos. 197 and 198), the mature protein contains 178 amino acids with a predicted molecular mass of 19 kD and pI 4.6. This data is in accordance with the two polypeptides detected at 19 kD in our study (spot nos. 1 and 2; Fig. 6). Previously, protease inhibitor activity of the purified mature BnD22, corresponding to a protein at 19 kD, has been studied by Ilami et al. (1997), but the purification by reverse-phase HPLC of this protein led to inactivation of BnD22 protease inhibitor activity. Using proteomics approaches, our study clearly demonstrated that these 19-kD mature proteins possess a TI activity. Interestingly, Reviron et al. (1992) have noted that drought also increased the synthesis of two polypeptides at 20 kD and pI 5.1, and another pair of BnD22 polypeptides at 22 kD and pI 5.1. It is likely that the accumulation of BnD22 isoforms was stress dependent. According to SDS-PAGE (Figs. 1 and 2) and 2-DE profiles (Figs. 5 and 6), 19-kD BnD22 protein abundance and BnD22 gene expression (Fig. 8) presented similar patterns in both treatments. These changes in transcript abundance suggest that mineral N availability and MeJA could play a direct or indirect part in the regulation pathway of the BnD22 gene, as previously observed under various stressful conditions. Indeed, similar transcriptional responses were observed in B. oleracea submitted to other constraints such as heat stress (Annamalai and Yanagihara, 1999) and leaf detachment (Nishio and Satoh, 1997). Moreover, both the BnD22 gene and protein have been shown to accumulate in the youngest leaves of oilseed rape subjected to salinity acclimation and abscisic acid treatment (Downing et al., 1992; Reviron et al., 1992; Ilami et al., 1997). From these data it can be suggested that BnD22 has a proteinase inhibitor role in the response of young leaves to several environmental constraints. Furthermore, in response to water deficit or osmotic stress, the endogenous levels of jasmonates markedly increase in plants such as maize (Zea mays; Xin et al., 1997), barley (Hordeum vulgare; Kramell et al., 1995), soybean (Glycine max; Creelman and Mullet, 1995), and pear (Pyrus bretschneideri; Gao et al., 2004). It has been shown that exogenous application of MeJA can improve the tolerance of plants to drought (Ghasempour et al., 1998). Increase in jasmonate levels can induce the expression of specific genes in plants that are involved in stress tolerance and several metabolic defense mechanisms such as protease inhibitors (Creelman and Mullet, 1997; Kessler and Baldwin, 2002; Mikkelsen et al., 2003; Sarosh and Meijer, 2007). It is interesting to point out that in oilseed rape (1) drought induced the BnD22 gene expression (Downing et al., 1992) and corresponding protein (Reviron et al., 1992), and (2) MeJA increased the BnD22 transcript up to 27-fold after 3 d of treatment (Fig. 8B). Thus, it could be hypothesized that the induction of BnD22 observed in leaves of water-stressed oilseed rape would be mediated by MeJA.
In a drought-resistant genotype of wheat (Triticum aestivum), the induction of a protein homologous to BnD22 identified in the thylakoid membrane showed a significant role for BnD22 during the stress response (Guseynova et al., 2006). In our study, BnD22 represented up to 9% of the leaf soluble protein content in plants treated with MeJA, whereas it represented approximately 1% of the soluble protein content in plants adapted to drought (Downing et al., 1992; Ilami et al., 1997). The accumulation of this inhibitor in response to diverse treatments suggested a physiological plant function for this protease inhibitor in plant responses to stresses. The accumulation of BnD22 in young leaves of oilseed rape subjected to abiotic stress is in accordance with the protective role that has been proposed for the accumulation of this TI undergoing desiccation (Reviron et al., 1992). TIs induced by dessication may protect enzymes susceptible to dehydration or contribute to the inhibition of proteases active during water stress (Lam et al., 1999). Because of the highly hydrophilic nature of TI, the accumulation of BnD22 in salt-stressed plants can also serve as an osmoprotectant (Dombrowski, 2003). Moreover, it is well known that the induction of TI expression by wounding and insect damage is dependent on increases in the wound-related hormone jasmonic acid (Ryan, 1990). The induction of TI activity, BnD22 protein, and corresponding mRNA by MeJA application suggests that BnD22 could be part of a cross-tolerance response acting as an antiherbivore compound by inhibiting pathogen proteases (Heath et al., 1997). Overall results indicate that the accumulation of the BnD22 in young leaves in response to pathogen attacks, wounding, osmotic stresses (drought and NaCl), and mineral N starvation was a means to preserve younger tissues from adverse conditions.
Abiotic stresses such as drought, heat stress, and N starvation, which led to the accumulation of BnD22, also led to a delay of leaf senescence in young leaves (Reviron et al., 1992; Annamalai and Yanagihara, 1999; Etienne et al., 2007). In this study, N-deprived young leaves retained their hardening and had the same morphology as untreated plants, although this was accompanied by an accumulation of TI activity for 7 d longer than in the control plants (Figs. 1 and 3). These results are consistent with those reported by Etienne et al. (2007) where the delay of senescence in N-deprived young leaves was concomitant with an accumulation of TI activity and high BnD22 gene expression. In recent studies, it has been suggested that leaf senescence can be regulated by protease inhibitors and especially TIs of Künitz type (Beers et al., 2000; Sugawara et al., 2002; Shatters et al., 2004; Mosolov and Valueva, 2005; Etienne et al., 2007). As previously suggested by Ilami et al. (1997), the hypothesis of involvement of BnD22 in regulating proteolysis during leaf senescence is reinforced by the fact that BnD22 accumulation in young expanding leaves of oilseed rape was concomitant with a lower proteolytic activity in leaves submitted to water stress. Moreover, as reported by Diop et al. (2004) in Vigna unguiculata Walp, there was a correlation between the level of protein and transcripts of an anti-protease (cystatin) and the degree of water stress tolerance. Consequently, in our study, it could be hypothesized that under N-deprived conditions, BnD22 induction in young leaves of oilseed rape could preserve the integrity of photosynthetic enzymes by inhibiting proteases to maintain metabolic activity and growth. In response to MeJA treatment, high levels of BnD22 protein accumulation (12-fold; Fig. 6) and BnD22 TI activity (7-fold; Fig. 3D) were maintained in young leaves until 7 d, while senescence-like symptoms such as yellowing caused by a large decline in chlorophyll are visible (Figs. 2, B and C, and 3B). Moreover, the hydrolysis of soluble protein content in young leaves treated with MeJA 3 d after starting the experiment (Fig. 2C) was accompanied by reduced levels of Rubisco (Fig. 2D), the most abundant protein in leaves and the main N reserves in the vegetative organs during leaf senescence (Lawlor, 2002). In many species, the Cys and Ser proteases are proteolytic systems that are the most frequently implicated in the N-mobilization processes associated with leaf senescence (Buchanan-Wollaston and Ainsworth, 1997; Coffeen and Wolpert, 2004; Roberts et al., 2006; Zhang et al., 2007). Pak and van Doorn (2005) have suggested that about 40% of the total protease activity in Iris could be governed by Ser proteases. Therefore, in our study, the high level of BnD22 and the corresponding anti-protease activity is not sufficient to avoid the protein breakdown associated with senescence. So, in response to MeJA treatment, it is likely the balance between the level of protease activities (induced during senescence) and anti-protease activities was in favor of proteolysis. This suggests that the ratio of proteases activities/protease inhibitor activities also modulates the rate of remobilization of protein-N compounds in leaves of oilseed rape. In addition, this could explain why, despite the high level of BnD22-TI activity (Fig. 3D), there is a decrease of protein amount per laminae (Fig. 2C) treated by MeJA.
Nishio and Satoh (1997) have clearly demonstrated that BnD22 belongs to class II WSCPs previously identified in B. oleracea. Using polyclonal antibodies against the WSCP of B. oleracea, six polypeptides were detected after 2-DE and western blots from total protein extracts of young leaves treated with MeJA (Fig. 6). ESI-LC MS/MS analysis revealed that anti-WSCP antibodies cross-reacted with two other protein WSCPs (WSCP1 and WSCP2) that are induced by MeJA (Fig. 7; Table II). WSCP2 was detected at 23 kD and pI 5.6, whereas three spots of WSCP1 exhibited similar molecular mass of 19 kD but with different pI 5.7, 5.8, and 6.4 (Fig. 6). As reported by Reviron et al. (1992), the fact that BnD22 and WSCP1 presented different pI could be explained by (1) the products of genes from the two different genomes (B. rapa × B. oleracea) of the amphidiploid oilseed rape or (2) posttranslational processing. For instance, from the data provided by NetPhos analysis (Fig. 7), it could be assumed that the different pI may correspond to a different degree of amino acid phosphorylation (Vilardell et al., 1990). Moreover, analysis of alignment between BnD22 and other WSCPs showed that these proteins are different (Table II; Figs. 6 and 7). Although the incomplete genome sequence information limited analysis of oilseed rape proteins, these data suggest that the oilseed rape proteome could contain several WSCPs as observed for B. oleracea. In this study, only BnD22 possesses a TI activity after 2-DE. In spite of the presence of a motif of the Künitz-type protease inhibitor, the lack of TI activity in WSCP1 (spot nos. 4, 5, and 6) could be explained by their low abundance (Figs. 4 and 6). Concerning protein WSCP2, the absence of a motif of the Künitz-type protease inhibitor explains that no TI activity is associated with this protein. WSCP2 was induced by MeJA at 23 and 25 kD (Fig. 6). These differences of molecular mass could be due to the cleavage of N-signal peptide between amino acid numbers 24 and 25 (Fig. 7). These posttranslational modifications explain why WSCP2 at 25 kD is not recognized by anti-WSCP antibodies contrary to WSCP2 at 23 kD.
Interestingly, BnD22 is a class II WSCP that is able to bind chlorophyll (Chl a/b) and chlorophyll precursors such as chlorophyllides a and b in tetrameric forms (Nishio and Satoh, 1997; Schmidt et al., 2003; Horigome et al., 2007). It has been proposed that WSCP as a chlorophyll carrier implied either chlorophyll degradation (Satoh et al., 1998) or chlorophyll biosynthesis (Schmidt et al., 2003; Reinbothe et al., 2004). However, recent studies have indicated that during leaf senescence, BnD22/WSCP could form a tetrameric BnD22-Chl complex to protect chlorophylls against photodegradation (Horigome et al., 2007). During leaf senescence, the chlorophyll degradation produces radical oxygen species such as singlet oxygen that are able to propagate cellular damage including decomposition of chlorophyll (Mittler et al., 2004). Schmidt et al. (2003) have shown that unbound Chl a was rapidly degraded, whereas WSCP-bound Chl a was decomposed at a significantly slower rate. Moreover singlet-oxygen production was lower for WSCP-bound Chl than for unbound Chl, suggesting that the BnD22-Chl complex could protect against photodegradation of Chl in stressed young leaves (Schmidt et al., 2003; Horigome et al., 2007). The tetrameric assembly of the BnD22-Chl complex reduces contact between the Chl and molecular oxygen and may cause the fluorescence quenching, leading to energy dissipation (Horigome et al., 2007). It is consistent with nonphotochemical quenching identified at the beginning of leaf senescence (Wingler et al., 2004). On the other hand, Park et al. (2007) have shown that during leaf senescence, a senescence-associated gene called Stay-green (Sgr) gene and corresponding protein are highly induced. In fact, Sgr protein regulates chlorophyll degradation by inducing Chl-LHCPII complex disassembly, leading to the degradation of free chlorophylls by catabolic enzymes such as chlorophyllases (Park et al., 2007). In our study, in leaves treated by MeJA, chlorophyll contents decline (especially at day 7; Fig. 2B) in spite of a high induction of BnD22/WSCP proteins (Fig. 6B). Consequently, it could be assumed that in response to MeJA treatment, the presence of WSCPs seems to be insufficient to avoid the chlorophyll breakdown by senescence-induced protein chlorophyll degradation such as Sgr protein.
Despite N-starvation conditions having reduced the growth at whole-plant level, the young leaves showed a significant growth (+362.6 ± 0.056 mg of dry matter versus +553.1 ± 0.024 mg of dry matter in control between days 14 and 28; data not shown) and a similar chlorophyll content as compared to control (Fig. 1B). These data are in accordance with results recently reported by Etienne et al. (2007) and suggest that CO2 assimilation is maintained in young leaves despite N starvation. Interestingly, this maintenance of growth in young leaves of N-deprived plants was accompanied by changes in protein pattern, including the accumulation of BnD22 (Fig. 1D). This suggests that, under stress conditions, the presence of BnD22-Chl complex may act as a photoprotection mechanism to delay leaf senescence in young leaves (Schmidt et al., 2003; Etienne et al., 2007). Altogether, results suggest that BnD22 possesses dual functions (WSCP and TI) that lead to protect younger tissues from adverse conditions by maintaining metabolism (protein integrity and photosynthesis).
CONCLUSION
Using proteomics approaches, this study clearly reveals that the 19-kD TI activity induced in young leaves of oilseed rape in response to stress provoked by N starvation or MeJA is identical to BnD22, a protein that belongs to class II WSCPs. The level of gene expression, protein abundance, and TI activity of BnD22 observed in young leaves present similar patterns and are associated with the maintenance of growth, chlorophyll content, and concentration of soluble proteins. This leads to maintenance of sink strength and may have important consequences for N-use efficiency. The dual WSCP/TI functions of BnD22 suggest that this protein is strongly involved in the maintenance of metabolism (protein integrity and photosynthesis capacity) of young leaves especially when plants are confronted with adverse conditions. In a sense, it could be hypothesized that BnD22 may have an important role in plant resistance to diverse forms of stress. Future investigations using immunolocalization at subcellular level in oilseed rape leaf and/or GFP technologies in Arabidopsis (Arabidopsis thaliana) will attempt to determine the intracellular localization and confirm the physiological functions of BnD22 in the maintenance/protection of the young leaves of plants exposed to various stresses. Additionally, to estimate the contribution of BnD22 to N-use efficiency, it could be interesting to study genotypes of oilseed rape characterized by contrasting leaf N remobilization, leaf lifespan, or by their resistance to low mineral N availability.
MATERIALS AND METHODS
Plant Material
Seeds of oilseed rape (Brassica napus ‘Capitol’) were surface sterilized by exposure to 80% ethanol for 30 s followed by 20% sodium hypochlorite for 20 min. After 10 washes in demineralized water, seeds were germinated on foam rubber (Oasis growing pinpot; Agrimedia). Just after emergence of the third leaf, seedlings were transplanted into 2-L pots filled with attapulgite (one plant per pot) and were grown under greenhouse conditions with a thermoperiod of 20°C (day) and 18°C (night) and a photoperiod of 16 h. Natural light was supplemented with Neon tubes (Philips TLD 36W) supplying an average photosynthetically active radiation of 200 μmol photons m−2 s−1 at the top of the canopy. Plants were watered every 2 d with 100 mL of nutrient solution containing 2 mm KNO3, 3 mm CaCl2, 1 mm K2SO4, 0.5 mm MgSO4, 0.4 mm KH2PO4, 0.15 mm K2HPO4, 0.2 mm Fe-Na-EDTA, 14 μm H3BO3, 5 μm MnSO4, 3 μm ZnSO4, 0.7 μm (NH4)6Mo7O24, 0.7 μm CuSO4, and 0.1 μm CoCl2. After 85 d of growth corresponding to the rosette stage, plants were split into four sets for nitrate starvation and corresponding control or foliar spraying of MeJA and the corresponding control (water spraying).
Application of Treatments (Mineral N Starvation or MeJA Spraying) and Tissue Sampling
Rosette plants grown with nutrient solution containing 3 mm KNO3 were split into two sets of plants. Plants were supplied every 2 d with nutrient solution without KNO3 (N starvation) and 3 mm of KNO3 (control plants). These mineral N treatments were applied during 28 d. Plants were sampled in triplicate after 14, 21, and 28 d of treatment.
For MeJA treatment, one set of plants was sprayed with MeJA solution (100 μm MeJA, 0.05% Tween 20, 6.5 mL plant−1 d−1) applied to the leaves. The second set of plants, corresponding to the control, was sprayed with water (containing 0.05% Tween 20, 6.5 mL plant−1 d−1) on the same date. Control and MeJA-treated plants were sampled in triplicate on the first day (day 0 of experiment) and after 1, 3, and 7 d of treatment.
For all treatments, each leaf rank was separated based on the date of their appearance from the oldest to youngest. The chlorophyll content was measured using a SPAD-502 chlorophyll meter (Minolta). Thereafter, leaves were weighed and the laminae was separated from the petiole, frozen in liquid N, and stored at −80°C until further analysis.
Soluble Protein Extraction and SDS-PAGE
Frozen laminae samples (200 mg fresh weight) from each triplicate were ground in a mortar with liquid N in the presence of 150 mg polyvinylpolypyrrolidone. Soluble protein samples were extracted in citrate Na-phosphate buffer (20 mm citrate and 160 mm Na2HPO4, pH 6.8). The homogenate was centrifuged at 12,000g at 4°C for 1 h and the resulting supernatant was used for the determination of soluble protein concentration by protein-dye staining (Bradford, 1976) using bovine serum albumin as standard. For SDS-PAGE, 3 μg of soluble proteins was prepared in 2× Laemmli lysis buffer containing β-mercaptoethanol (5% v/v). SDS-PAGE was performed as described by Laemmli (1970) in a 5.5% polyacrylamide (w/v) stacking gel and a 15% polyacrylamide (w/v) resolving gel. Gels were stained with the silver staining procedure described by Blum et al. (1987).
Total Protein Extraction
Frozen laminae samples (200 mg fresh weight) from each triplicate were ground in a mortar using liquid N and resuspended in 2 mL of cold acetone containing 10% TCA. After centrifugation at 16,000g for 3 min at 4°C, the supernatant was discarded and the pellet was rinsed as previously described by Wang et al. (2003). The pellet was resuspended in 200 μL of rehydration R2D2 buffer [5 m urea, 2 m thiourea, 2% CHAPS, 2% N-decyl-N,N-dimethyl-3-ammonio-1-propanesulfonate, 20 mm dithiothreitol, 5 mm Tris(2-carboxyethyl)phosphine, 0.5% IPG buffer (GE Healthcare), pH 4 to 7; Mechin et al., 2003]. The total protein concentration was determined by the method of Bradford (1976) using bovine serum albumin as standard.
2-DE
For 2-DE of soluble proteins, 100 μL of protein was purified using a ReadyPrep 2-DE clean-up kit (Bio-Rad). The final pellet was resuspended in rehydration R2D2 buffer. The extract of total or soluble proteins was first separated according to charge in the electrofocusing PROTEAN IEF system (Bio-Rad) at 20°C, using 18-cm gel strips forming an immobilized linear pH gradient from 4 to 7 (GE Healthcare). Each strip was rehydrated at 50 μA/gel for 14 h in the presence of 330 μL of R2D2 buffer containing 125 μg of total or soluble proteins. Isoelectric focusing ran for 15 min at 250 V, 2 h at 500 V, and then until 50 kV at 10,000 V. After electrofocusing, the strips were immediately equilibrated in the equilibration buffer (75 mm Tris-HCl, 3% [w/v] SDS, 300 mm Tris base) containing dithiothreitol (65 mm), followed by a second incubation in equilibration buffer containing iodoacetamide (50 mm) and bromphenol blue (0.5%). 2-DE electrophoresis was carried out on 12% polyacrylamide (w/v) gels (20 × 20 cm) using an Investigator system (Millipore) at 300 mV. Gels were stained using the silver-staining procedure described by Blum et al. (1987). Gels were scanned with the ProXPRESS 2D proteomic imaging system (Perkin-Elmer) before image analysis.
Image Analysis of 2-DE Gels
Images of the 2-DE gels were acquired with the ProXPRESS 2D proteomic Imaging system and analyzed using the Progenesis SameSpots software v3.0 (Nonlinear Dynamics) according the manufacturer's protocol. Gels from three independent biological replicates were used. Spot detection, warping, and matching were performed automatically by the software. Matching was automatic but verified manually; artifacts, or spots that could not be confidently verified as true matches, were disregarded rather than manually edited, and misalignments were corrected by manual warping when appropriate. Molecular mass and pI were calculated using Samespots software calibrated with commercial molecular mass standards (precision protein standards unstained; Bio-Rad) run in separate marker lane on 2-DE gel.
Detection of TI Activity after SDS-PAGE and 2-DE
For SDS-PAGE, one volume of each triplicate was mixed together and prepared in 2× lysis buffer without β-mercaptoethanol. SDS-PAGE was performed using a 5.5% polyacrylamide (w/v) stacking gel and an 18% polyacrylamide (w/v) resolving gel as described by Etienne et al. (2007). Seventy-five micrograms of soluble proteins was loaded per lane and separated by SDS-PAGE. For the 2-DE gel, 340 μg of soluble proteins was purified using a ReadyPrep 2-D clean-up kit (Bio-Rad). After solubilization in R2D2 buffer, proteins were separated by 2-DE gels as described above. For detection of TI activity, gels were stained as described by Etienne et al. (2007) using a procedure adapted from Yeh et al. (1997). Briefly, gels were washed for 30 min with 25% 2-propanol (v/v) and 30 min with Tris buffer (20 mm, pH 6). Thereafter, gels were incubated 30 min in Tris-CaCl2 buffer (50 mm Tris base, 50 mm CaCl2, pH 7.6) containing 40 mg L−1 bovine trypsin type I (EC 3.4.21.4). The detection of TI was observed after 45 min in staining solution (0.6 mm N-acetyl-dl-Ala-2-naphthyl ester and 0.84 mm O-dianisidine tetrazotized in Tris-HCl-CaCl2 buffer [50 mm Tris base, 50 mm CaCl2, pH 7.6]). Gels were finally washed in 2% acetic acid (v/v) and were scanned with the ProXPRESS 2D proteomic imaging system. TI activity was estimated with the Millipore BioImage computerized image analysis system (Millipore) by measurement of integrated intensity. For each leaf rank, gels were made for each biological repetition (n = 3). Each gel was replicated two times and the data relating to the specific TI activity were expressed as integrated intensity per milligram of proteins. Gels presented in the manuscript corresponded to the gels obtained with the mix of the three biological repetitions.
Immunoblot Analysis
The proteins samples were first separated by 2-DE as described above except that 250 μg of proteins was loaded in the strip. Electrophoretic transfer of polypeptides from 2-DE gels onto polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Proteigene) was conducted by semidry electroblotting (2.5 mA for 20 min; Milli Blot system; Proteigene), according to the protocol described by Towbin et al. (1979). After blotting, PVDF membranes were treated with polyclonal anti-cauliflower-WSCP (dilution 1/1,000) rabbit antibodies that cross-reacted with class II WSCP of Brassica olearacea (Nishio and Satoh, 1997). The antigen-antibody complex was visualized with alkaline phosphatase linked to goat anti-rabbit IgG (Bio-Rad, dilution 1/6,000) as described by Blake et al. (1984).
Protein Identification by ESI-LC MS/MS
Spots of interest were manually excised from gels and destained as described by Gharahdaghi et al. (1999). After digestion with 40 μL of sequencing grade modified trypsin solution (41 units per μL; Promega) overnight at 37°C, samples were resuspended in 10 μL of 0.1% formic acid in ultrapure water. An electrospray ion trap spectrometer (LCQ DecaXP; ThermoFinnigan) coupled online with HPLC (SurveyorLC) was used for peptide analysis. Peptides were separated by reverse-phase HPLC on a C18 capillary column (ThermoHyPurity C18 150 × 0.18). A linear 18-min gradient (flow rate, 3 μL min−1) from 5% to 80% B was used, where solvent A was 0.1% aqueous formic acid and solvent B was 0.1% formic acid in acetonitrile. The ESI parameters were as follows: spray voltage, 3.5 kV; sheath gas flow rate, 30; capillarity temperature, 200°C; capillarity voltage, 30 V; tube lens offset, 35 V. Mass spectrometry was acquired in a mode that alternated a full MS scan (mass range: 400–1,600) and a collision-induced dissociation MS/MS of the most abundant ion. The collision energy for the MS/MS scan was preset at the value of 35%. The LC MS/MS data were converted into DTA-format files that were further searched for proteins with MASCOT Daemon (Matrix Science). Proteins with two or more unique peptides matching the protein sequence were automatically considered as a positive identification. In the situation where proteins had a unique peptide sequence match, the difference between theoretical and experimental Mr/pI should not exceed ±25% variance to be considered a match. Moreover, only peptides matching an individual ion score >41 were accepted as a positive identification. For protein identification, two strategies were employed to mine the maximum information. Measured peptides were searched in the NCBInr-protein sequence database viridiplantae (green plants) and in the Brassica EST database (Brassica Genome Gateway 2007).
Alignment analysis of assigned proteins was predicted using ClustalW (npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html). The research of predicted cleavage sites of signal peptides and phosphorylation sites was performed using SignalP 3.0 (www.cbs.dtu.dk/services/SignalP/) and NetPhos 2.0 (www.cbs.dtu.dk/services/NetPhos/), respectively.
RNA Extraction
Total RNA was extracted from 250 mg of laminae fresh matter. Frozen samples were ground to a powder with a pestle in a mortar containing liquid N. The resulting powder was suspended in 750 μL extraction buffer (0.1 m Tris, 0.1 m LiCl, 0.01 m EDTA, 1% SDS [w/v], pH 8) and 750 μL of hot phenol (80°C, pH 4). This mixture was vortexed for 30 s and after addition of 750 μL of chloroform/isoamylalcohol (24:1), the homogenate was centrifuged at 15,000g (5 min, 4°C). The supernatant was transferred into 4 m LiCl solution (w/v) and incubated overnight at 4°C. After centrifugation (15,000g, 30 min, 4°C), the pellet was suspended in 250 μL of sterile water. Fifty microliters of 3 m sodium acetate (pH 5.6) and 1 mL of 96% ethanol were added to precipitate the total RNA for 1 h at −80°C. After centrifugation (15,000g, 20 min, 4°C), the pellet was washed with 1 mL of 70% ethanol, then centrifuged at 15,000g for 5 min at 4°C. The resulting pellet was dried for 5 min at room temperature and resuspended in sterile water containing 0.1% SDS and 20 mm EDTA. Quantification of total RNA was performed by spectrophometre at 260 nm (BioPhotometer) before reverse transcription (RT) and Q-PCR analyses.
RT and Q-PCR Conditions
For RT, 1 μg of total RNA was converted to cDNA with an iScript cDNA synthesis kit using the manufacturer's protocol (Bio-Rad). For Q-PCR amplification, primers of BnD22 were designed after multiple alignment of nucleotidic sequences between BnD22 (accession no. X65637), encoding an oilseed rape TI, WSCP1 (accession no. AB012699) and WSCP2 (accession no. AB078330), encoding B. oleracea WSCP1 and WSCP2, respectively, and HSIP (accession no. AJ007414), encoding a B. oleracea HSIP. To target BnD22 cDNA, specific primers were designed in the nonconserved region. Thus, specific primers selected for the Q-PCR analysis were: BnD22 forward primer 5′-CCGGTTAGCTTCGGATATGA-3′ and reverse primer 5′-AGCTATTTGGGGCCGTACTT-3′. For Q-PCR analysis, EF1-α gene (accession no. DQ312264), encoding an oilseed rape elongation factor, was used as an internal control gene (Nicot et al., 2005) and was amplified using the following specific primers: EF1-α forward primer 5′-TTTCGAGGGTGACAACATGA-3′ and reverse primer 5′-CCGTTCCAATACCACCAATC-3′. Q-PCR reactions were performed with 4 μL of 200× diluted cDNA, 500 nm of primers, and 1× SYBR Green PCR Master Mix (Bio-Rad) in a ChromoFour system (Bio-Rad). For each pair of primers, a threshold value and PCR efficiency have been determined using a cDNA preparation diluted over 10-fold. For both pairs of primers, PCR efficiency was around 100%. The specificity of PCR amplification was examined by monitoring the presence of the single peak in the melting curves of Q-PCR. After sequencing (Biofidal) and BLASTn analysis (www.ncbi.nlm.nih.gov/blast/Blast.cgi) to check the correct amplification of the target cDNA, the BnD22 amplicon (199 bp of length) was 100% homologous to the BnD22 cDNA sequence. For each sample, the subsequent Q-PCR reactions were realized in triplicate and the relative expression of the BnD22 genes in each sample was referred to a control sample (control plant at day 14 for N-starvation experiment and control plant at day 0 for MeJA treatment) and was determined with the ΔΔCt method using the following equation: relative expression = 2−[ΔCt sample − ΔCt control], with ΔCt = CtBnD22 − CtEF1-α where Ct refers to the threshold cycle determined for each gene in the exponential phase of PCR amplification. Using this analysis method, relative expression of the BnD22 gene in the control sample was equal to one (20) by definition (Livak and Schmittgen, 2001).
Statistics
Results are presented as mean values for the three plant material (triplicate) batches with ses. The effects of mineral N starvation or MeJA treatment were assessed by ANOVA, and mean separation was performed using Fisher-Snedecor test. Statistical significance was postulated at P < 0.05.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers X65637, AB078330, AB012699, AJ007414, U18995, AB078330, NM112518, and DQ312264.
Acknowledgments
The authors would like to thank Dr. Laurent Coquet, Dr. Philippe Laîné, Dr. Aurélie Verneuil, and Dr. Vianney Pichereau for their valuable help in ESI-LC MS/MS analyses, and Sandrine Rezé for her technical help in Q-PCR analyses. We would like to thank the three anonymous reviewers for their valuable comments.
This work was supported by a Ph.D. grant from the Institut National de la Recherche Agronomique and the Conseil Régional de Basse-Normandie (to M.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jean-Christophe Avice (jean-christophe.avice@unicaen.fr).
References
- Aerts R, Chapin FS (2000) The mineral nutrition of wild plants revisited: a re-evaluation of processes and patterns. Adv Ecol Res 30 402–407 [Google Scholar]
- Annamalai P, Yanagihara S (1999) Identification and characterization of a heat-stress induced gene in cabbage encodes a Kunitz type protease inhibitor. J Plant Physiol 155 226–233 [Google Scholar]
- Beers EP, Woffenden BJ, Zhao CS (2000) Plant proteolytic enzymes: possible roles during programmed cell death. Plant Mol Biol 44 399–415 [DOI] [PubMed] [Google Scholar]
- Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC (1984) A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem 13 6175–6179 [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamid gel. Electrophoresis 8 93–99 [Google Scholar]
- Bradford MM (1976) A rapid method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C (1997) Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridisation. Plant Mol Biol 33 821–834 [DOI] [PubMed] [Google Scholar]
- Cipollini DF, Bergelson J (2000) Environmental and developmental regulation of trypsin inhibitor activity in Brassica napus. J Chem Ecol 26 1411–1422 [Google Scholar]
- Cipollini DF, Sipe M (2001) Jasmonic acid treatment and mammalian herbivory differentially affect chemical defenses and growth of wild mustard (Brassica kaber). Chemoecology 11 137–143 [Google Scholar]
- Coffeen WC, Wolpert TJ (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1995) Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA 92 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1997) Oligosaccharides, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell 9 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejoux JF, Recous S, Meynard JM, Trinsoutrot I, Leterme P (2000) The fate of nitrogen from winter-frozen rapeseed leaves: mineralization, fluxes to the environment and uptake by rapeseed crop in spring. Plant Soil 218 257–272 [Google Scholar]
- Diop NN, Kidric M, Repellin A, Gareil M, d'Arcy-Lameta A, Pham Thi AT, Zuily-Fodil Y (2004) A multicystatin is induced by drought stress in cowpea (Vigna unguiculata (L.) Walp.) leaves. FEBS Lett 577 545–550 [DOI] [PubMed] [Google Scholar]
- Dombrowski JE (2003) Salt stress activation of wound-related genes in tomato plants. Plant Physiol 132 2098–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing WL, Mauxion F, Fauvarque MO, Reviron MP, Devienne D, Vartanian N, Giraudat J (1992) A Brassica-napus transcript encoding a protein related to the Kunitz protease inhibitor family accumulates upon water-stress in leaves, not in seeds. Plant J 2 685–693 [PubMed] [Google Scholar]
- Etienne P, Desclos M, Le Gou L, Gombert J, Bonnefoy J, Maurel K, Le Dily F, Ourry A, Avice JC (2007) N-protein mobilisation associated with the leaf senescence process in oilseed rape is concomitant with the disappearance of trypsin inhibitor activity. Funct Plant Biol 34 895–906 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA (1992) Octadecanoid precursor of jasmonic acid activates the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XP, Wang XF, Lu YF, Zhang LY, Shen YY, Liang Z, Zhang DP (2004) Jasmonic acid is involved in the water-stress-induced betaine accumulation in pear leaves. Plant Cell Environ 24 497–507 [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM (1999) Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20 601–605 [DOI] [PubMed] [Google Scholar]
- Ghasempour HR, Anderson EM, Gianello RD, Gaff DF (1998) Growth inhibitor effects on protoplasmic drought tolerance and protein synthesis in leaf cells of the resurrection grass, Sporobolus stapfianus. Plant Growth Regul 24 179–183 [Google Scholar]
- Gombert J, Etienne P, Ourry A, Le Dily F (2006) The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. J Exp Bot 57 1949–1956 [DOI] [PubMed] [Google Scholar]
- Good A, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9 597–605 [DOI] [PubMed] [Google Scholar]
- Guseynova IM, Sueleymanov SY, Aliyev JA (2006) Protein composition and native state of pigments of thylakoid membrane of wheat genotypes differently tolerant to water stress. Biochemistry 71 173–177 [DOI] [PubMed] [Google Scholar]
- Heath I, McDonald G, Christeller JT, Lee M, Bateman K, West J, van Heesjwick R, Anderson MA (1997) Proteinase inhibitors from Nicotiana alata enhance plant resistance to insect pests. J Insect Physiol 43 833–842 [DOI] [PubMed] [Google Scholar]
- Horigome D, Satoh H, Itoh N, Mitsunaga K, Oonishi I, Nakagawa A, Uchida A (2007) Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein. J Biol Chem 282 6525–6531 [DOI] [PubMed] [Google Scholar]
- Horn M, Patankar AG, Zavala JA, Wu J, Dolecková-Maresová L, Vujtechová M, Mares M, Baldwin IT (2005) Differential elicitation of two processing proteases controls the processing pattern of the trypsin proteinase inhibitor precursor in Nicotiana attenuata. Plant Physiol 139 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53 927–937 [DOI] [PubMed] [Google Scholar]
- Ilami G, Nespoulous C, Huet JC, Vartanian N, Pernollet JC (1997) Characterization of BnD22, a drought-induced protein expressed in Brassica napus leaves. Phytochemistry 45 1–8 [Google Scholar]
- Jiang JF, Han Y, Xing LJ, Xu YY, Xu ZH, Chong K (2006) Cloning and expression of a novel cDNA encoding a mannose-specific jacalin-related lectin from Oryza sativa. Toxicon 47 133–139 [DOI] [PubMed] [Google Scholar]
- Jiang WB, Lers A, Lomaniec E, Aharoni N (1999) Senescence-related serine protease in parsley. Phytochemistry 50 377–382 [Google Scholar]
- Kang SG, Choi JH, Suh SG (2002) A leaf-specific 27 kDa protein of potato künitz-type proteinase inhibitor is induced in response to abscisic acid, ethylene, methyl jasmonate, and water deficit. Mol Cells 13 144–147 [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53 299–328 [DOI] [PubMed] [Google Scholar]
- Kramell R, Atzorn R, Schneider G, Miersch O, Bruckner C, Schmidt J, Sembdner G, Parthier B (1995) Occurrence and identification of jasmonic acid and its amino acid conjugates induced by osmotic stress in barley leaf tissue. J Plant Growth Regul 14 29–36 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the heat bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lam E, Pontier D, del Pozo O (1999) Die and let live-programmed cell death in plants. Curr Opin Plant Biol 2 502–507 [DOI] [PubMed] [Google Scholar]
- Lawlor DW (2002) Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. J Exp Bot 53 789–799 [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Malagoli P, Laine P, Rossato L, Ourry A (2005) Dynamics of nitrogen uptake and mobilization in field-grown winter oilseed rape (Brassica napus) from stem extension to harvest. II. An N-15-labelling-based simulation model of N partitioning between vegetative and reproductive tissues. Ann Bot (Lond) 95 1187–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechin V, Consoli L, Le Guilloux M, Damerval C (2003) An efficient solubilization buffer for plant proteins focused in immobilized pH gradients. Proteomics 3 1299–1302 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Larsen Petersen B, Glawischnig E, Bøgh Jensen A, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Mosolov VV, Valueva TA (2005) Proteinase inhibitors and their function in plants: a review. Appl Biochem Microbiol 41 227–246 [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56 2907–2914 [DOI] [PubMed] [Google Scholar]
- Nishio N, Satoh H (1997) A water-soluble chlorophyll protein in cauliflower may be identical to BnD22, a drought-induced, 22 kilodalton protein in rapeseed. Plant Physiol 115 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Waternac C, Leyser HMO, Bowles DJ (1996) Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1918 [DOI] [PubMed] [Google Scholar]
- Pak C, van Doorn WG (2005) Delay of Iris flower senescence by protease inhibitors. New Phytol 165 473–480 [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peňa-Cortés H, Willmitzer L, Sànchez-Serrano JJ (1991) Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell 3 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, Satoh H, Alcaraz JP, Reinbothe S (2004) A novel role of water-soluble chlorophyll proteins in the transitory storage of chorophyllide. Plant Physiol 134 1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reviron MP, Vartanian N, Sallantin M, Huet JC, Pernollet JC, De Vienne D (1992) Characterization of a novel protein induced by progressive or rapid drought and salinity in Brassica napus leaves. Plant Physiol 100 1486–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M (1991) Seed storage proteins: the enzyme inhibitors. In PM Dey, JB Harborne, eds, Methods in Plant Biochemistry, Amino Acids, Proteins and Nucleic Acids, Vol 5. Academic Press, New York, pp 259–305
- Roberts IN, Passeron S, Barneix AJ (2006) The two main endoproteases present in dark-induced senescent wheat leaves are distinct subtilisin-like proteases. Planta 224 1437–1447 [DOI] [PubMed] [Google Scholar]
- Rossato L, Laine P, Ourry A (2001) Nitrogen storage and remobilization in Brassica napus L. during the growth cycle: nitrogen fluxes within the plant and changes in soluble protein patterns. J Exp Bot 52 1655–1663 [PubMed] [Google Scholar]
- Rossato L, Macduff JH, Lainé P, Le Deunff E, Ourry A (2002) Nitrogen storage and remobilization in Brassica napus L. during the cycle: effects of methyl jasmonate on nitrate uptake, senescence, growth, and VSP accumulation. J Exp Bot 53 1131–1141 [DOI] [PubMed] [Google Scholar]
- Ryan CA (1990) Protease inhibitor in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28 425–449 [Google Scholar]
- Sanchez-Hernandez C, Martinez-Gallardo N, Guerrero-Rangel A, Valdes-Rodriguez S, Delano-Frier J (2004) Trypsin and alpha-amylase inhibitors are differentially induced in leaves of amaranth (Amaranthus hypochondriacus) in response to biotic and abiotic stress. Physiol Plant 122 254–264 [Google Scholar]
- Sarosh BR, Meijer J (2007) Transcriptional profiling by cDNA-AFLP reveals novel insights during methyl jasmonate, wounding and insect attack in Brassica napus. Plant Mol Biol 64 425–438 [DOI] [PubMed] [Google Scholar]
- Satoh H, Nakayama K, Okada M (1998) Molecular cloning and functional expression of a water-soluble chlorophyll protein, a putative carrier of chlorophyll molecules in cauliflower. J Biol Chem 273 30568–30575 [DOI] [PubMed] [Google Scholar]
- Satoh H, Uchida A, Nakayama K, Okada M (2001) Water-soluble chlorophyll protein in Brassicaceae plants is a stress-induced chlorophyll-binding protein. Plant Cell Physiol 42 906–911 [DOI] [PubMed] [Google Scholar]
- Schjoerring JK, Bock JGH, Gammelvind L, Jensen CR, Mogensen VO (1995) Nitrogen incorporation and remobilization in different shoot components of field-grown winter oilseed rape (Brassica napus L.) as affected by rate of nitrogen application and irrigation. Plant Soil 177 255–264 [Google Scholar]
- Schmidt K, Fufezan C, Krieger-Liszkay A, Satoh H, Paulsen H (2003) Recombinant water-soluble chlorophyll protein from Brassica oleracea var. Botrys binds chlorophyll derivates. Biochemistry 42 7427–7433 [DOI] [PubMed] [Google Scholar]
- Shatters RG, Bausher MG, Hunter WB, Chaparro JX, Dang PM, Niedz RP, Mayer RT, McCollum TG, Sinisterra X (2004) Putative protease inhibitor gene discovery and transcript profiling during fruit development and leaf damage in grapefruit (Citrus paradisi Macf.). Gene 326 77–86 [DOI] [PubMed] [Google Scholar]
- Sin SF, Chye ML (2004) Expression of proteinase inhibitor II proteins during floral development in Solanum americanum. Planta 219 1010–1022 [DOI] [PubMed] [Google Scholar]
- Sugawara H, Shibuya K, Yoshioka T, Hashiba T, Satoh S (2002) Is a cysteine proteinase inhibitor involved in the regulation of petal wilting in senescing carnation (Dianthus caryophyllus L.) flowers? J Exp Bot 53 407–413 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Kassner N, Struck C, Lohaus G (2005) Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 221 328–338 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell J, Goday A, Freire MA, Torrent M, Martinez MC, Torne JM, Pages M (1990) Gene sequence, developmental expression, and protein phosphorylation of RAB-17 in maize. Plant Mol Biol 14 423–432 [DOI] [PubMed] [Google Scholar]
- Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresto M (2003) Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24 2369–2375 [DOI] [PubMed] [Google Scholar]
- Wingler A, Marès M, Pourtau N (2004) Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol 161 781–789 [DOI] [PubMed] [Google Scholar]
- Xin ZY, Yhou X, Pilet PE (1997) Level changes of jasmonic, abscisic, and indole-3yl-acetic acids in maize under desiccation stress. J Plant Physiol 151 120–124 [Google Scholar]
- Yasumura Y, Hikosaka K, Tadaki Hirose T (2007) Nitrogen resorption and protein degradation during leaf senescence in Chenopodium album grown in different light and nitrogen conditions. Funct Plant Biol 34 409–417 [DOI] [PubMed] [Google Scholar]
- Yeh KW, Chen JC, Lin MI, Chen YM, Lin CY (1997) Functional activity of sporamin from sweet potato (Ipomoea batatas Lam): A tuber storage protein with trypsin inhibitory activity. Plant Mol Biol 33 565–570 [DOI] [PubMed] [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6 79–84 [DOI] [PubMed] [Google Scholar]
- Zhang P, Wang F, Zhang LF, Rui Z, Xu LL (2007) Endopeptidase isoenzyme characteristics in Cucumis sativus leaves during dark-induced senescence. Journal of Integrative Plant Biology 49 507–514 [Google Scholar]
- Zimmermann P, Zentgraf U (2005) The correlation between oxidative stress and leaf senescence during plant development. Cell Mol Biol Lett 10 515–534 [PubMed] [Google Scholar]