Abstract
Secondary xylem (wood) formation in gymnosperms requires that the tracheid protoplasts first build an elaborate secondary cell wall from an array of polysaccharides and then reinforce it with lignin, an amorphous, three-dimensional product of the random radical coupling of monolignols. The objective of this study was to track the spatial distribution of monolignols during development as they move from symplasm to apoplasm. This was done by feeding [3H]phenylalanine ([3H]Phe) to dissected cambium/developing wood from lodgepole pine (Pinus contorta var latifolia) seedlings, allowing uptake and metabolism, then rapidly freezing the cells and performing autoradiography to detect the locations of the monolignols responsible for lignification. Parallel experiments showed that radioactivity was incorporated into polymeric lignin and a methanol-soluble pool that was characterized by high-performance liquid chromatography. [3H]Phe was incorporated into expected lignin precursors, such as coniferyl alcohol and p-coumaryl alcohol, as well as pinoresinol. Coniferin, the glucoside of coniferyl alcohol, was detected by high-performance liquid chromatography but was not radioactively labeled. With light microscopy, radiolabeled phenylpropanoids were detected in the rays as well as the tracheids, with the two cell types showing differential sensitivity to inhibitors of protein translation and phenylpropanoid metabolism. Secondary cell walls of developing tracheids were heavily labeled when incubated with [3H]Phe. Inside the cell, cytoplasm was most strongly labeled followed by Golgi and low-vacuole label. Inhibitor studies suggest that the Golgi signal could be attributed to protein, rather than phenylpropanoid, origins. These data, produced with the best microscopy tools that are available today, support a model in which unknown membrane transporters, rather than Golgi vesicles, export monolignols.
The processes of secondary xylem development in gymnosperms, from cambial divisions to maturation of tracheids by programmed cell death, have been defined using ultrastructural (Samuels et al., 2002), molecular (Allona et al., 1998; Kirst et al., 2003), and biochemical (Savidge, 1989) approaches. The most prominent feature of conifer xylogenesis is the development of the lignified secondary cell wall in tracheids, which takes place in two sequential steps: polysaccharide synthesis followed by lignification.
Lignification results from the dehydrogenative polymerization of monolignols, which are synthesized from Phe via the phenylpropanoid pathway. Detailed characterization of genes encoding phenylpropanoid biosynthetic enzymes has led to revisions of the pathway for monolignol biosynthesis (Humphreys and Chapple, 2002; Boerjan et al., 2003; Goujon et al., 2003). In gymnosperms, the major monolignol, coniferyl alcohol, accumulates in its glucosylated form as coniferin, and its concentration in the inner bark has been positively correlated with active growth (Marcinowski and Grisebach, 1977; Savidge, 1988). The abundance of coniferin led to the hypothesis that, first, coniferin is exported during xylem development, then coniferin β-glucosidases in the secondary cell wall cleave off the Glc, generating coniferyl alcohol for lignification in the apoplast (Freudenberg, 1959). However, the mechanism and form of monolignols being exported from their site of synthesis in the cytoplasm to their site of polymerization in the apoplast are still poorly understood (Whetten and Sederoff, 1995; Fukushima et al., 1997; Whetten et al., 1998; Ehlting et al., 2005).
During secondary cell wall deposition in lodgepole pine (Pinus contorta var latifolia), the protoplasts of the developing tracheids have prominent Golgi with grape-like clusters of vesicles being shed from the trans-Golgi network (Samuels et al., 2002). The predominant hemicelluloses of Pinus secondary cell wall, galactoglucomannans, have been localized to these vesicles using enzyme-gold probes (glucomannan-specific mannanase linked to colloidal gold; Samuels et al., 2002). This work led to the following question: do these abundant Golgi vesicles also carry monolignols to the developing secondary cell wall?
The objective of this study was to localize monolignols in intact developing tracheids during lignification using transmission electron microscopy (TEM). Since antibodies specific to lignin detect only polymeric forms with varying degrees of condensation (Müsel et al., 1997), they are unable to detect monolignols prior to polymerization. Therefore, we employed a novel autoradiography approach to localize monolignols. Autoradiography has been used in the past to localize soluble radioactive phenylpropanoids, and it has been reported that monolignols are secreted via Golgi-mediated vesicle fusion with the plasma membrane (Pickett-Heaps, 1968; Wooding, 1968; Takabe et al., 1985). In these studies, following the uptake of radiolabeled precursors, developing xylem samples for autoradiography were prepared for light microscopy and TEM using chemical fixatives. The slow penetration of chemical fixatives during this process has been shown to be particularly detrimental to developing wood cells (Rensing, 2002), and during fixation, solute migration and membrane vesiculation can occur (Gilkey and Staehelin, 1986). In contrast, cryofixation, such as high-pressure freezing, fixes cellular constituents within milliseconds; consequently, the cell contents remain immobile during freeze substitution at −80°C, while frozen cellular water is substituted with solvent such as acetone prior to embedding in plastic resin for sectioning (Kiss and Staehelin, 1995).
Prior to cryofixation, we fed [3H]Phe to dissected lodgepole pine cambium and developing secondary xylem and then localized the incorporated phenylpropanoids by autoradiography. To verify that [3H]Phe was incorporated into the phenylpropanoid pathway, chemical analyses were performed to test [3H]Phe incorporation into lignin as well as methanol-soluble precursors. Quantitative TEM autoradiography of [3H]phenylpropanoid incorporation during inhibition of protein synthesis showed that wall lignification was not changed but Golgi label dropped dramatically. In samples in which phenylpropanoid metabolism was inhibited, wall lignification was strongly decreased but there was no change in Golgi label. These data support a membrane transporter-mediated, rather than Golgi-mediated, export of monolignols.
RESULTS
In order to interpret autoradiography results showing where [3H]Phe and its derivatives are found in developing wood, it was necessary to determine the biochemical fate of the [3H]Phe (i.e. did [3H]Phe remain as unincorporated amino acid or did it enter the phenylpropanoid pathway and ultimately get incorporated into lignin?). Dissected pine cambium and associated developing xylem were incubated for 4 h in medium containing [3H]Phe, which were also the conditions used for cryofixation and microscopy. Samples were then processed to detect radiolabel in both polymerized lignin and in soluble components. Following depolymerization and solubilization of lignin by thioacidiolysis, lignin polymer breakdown products were consistently shown to be radioactive, indicating incorporation of [3H]Phe into the lignin polymer (Supplemental Fig. S1).
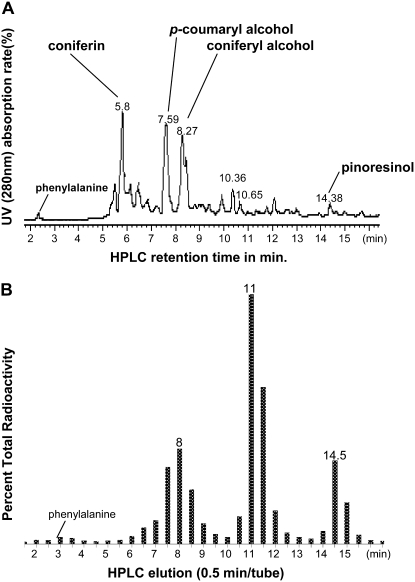
To investigate the nature of the soluble metabolites that were labeled during the 4-h incubation with [3H]Phe, methanol-soluble phenolic compounds were separated and identified using HPLC and liquid chromatography-mass spectrometry (LC-MS). Coniferin, coniferyl alcohol, and p-coumaryl alcohol peaks were prominent, as detected with absorption of UV light at 280 nm (Fig. 1A). Identification of the peaks was first by coelution with standard compounds and then confirmed by LC-MS. Minor peaks included the coniferyl alcohol dimer, pinoresinol, and Phe. To test which of these peaks were radioactive, fractions were collected every 30 s for 75 min and tested for radioactivity using scintillation counting. Phe represented only a small fraction of the total soluble radiolabel, suggesting that most of the [3H]Phe was metabolized (Fig. 1B). The expected monolignols, coniferyl alcohol and p-coumaryl alcohol, had retention times of 7.8 and 8.7 min. All fractions during these time periods were radiolabeled (Fig. 1B). Coniferin, the glucoside of coniferyl alcohol, although abundantly detected by HPLC, was not radioactive after the 4-h incubation period. There was, however, a strong radioactive signal correlated with pinoresinol. In addition, there was a large radioactive peak that eluted at 11 min. There were three peaks detected by UV absorption from 10 to 11 min that gave complex MS fragmentation patterns, including coniferyl alcohol and p-coumaryl alcohol fragments. These peaks did not coelute with the 8-O-4 coniferyl alcohol dimer (elution time, 12.0 min), the 8-5 coniferyl alcohol dimer (elution time, 12.4 min), or the 8-5 dimer of p-coumaryl alcohol or p-coumarylresinol. Overall, these data indicate that treatment of pine developing secondary xylem with [3H]Phe leads to the incorporation of radioactivity into lignin and monolignols, lignans, and additional compounds. Knowing that the [3H]Phe was entering the monolignol biosynthetic pathway, we could then use cryofixation to immobilize these metabolites in situ and visualize them using light and electron microscopy.
Figure 1.
Analysis of radioactive compounds in methanolic extracts of dissected cambium and associated developing secondary xylem from lodgepole pine following 4 h of incubation in [3H]Phe. A, HPLC results show separation of phenolic compounds, detected by their UV absorption at 280 nm and identified by coelution with standards and LC-MS. B, Radioactivity from each HPLC fraction above was collected every 30 s, and the amount of radiolabel in the fraction was measured by scintillation counting. Peaks correspond to the monolignols p-coumaryl alcohol and coniferyl alcohol at 8 min of elution, pinoresinol at 14.5 min, and a large peak at 11 min corresponding to three peaks at 10 to 11 min that gave complex MS fragmentation patterns, including coniferyl alcohol and p-coumaryl alcohol fragments.
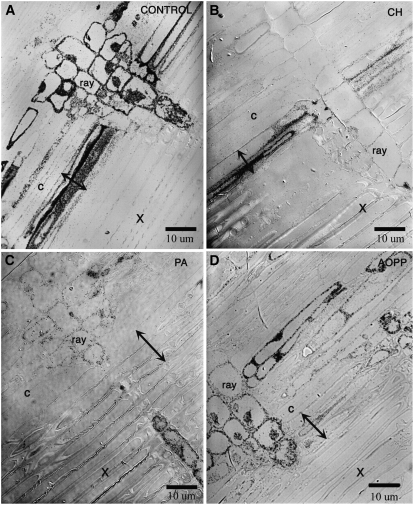
To track the distribution of [3H]Phe derivatives in developing pine wood, dissected cambium and associated developing xylem were incubated in [3H]Phe prior to high-pressure freezing, freeze substitution, and autoradiography. Light microscopy was used to establish a general overview of the uptake and incorporation of radiolabeled phenylpropanoids in developing secondary xylem as dark deposits in the tracheids, rays, and phloem of cryofixed samples (Fig. 2). In the control samples, the radioactivity of developing secondary xylem was concentrated in the secondary cell walls of tracheids, including the bordered pits. It is difficult to distinguish the secondary cell wall from the cytoplasm using a light microscope, because the developing tracheids have such large central vacuoles that only a thin layer of cytoplasm is pressed against the longitudinal walls. When the plane of the section passed through this cortical cytoplasm and secondary cell wall, the label pattern was most intense (Fig. 2A, double-headed arrow). In the cytoplasm-rich rays, radioactivity was found throughout the cytoplasm, including the nucleus. Radioactivity was relatively low in the vacuoles of all cell types.
Figure 2.
Light autoradiographs of pine stem sections through the cambium (c) and secondary xylem (X) following the incorporation of [3H]Phe. In each panel, the region of developing xylem where cells are lignifying is indicated by double-headed arrows. The black areas are the result of silver grain deposition over areas of radioactivity. A, Controls treated with [3H]Phe alone have heavy incorporation. B, Addition of cycloheximide (CH), an inhibitor of protein synthesis, diminished [3H]Phe labeling in rays. C, Inhibitors of the phenylpropanoid pathway such as the C4H inhibitor PA had the opposite effect: labeling over the developing xylem was strongly reduced, while that in the rays and cortical parenchyma remained. D, Inhibition of phenylpropanoid metabolism with the PAL inhibitor AOPP.
To evaluate the extent of [3H]Phe incorporation resulting from protein synthesis, developing wood samples were incubated with both [3H]Phe and cycloheximide to block protein translation (Fig. 2B). Cycloheximide treatment decreased the radiolabel incorporated into rays but developing tracheids were still labeled, indicating that [3H]Phe incorporation in these cells represents other metabolic pathways than translation (i.e. phenylpropanoid metabolism). Inhibition of protein synthesis could potentially inhibit the translation of monolignol biosynthetic enzymes in tracheids or rays; however, following cycloheximide treatment, radioactive label was still strongly incorporated into the secondary cell wall of the tracheids, suggesting that monolignol production was not impaired. As an additional control, [3H]Leu was used instead of [3H]Phe and the incorporation of radioactivity into these samples was much lower and uniform across cell types than the incorporation of [3H]Phe (Supplemental Fig. S2); thus, general amino acid incorporation due to protein synthesis cannot account for the strong signal seen in [3H]Phe samples.
In complementary experiments, phenylpropanoid biosynthesis was decreased either by inhibition of cinnamate-4-hydroxylase (C4H) using piperonylic acid (PA; Chong et al., 2001) or by inhibition of Phe ammonia-lyase (PAL) using 2-aminoindan-2-phosphonic acid (AIP) or l-α-aminooxy-β-phenylpropionic acid (AOPP; Appert et al., 2003). When inhibitors were applied with [3H]Phe, the label in the developing tracheids' secondary cell wall was diminished (Fig. 2, B–D). The ray cells were still labeled; however, the intensity was lowered. This effect was difficult to judge because there was strong variability in label density between sections, even within one treatment. Therefore, the label density was quantified by measuring gray levels, and these levels were corrected to the mean white of the background, to obtain average density values in defined cell types (secondary cell wall/cortical cytoplasm of developing tracheids, cytoplasm of rays) under different inhibitory conditions (Fig. 3). In developing tracheids, inhibition of C4H by 10 μm PA decreased the average label to 19% of the control density, suggesting that a large component of [3H]Phe incorporation into the these cells was phenylpropanoid in nature. Since treatment with 100 μm PA led to changes in Golgi ultrastructure (Supplemental Fig. S3), an additional enzyme in the phenylpropanoid pathway, PAL, was inhibited with 50 μm AIP or AOPP (Appert et al., 2003). As with PA inhibition of C4H, treatment with AOPP or AIP led to decreases in label in the tracheids' secondary cell wall. Label levels from samples treated with either AOPP or AIP were not statistically different from each other, so all PAL inhibitor data were pooled. As with PA inhibition of C4H, treatment with PAL inhibitors led to decreases in label in the tracheids' secondary cell walls to 31% of control levels (Fig. 3). In comparison, after treatment with cycloheximide, tracheid walls were still labeled at 75% of control levels, suggesting that the inhibition of protein biosynthesis had little effect on the lignification of tracheids.
Figure 3.
Quantification of gray levels in light microscopy autoradiographs of pine developing secondary xylem. Dissected cambium and associated tissue samples were fed [3H]Phe and various inhibitors for 4 h. To inhibit the phenylpropanoid pathway, AIP and AOPP were used as inhibitors of PAL, and PA was used as an inhibitor of C4H. Cycloheximide (CH) was used to block translation and inhibit the incorporation of Phe into proteins. Gray levels, where 0 is black and 1,024 is white, were inverted and scaled to mean white background to give positive mean ± se values.
The parenchymous rays of conifers remain active in phenylpropanoid metabolism after tracheids finish their maturation (Davin and Lewis, 2000), and a role for rays in contributing monolignols for tracheid lignification can be envisioned. When inhibitors of phenylpropanoid metabolism were applied with [3H]Phe, the radioactivity incorporated into the rays was decreased. For AOPP inhibition of PAL, the levels were decreased to 36% of control levels, while for PA inhibition of C4H, the levels were decreased to 60% of control levels, suggesting that phenylpropanoid metabolism is active in these cells. Cycloheximide inhibition of protein synthesis, which did not stop the lignification of tracheid cell walls, strongly decreased the incorporation of radiolabeled phenylpropanoids into rays to 16% of control levels.
These experiments provide verification that [3H]Phe is incorporated into lignin and monolignols in the samples under these experimental conditions. These experiments do not tell us where, in the lignifying cells, that monolignols are found (i.e. if they are localized in the Golgi). The effects of inhibitors on tracheids were predictable, while inhibitor effects on rays, plus the presence of radiolabeled pinoresinol in the chemical analysis, suggest that phenylpropanoid metabolism in the rays may be more complex. For this reason, subsequent experiments to determine the subcellular distribution of monolignols in lignifying cells were done on tracheids themselves. To determine the subcellular location of the monolignols, the radioactive samples were thin sectioned, coated with emulsion on grids, developed, and examined with TEM.
When radiolabeled developing tracheid cells were examined with TEM, the most heavily labeled area was the developing secondary cell wall, with relatively low label inside the protoplasts (Fig. 4). Tracheid ultrastructure was identical in samples frozen directly after dissection and in samples incubated for 2 to 6 h in medium containing 0.2 m Suc and [3H]Phe (data not shown; see Samuels et al. [2002] for reference ultrastructure). Both [3H]Phe alone (Fig. 4A) and [3H]Phe plus cycloheximide (Fig. 4, B and C) treatments produced similar patterns of heavy label deposition in the secondary cell wall, while PA treatment led to decreased secondary wall label (Fig. 4D). Increasing the time of incubation (4 h, 6 h, or overnight) or exposure of sections to the emulsion did not increase the amount of radioactivity observed in the protoplast, only in the secondary cell wall. This suggests that the flux of phenylpropanoids through these cells to the secondary cell wall is very rapid and that phenylpropanoids are not pooling inside the tracheid cells.
Figure 4.
TEM autoradiographs of pine tracheids treated with [3H]Phe and either an inhibitor of protein synthesis (cycloheximide [CH]) or an inhibitor of phenylpropanoid metabolism (PA). Silver grains indicating sites of incorporation of radioactive phenylpropanoid are most abundant over the secondary cell wall (CW) but are also associated with the Golgi (G) in control and PA samples. Label over vacuole (V) is relatively low. PM, Plasma membrane.
Radiolabeling was observed in the protoplast of developing tracheid cells and in the cytoplasm, Golgi, and vacuole in both [3H]Phe alone (Figs. 4A and 5) and cycloheximide-treated (Fig. 4, B and C) samples. The radioactive decay pathway into the autoradiographic emulsion is not necessarily perpendicular to the plane of the section but can radiate out from a point source in all directions; thus, the source-detector geometry means that the reduced silver grains in the emulsion are located in a pattern decreasing exponentially from the radioactive source. For 3H detection by Ilford L4 emulsion, the half-distance was determined to be 145 nm (e.g. 50% of the radioactivity radiating from a point source was detected experimentally within a circle of this radius; Salpeter and Bachmann, 1972). By counting a “probability circle” of a 500-nm circle around a silver grain, we can estimate that 90% of decay events exposing that grain would come from within that circle. Therefore, all organelles within the circle were scored as potential sources (Fig. 5). The area of each organelle was outlined as illustrated in Figure 5, and where the probability circle covered a portion of this area, a percentage of the probability circle was attributed to that organelle. For example, for a decay event in which one-third of the probability circle covers the Golgi area, 0.33 of a probability circle/decay event would be attributed to the Golgi. The sum of decay events divided by total organelle area measured how much radioactivity can be attributed to that organelle.
Figure 5.
Quantification of radioactive label in developing tracheids of pine following cryofixation, autoradiography, and TEM. A TEM autoradiography quantification example is shown (control, [3H]Phe only). The density of the decay events was measured by drawing probability circles with radii of 500 nm, which would capture 90% of possible sources around a decay event. The area of the underlying organelle that intersected the probability circle was calculated. CW, Cell wall; G, Golgi; V, vacuole. Bar = 2 μm.
Quantification of decay events, detected in TEM thin sections, showed that the secondary cell wall had the highest number of decay events per square micrometer. As with the coarser light microscopy measurements, this cell wall deposition was strongly inhibited with PA but not cycloheximide (Fig. 6A). This is important because it demonstrates that during the incubation period, cycloheximide treatment did not lead to inhibition of phenylpropanoid metabolism due to decreased biosynthetic enzyme production. In the cytoplasm of control samples treated with [3H]Phe alone, the majority of label was found in the cytoplasmic compartment, which would include the endoplasmic reticulum in our samples, as well as in the Golgi and the vacuole. By comparing samples in which phenylpropanoid metabolism or protein incorporation was inhibited with controls, label in the Golgi could be attributed to either primarily phenylpropanoid or primarily protein. Inhibition of protein synthesis with cycloheximide decreased the Golgi label, while inhibition of phenylpropanoid metabolism with PA did not (Fig. 6B). This implies that the signal in the Golgi can be attributed to protein rather than to phenylpropanoids and makes Golgi-mediated exocytosis an unlikely route for the export of monolignols.
Figure 6.
Quantification of radioactive label in developing tracheids of pine following cryofixation, autoradiography, and TEM. Values (mean ± se) indicate the density of decay events associated with radiolabel in the lignifying secondary cell wall of tracheids (A) and radiolabel associated with intracellular structures of tracheids (B). Total decays divided by the summed area for each contributing organelle gave a density metric of where radioactivity was located. Inhibition of protein synthesis with cycloheximide (CH) did not alter lignin deposition in the cell wall but produced significantly decreased incorporation of radioactivity in the Golgi (Mann-Whitney test, * P < 0.05). In contrast, inhibition of phenylpropanoid metabolism by the C4H inhibitor PA significantly decreased label in cell wall, but Golgi label was not different from the control value.
Since the vacuoles were not strongly labeled and the cytoplasm of the developing xylem had low label levels, we were concerned that soluble metabolites were being extracted during processing (i.e. by the freeze substitution medium and embedding resin). The substitution medium and resins were found to contain measurable, but low, radioactivity (Supplemental Fig. S4). In particular, the extraction, as assessed by scintillation counts, increased with the concentration of epoxy resin during embedding. We utilized low-temperature embedding at −70°C using Lowicryl HM20 to reduce the extent of extraction. Scintillation counts showed no measurable extraction by the resin. In low-temperature-embedded samples, preservation of endoplasmic reticulum was improved but abundant labeling was still not observed in the cytoplasm of developing xylem. Consistent with the earlier experiments, the radioactivity was found throughout the cytoplasm, with lower label in the Golgi and vacuoles (Supplemental Fig. S4).
DISCUSSION
The combination of autoradiography and cryofixation has allowed us to reexamine the question of how monolignols are exported during tracheid lignification in pine. In diverse species, phenylpropanoid biosynthetic enzymes such as PAL have been localized in the cytosol (Smith et al., 1994; Takabe et al., 2001), while cytochrome P450 enzymes such as C4H and p-coumarate-3-hydroxylase have been shown to be associated with the endoplasmic reticulum (Ro et al., 2001; Raes et al., 2003). The location of these enzymes suggests that monolignols are synthesized in the cytosolic compartment and, to be transported to the apoplast, must cross the plasma membrane. In a Golgi-vesicle model of transport, monolignols would have to accumulate in the endomembrane system, either at the endoplasmic reticulum or the Golgi. Such accumulation was not observed in this study, in which [3H]Phe incorporated into the Golgi of lignifying tracheids was due to protein synthesis rather than phenylpropanoid metabolism. This result does not support the Golgi-mediated export of monolignol hypothesis and suggests that other mechanisms are used by the plant to export monolignols into the secondary cell wall.
If monolignols were carried within the endomembrane system, they would move from the cell with kinetics determined by the rate of vesicle budding and fusion. The time of transit of a cargo protein from synthesis on the rough endoplasmic reticulum to secretion at the plasma membrane in classical autoradiographic studies of mammalian cells is around 120 min (Palade, 1975), a figure that has been supported more recently in live mammalian cell studies using GFP (Hirschberg et al., 1998). Assuming that phenylpropanoid-containing vesicles moved with the same kinetics, they would be easily “trapped” using cryofixation during their transit. While abundant Golgi and Golgi-vesicle clusters were observed in this and earlier (Samuels et al., 2002) studies, they did not appear loaded with phenylpropanoids.
Our results are consistent with an early study of autoradiography in sycamore (Acer pseudoplatanus) wood, in which [3H]Glc labeled the Golgi of the developing xylem, presumably due to polysaccharide biosynthesis, but [3H]Phe treatment gave sparse, random distribution of label over the cytoplasm and strong label of the secondary cell wall (Wooding, 1968). A review of the literature supporting the Golgi-vesicle paradigm of monolignol export clearly indicates that there is no experimental evidence that directly contradicts our findings. The classic autoradiography study by Pickett-Heaps (1968) showed incorporation of [3H]cinnamic acid into developing primary xylem of wheat coleoptiles, with label often occurring at the cell periphery, where “Golgi-derived” vesicles were fusing with the cell wall “particularly when the plane of section approached the wall thickenings tangentially” (p. 194). Other autoradiography studies that employed [3H]Phe to label developing xylem found the label associated with rough endoplasmic reticulum, the Golgi, and vesicles fusing with the plasma membrane (Fujita and Harada, 1979; Takabe et al., 1985). In these studies, vesicles close to or fusing with the plasma membrane were assumed to be Golgi vesicles, which is a fair assumption given the prominent Golgi structures in developing xylem cells with high polysaccharide production. Unlike cryofixed cells, in which the plasma membrane retained a smooth profile typical of cells under turgor, cells that are chemically fixed show wavy, vesiculated membrane profiles. If the region of cytoplasm adjacent to the plasma membrane contained label, and if it was vesiculated, it is easy to see why previous studies concluded that “Golgi” vesicles were labeled as they fused with the plasma membrane. In addition, these studies lacked controls for protein incorporation, and our data here show that Phe treatment results in radiolabel in the Golgi that is more likely to be protein than monolignol in nature.
The demonstration that [3H]Phe fed to dissected developing xylem is incorporated into lignin is not surprising. Similar results were found in earlier studies, in which [3H]Phe or [3H]cinnamic acid fed to developing xylem was recovered as lignin constituents following alkaline nitrobenzene oxidation (Pickett-Heaps, 1968; Wooding, 1968). Given that radioactivity was found in the thioacidolysis breakdown products and the coniferyl and p-coumaryl alcohol fractions, the simplest interpretation is that both monomeric lignin precursors and polymer were radioactively labeled during the experimental incubation period.
What is more surprising is that no radiolabel was shown to be associated with coniferin in these samples. However, this result is consistent with the low phenylpropanoid label found inside the central vacuole, where coniferin would be expected to be stored. Using HPLC, it was clear that coniferin was present in our samples, and qualitatively, the levels were consistent with reports of coniferin levels present in gymnosperms (Marcinowski and Grisebach, 1977; Savidge, 1988; Leinhos and Savidge, 1994). Historically, feeding experiments using radiolabeled coniferin showed incorporation of radiolabel into lignin, and introducing radiolabeled Phe into spruce (Picea abies) seedlings led to labeled coniferin being detected in 1 to 2 d (Freudenberg, 1959). Pulse-chase experiments showed that when spruce seedlings were fed a pulse of radiolabeled Phe from 0 to 22 h, coniferin levels peaked during the chase period at about 100 h (Marcinowski and Grisebach, 1977). Using microtome sections of Pinus thunbergii, Fukushima and coworkers (1997) determined that the spatial and temporal distributions of coniferin were highest early during xylem differentiation and early in the growing season. The relatively slow turnover of coniferin and the discrepancy between coniferin levels and the amount of lignin deposited led to the suggestion that the direct pathway from coniferyl alcohol to lignin makes a larger contribution than flow via coniferin (Fukushima et al., 1997). It is possible that distinct spatially and temporally regulated pools of coniferin exist: one that turns over rapidly and is directly associated with metabolite channeling toward lignification, and a slowly exchanging storage pool of coniferin, associated with other phenylpropanoid metabolism such as pinoresinol production in the rays. If coniferin is not the form of monolignol transported during tracheid development, than the predicted transport substrates would be p-coumaryl alcohol in the early stages of lignification and coniferyl alcohol subsequently (Terashima and Fukushima, 1988).
If Golgi-mediated export of monolignols is not operating, then alternative mechanisms are diffusion or transporter-mediated export. Studies of model membranes using liposomes or lipid bilayer discs demonstrate partitioning of model phenolic substrates into the membrane (Boija and Johansson, 2006; Boija et al., 2007). It is not clear how the desorption of these compounds from that hydrophobic environment into the apoplast would occur in this model.
Therefore, a transporter model of monolignol export is attractive, but the nature of this transporter remains elusive. Such a transporter could be powered either by ATP hydrolysis directly (e.g. ATP-binding cassette [ABC] transporters; Yazaki, 2006; Verrier et al., 2008) or indirectly via the proton gradient. ABC transporters have been proposed to play a role exporting monolignols across the plasma membrane into the developing secondary cell wall, but there is no direct evidence for this model. In several studies of gene expression during wood formation, ESTs for ABC transporters have been reported (Allona et al., 1998; Hertzberg et al., 2001; Kirst et al., 2003). A microarray study of gene expression during lignification of the bolting Arabidopsis primary inflorescence stem also clearly identified several ABC transporter genes that were coordinately expressed with known phenylpropanoid metabolic genes (Ehlting et al., 2005), but inhibiting the most highly coordinately regulated ABC transporter genes produced only altered auxin transport and not lignin-deficient phenotypes (M. Kaneda and A.L. Samuels, unpublished data). The loss of a monolignol exporter could lead to increased intracellular substrate concentrations, which in turn could induce ABC transporters for xenobiotic export. This could make detection of transport phenotype difficult, unlike the situation in epidermal cells, where the loss of ABCG subfamily members blocks cuticular lipid export and lipids accumulate in large inclusions (Pighin et al., 2004; Bird et al., 2007).
Gymnosperm wood provides a simple system to study wood formation and lignification, since unlike the multiple secondary xylem's cell types of angiosperms, the conifer secondary xylem consists primarily of axial tracheids and parenchymous rays. Strong inhibition of label into the ray by the protein translation inhibitor cycloheximide suggests that rays are metabolically active throughout tracheid differentiation and that proteins are actively turning over in ray cells. The observation that inhibition of protein synthesis did not inhibit lignification in the tracheid secondary cell wall while it did strongly decrease label in the ray's cytoplasm suggests that either monolignols move through the ray cells very quickly or the phenylpropanoids produced in the rays are not a major source of monolignols for lignification. This would be surprising, since it has often been suggested that neighboring cells could contribute to developing tracheary element lignification, as shown in the model Zinnia tracheary element culture system, in which neighboring cells contribute to the lignification of tracheary elements (Hosokawa et al., 2001; McCann et al., 2001). This paradigm is also based on reports of phenylpropanoid biosynthetic enzymes being produced in xylem parenchyma adjacent to lignifying cells, as detected by GUS assay (Hauffe et al., 1991), and immunolocalization of phenylpropanoid enzymes (Samaj et al., 1998). These enzymes could also/alternatively be involved in the production of diverse phenylpropanoids such as lignans. Inhibition of phenylpropanoid metabolism led to decreased label in the ray cytoplasm, suggesting that rays are active in phenylpropanoid metabolism. The presence of radiolabeled pinoresinol, the coupling product of coniferyl alcohol, in the methanol-soluble extract would be consistent with lignan production in rays. This production has been linked to both sapwood and heartwood formation and was suggested to be the result of dirigent protein activity (Davin and Lewis, 2000).
CONCLUSION
During secondary xylem development in pine, derivatives of [3H]Phe could be tracked in situ using a combination of cryofixation and autoradiography. The radiolabeled phenylpropanoids did not accumulate inside developing tracheids but were quickly deposited in the lignifying secondary cell wall. Within the cell, cytoplasm and Golgi were labeled following treatment with [3H]Phe. Using a combination of inhibitors of phenylpropanoid metabolism or protein synthesis, the Golgi signal was revealed to be protein, rather than monolignol, in nature. These data are more consistent with a transporter-mediated export of monolignols than a Golgi-mediated export hypothesis.
MATERIALS AND METHODS
Dormant lodgepole pine (Pinus contorta var latifolia) seedlings were transplanted, three per 15-cm square planters, and grown in a growth chamber at 24°C under 24 h of light. When hand sections showed evidence of wood formation, typically after 7 to 10 d, seedlings were destructively sampled. To sample wood later in the growing season, seedlings were maintained in the growth chamber for up to 3 months. Five-centimeter-long stem segments, cut from the seedlings 2 cm below the base of the terminal bud, were radially bisected. Approximately 0.5-mm-thick radial longitudinal slices containing portions of xylem, cambium, and phloem were hand cut from each half and immediately immersed in 0.2 m Suc, after which the cortex was removed.
For autoradiography, the slices were transferred to 0.25 mL of 0.2 m Suc containing 25 μCi of l-[2,6-3H]Phe (1.8 μm Phe) in 2-mL Eppendorf tubes. Separate solutions containing labeled Phe were prepared with various inhibitors. PA (Sigma) and cycloheximide (Sigma) were used at 10 μm. For inhibition of PAL, both AIP and AOPP (gifts from Dr. N. Amrhein, Institute of Plant Sciences, Swiss Federal Institute of Technology) were used at 50 μm. The tissues were incubated for the prescribed time at room temperature, after which they were processed either for autoradiography by high-pressure freezing or for biochemical analysis. Results from AOPP (4 h) and AIP (4 or 6 h) were not significantly different, so results for all PAL inhibitors were pooled.
High-Pressure Freezing
Slices were high-pressure frozen in 0.2 m Suc (as cryoprotectant) using a Bal-Tec HPM 010. For standard processing, frozen samples were freeze substituted with 2% osmium tetroxide (Electron Microscopy Sciences) and 8% dimethoxypropane (Aldrich) in acetone for 120 h using a dry ice-acetone bath that equilibrated at −80°C. The tissues were then warmed to −20°C in a freezer for 4 h and to 4°C in a refrigerator for 4 h, after which time they were brought to room temperature. The tissues were transferred to fresh dry HPLC-grade acetone, and then Spurr's resin was gradually added over 2 h to bring the concentration to approximately 25%. The slices were then transferred to 50% resin for 2 h, to 75% resin in open vials for 12 h, and finally to 100% resin with changes two times per day for 3 d. The infiltrated samples were polymerized in fresh Spurr's resin at 60°C. Low-temperature-embedded samples were transferred from high-pressure freezing to uranyl acetate-saturated acetone in a Leica AFS (Leica Microsystems) precooled to −80°C and allowed to substitute for 120 h. They were subsequently warmed to −70°C and gradually infiltrated with Lowicryl HM20. Samples in 100% Lowicryl were polymerized by UV light at −50°C and then allowed to warm to room temperature. Extraction of radiolabeled compounds during fixation and embedding was assessed by adding 10 μL from each mixture to 3 mL of Fisher ScintiVerse scintillation cocktail and counting the decays on a Beckman LS600IC liquid scintillation counter.
Light Microscopy
Sections (0.3 μm) were mounted on glass slides by heating. All of the blocks from each treatment were present on each slide to control for variability of emulsion thickness between slides and to facilitate easier comparisons. One slide was stained with toluidine blue for structural reference. Slides for autoradiography were coated in Ilford L4 emulsion mixed with an equal amount of distilled water at 40°C under sodium safelight with closed filters (Thomas Duplex Super Safelight). The slides were dipped vertically into the emulsion and placed vertically on a paper towel to dry. The slides were stored in a black plastic slide box, wrapped in two photographic storage bags, and stored at 4°C for 2 weeks. Emulsions were developed for 2 min in Kodak D19 developer diluted with one part distilled water, rinsed in distilled water, and then fixed for 1 min in Ilford Multigrade Paper Fixer mixed with four parts water. They were dried after a 20-min rinse in cold water. The autoradiographs were observed with differential interference contrast using a Zeiss Axioplan light microscope (Carl Zeiss). Images were captured using a Q-CAM digital camera (Q-Imaging) and photographed unstained. To determine the labeling intensity of different treatments, the gray levels of the tissues were quantified using Kohler illumination only (Openlab; Improvision). The darkness level, expressed as the inverse of the pixel value, was measured as an indication of labeling intensity after normalization against the mean background white level for each image. Deposits were measured as gray levels of a monochrome image, using unstained sections of cryofixed material for 977 observations from five independent experiments.
Electron Microscopy
Sections (70 nm thick) containing the labeled material were mounted on formvar-coated grids, stained for 30 min with uranyl acetate and 10 min with lead citrate, and then carbon coated. The grids were observed and photographed prior to coating with emulsion for observation of cell structure at high resolution. Grids were coated with Ilford L4 emulsion under sodium safelight with closed filters. Seven-millimeter-diameter wire loops were dipped in Ilford L4 emulsion at 40°C diluted with two parts water, and the excess was allowed to drain off. Loops were placed in the horizontal position and allowed to dry only until the emulsion started to get shiny. The grid was then placed onto film, section side down, and the emulsion was allowed to dry completely. Coated grids were exposed in a standard grid storage box inside two black photographic storage bags for 3 weeks at 4°C. Emulsions were developed as above for light microscopy. For quantification, the density of the decay events was measured by drawing probability circles with radii of 500 nm, which would capture 90% of possible sources around a decay event (Salpeter and Bachmann, 1972). The area of each organelle was outlined, and where the probability circle covered a portion of this area, a percentage of the probability circle was attributed to that organelle. The total area of the underlying organelles was calculated to express the radiolabel as decay events per square micrometer. Heavy deposits in the secondary cell wall were not quantified. Nine to 12 cells from two to three independent experiments were quantified.
Detection of Radiolabel in Tissues and Solutions
After the incorporation of [3H]Phe, tissues were removed from the radiolabeling solutions, rinsed twice in 0.2 m Suc, and placed on several layers of filter paper to absorb excess apoplastic fluid. Tissues were then frozen in liquid nitrogen and ground with a mortar and pestle. The ground tissue was extracted three times with 1 mL of room temperature HPLC-grade methanol. The methanol fractions were combined and centrifuged at 13,000 rpm for 2 min, and supernatant was collected and evaporated to dryness under a stream of nitrogen. To the methanolic fraction, an equal volume of ethyl acetate was added, mixed thoroughly, and allowed to phase partition. The ether phase was then removed and retained, while the extract was again extracted with a second volume of ethyl acetate, removed, and pooled. The ether phase was concentrated to dryness in a SpeedVac, resuspended in 100 μL of methanol, and analyzed by HPLC via a Summit HPLC system (Dionex) fit with a C18 Luna column (150 × 2.1 mm, 3 μm; Phenomenex); an autosampler and a photodiode array detector were employed for all HPLC analysis. Fractions were collected continuously every 0.5 min. For measurements of radioactivity, 100 μL from each fraction was mixed in 3 mL of Fisher ScintiVerse scintillation cocktail, and decays were counted on a Beckman LS600IC liquid scintillation counter.
LC-MS detection was achieved by injecting 10 μL onto a C18 Luna column (150 × 2.1 mm, 3 μm) using a Waters 2695 Separations module. Separation was performed with a mobile phase gradually changing from 83% solvent A (water:acetonitrile:formic acid [100:1:0.1, v/v/v], pH 2.5) to 77% solvent B (acetonitrile:water:formic acid [100:1:0.1, v/v/v], pH 2.5) within 21 min, at a flow rate of 0.3 mL min−1, and a column temperature of 40°C. Detection was done using negative ionization on a Micromass Quattro Micro API triple quadrupole mass spectrometer with an atmospheric pressure chemical ionization source. The instrument was operated under the following conditions: source temperature, 130°C; atmospheric pressure chemical ionization probe temperature, 500°C; corona current, 5.0 μA; cone voltage, 25 V; extractor voltage, 5 V; radio frequency lens, 0.0 V. Nitrogen from a nitrogen generator (Domnick Hunter) was used as both the cone gas (50 L h−1) and the desolvation gas (200 L h−1). Quadrupole-1 parameters were as follows: low mass resolution, 14; high mass resolution, 14; ion energy, 0.5 V. Quadrupole-2 parameters were as follows: low mass resolution, 14; high mass resolution, 14; ion energy, 3.0 V. Collision cell entrance and exit potential were set at 50 V. Multipliers were set at 650 V. Scan time was 1 s, and interscan delay was 0.02 s. Data were acquired in continuous mode. Data acquisition and instrument control were performed using Masslynx 4.0.
Thioacidolysis
Residual wood following methanolic extraction was subjected to lignin degradation and solubilization by thioacidolysis (Rolando et al., 1992). A 100-μL aliquot of solution containing degradation products was sampled and mixed in 3 mL of Fisher ScintiVerse scintillation cocktail. Decays were counted on a Beckman LS600IC liquid scintillation counter.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. [3H]Phe incorporation into lignin demonstrated by radioactive thioacidolysis products.
Supplemental Figure S2. Quantification of autoradiographs comparing the incorporation of [3H]Phe with [3H]Leu into developing secondary xylem of pine.
Supplemental Figure S3. Comparison of Golgi and associated vesicles in precursor (cambium) or differentiating tracheids during secondary wall development.
Supplemental Figure S4. Extraction of [3H]Phe and derivatives in electron microscopy preparation medium by either conventional resin-embedding technique at room temperature or low-temperature resin embedding.
Supplementary Material
Acknowledgments
We thank Russell Chidgey, Kyu-Young Kang, and Rebecca Dauwe in the Mansfield laboratory (Department of Wood Sciences, University of British Columbia, Vancouver) for assistance with chemical analyses. We also thank Brian Ellis, Carl Douglas, and David Bird for critical readings of the manuscript. The support of the University of British Columbia BioImaging Facility staff is gratefully acknowledged.
This work was supported by the Canadian Natural Sciences and Engineering Research Council (grant no. 33780 to A.L.S. and grant no. 238354 to S.D.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: A. Lacey Samuels (lsamuels@interchange.ubc.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allona I, Quinn M, Shoop E, Swope K, Cyr SS, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, et al (1998) Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA 95 9693–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appert C, Zon J, Amrhein N (2003) Kinetic analysis of the inhibition of phenylalanine ammonia-lyase by 2-aminoindan-2-phosphonic acid and other phenylalanine analogues. Phytochemistry 62 415–422 [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52 485–498 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Physiol Plant 54 519–546 [DOI] [PubMed] [Google Scholar]
- Boija E, Johansson G (2006) Interactions between model membranes and lignin-related compounds studied by immobilized liposome chromatography. Biochim Biophys Acta 1758 620–626 [DOI] [PubMed] [Google Scholar]
- Boija E, Lundquist A, Edwards K, Johansson G (2007) Evaluation of bilayer disks as plant cell membrane models in partition studies. Anal Biochem 364 145–152 [DOI] [PubMed] [Google Scholar]
- Chong J, Pierrel MA, Atanassova R, Werck-Reichhart D, Fritig B, Saindrenan P (2001) Free and conjugated benzoic acid in tobacco plants and cell cultures: induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol 125 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Lewis NG (2000) Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol 123 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42 618–640 [DOI] [PubMed] [Google Scholar]
- Freudenberg K (1959) Biosynthesis and constitution of lignin. Nature 163 1152–1155 [DOI] [PubMed] [Google Scholar]
- Fujita M, Harada H (1979) Autoradiographic investigations of cell wall development. II. Tritiated phenylalanine and ferulic acid assimilation in relation to lignification. Mokuzai Gakkaishi 25 89–94 [Google Scholar]
- Fukushima K, Taguuchi S, Matsui N, Yasuda S (1997) Distribution and seasonal changes of monolignol glucosides in Pinus thunbergii. Mokuzai Gakkaishi 43 254–259 [Google Scholar]
- Gilkey JC, Staehelin LA (1986) Advances in ultrarapid freezing for the preservation of cellular ultrastructure. J Electron Microsc Tech 3 177–210 [Google Scholar]
- Goujon T, Sibout R, Eudes A, Mackay J, Jouanin L (2003) Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol Biochem 41 677–687 [Google Scholar]
- Hauffe K, Paszkowski U, Schulze-Lefert P, Hahlbrock K, Dangl J, Douglas C (1991) A parsley 4CL-1 promoter fragment specifies complex expression patterns in transgenic tobacco. Plant Cell 3 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri T, Lundeberg J, et al (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J (1998) Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J Cell Biol 143 1485–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M, Suzuki S, Umezawa T, Sato Y (2001) Progress of lignification mediated by intercellular transportation of monolignols during tracheary element differentiation of isolated zinnia mesophyll cells. Plant Cell Physiol 42 959–968 [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5 224–229 [DOI] [PubMed] [Google Scholar]
- Kirst M, Johnson AF, Baucom C, Ulrich E, Hubbard K, Staggs R, Paule C, Retzel E, Whetten R, Sederoff R (2003) Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda) with Arabidopsis thaliana. Proc Natl Acad Sci USA 100 7383–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss JZ, Staehelin LA (1995) High pressure freezing. In NJ Severs, DM Shotton, eds, Rapid Freezing, Freeze Fracture and Deep Etching. Wiley-Liss, New York, pp 89–104
- Leinhos V, Savidge R (1994) Investigation of coniferin compartmentation in developing xylem of conifers during lignification. Acta Hortic 381 97–102 [Google Scholar]
- Marcinowski S, Grisebach H (1977) Turnover of coniferin in pine seedlings. Phytochemistry 16 1665–1667 [Google Scholar]
- McCann MC, Stacey NJ, Dahiya P, Milioni D, Sado PE, Roberts K (2001) Zinnia: everybody needs good neighbors. Plant Physiol 127 1380–1382 [PMC free article] [PubMed] [Google Scholar]
- Müsel G, Schindler T, Bergfeld R, Ruel K, Jacquet G, Lapierre C, Speth V, Schopfer P (1997) Structure and distribution of lignin in primary and secondary cell walls of maize coleoptiles analyzed by chemical and immunological probes. Planta 201 146–159 [Google Scholar]
- Palade G (1975) Intracellular aspects of the process of protein synthesis. Science 189 347–358 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD (1968) Xylem wall deposition: radioautographic investigations using lignin precursors. Protoplasma 65 181–205 [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL (2004) Plant cuticular lipid export requires an ABC transporter. Science 306 702–704 [DOI] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing KH (2002) Chemical and cryo-fixation for transmission electron microscopy of gymnosperm cambial cells. In NJ Chaffey, ed, Wood Formation in Trees: Cell and Molecular Biology Techniques. Taylor and Francis, New York, pp 65–81
- Ro DK, Mah N, Ellis BE, Douglas CJ (2001) Functional characterization and subcellular localization of poplar (Populus trichocarpa × Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol 126 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando C, Monties B, Lapierre C (1992) Thioacidolysis. In SY Lin, CW Dence, eds, Methods in Lignin Chemistry. Springer-Verlag, Berlin, pp 334–349
- Salpeter M, Bachmann L (1972) Autoradiography. In M Hayat, ed, Principles and Techniques of Electron Microscopy, Vol 2. Van Nostrand Reinhold, New York, pp 221–278
- Samaj J, Hawkins S, Lauvergeat V, Grima-Pettenati J, Boudet A (1998) Immunolocalization of cinnamyl alcohol dehydrogenase 2 (CAD2) indicates a good correlation with cell-specific activity of CAD2 promoter in transgenic poplar shoots. Planta 204 437–443 [DOI] [PubMed] [Google Scholar]
- Samuels AL, Rensing KH, Douglas CJ, Mansfield SD, Dharmawardhana DP, Ellis BE (2002) Cellular machinery of wood production: differentiation of secondary xylem in Pinus contorta var latifolia. Planta 216 72–82 [DOI] [PubMed] [Google Scholar]
- Savidge RA (1988) A biochemical indicator of commitment to tracheid differentiation in Pinus contorta. Can J Bot 66 2009–2102 [Google Scholar]
- Savidge RA (1989) Coniferin a biochemical indicator of commitment to tracheid differentiation in conifers. Can J Bot 67 2663–2668 [Google Scholar]
- Smith CG, Rodgers MW, Zimmerlin A, Ferdinando D, Bolwell GP (1994) Tissue and subcellular immunolocalisation of enzymes of lignin synthesis in differentiating and wounded hypocotyl tissue of French bean (Phaseolus vulgaris L.). Planta 192 155–164 [Google Scholar]
- Takabe K, Fujita M, Harada H, Saiki H (1985) Autoradiographic investigations of lignification in the cell walls of cryptomeria (Cryptomeria japonica d. Don). Mokuzai Gakkaishi 31 613–619 [Google Scholar]
- Takabe K, Takeuchi M, Sato T, Ito M, Fujita M (2001) Immunocytochemical localization of enzymes involved in lignification of the cell wall. J Plant Res 114 509–515 [Google Scholar]
- Terashima N, Fukushima K (1988) Heterogeneity in formation of lignin. XI. An autoradiographic study of the heterogeneous formation and structure of pine lignin. Wood Sci Technol 22 259–270 [Google Scholar]
- Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, et al (2008) Plant ABC proteins: a unified nomenclature and updated inventory. Trends Plant Sci 13 151–159 [DOI] [PubMed] [Google Scholar]
- Whetten R, Sederoff R (1995) Lignin biosynthesis. Plant Cell 7 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten RW, Mackay JJ, Sederoff RR (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 49 585–609 [DOI] [PubMed] [Google Scholar]
- Wooding FDP (1968) Radioautographic and chemical studies of incorporation into sycamore vascular tissue walls. J Cell Sci 3 71–80 [DOI] [PubMed] [Google Scholar]
- Yazaki K (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580 1183–1191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.