Abstract
We have used reverse genetics to identify genes involved in legume-rhizobium symbiosis in Lotus japonicus. We obtained the sequences of 20 putative transcription factors from previously reported large-scale transcriptome data. The transcription factors were classified according to their DNA binding domains and patterns of expression during the nodulation process. We identified two homologues of Medicago truncatula MtHAP2-1, which encodes a CCAAT-binding protein and has been shown to play a role in nodulation. The functions of the remaining genes in the nodulation process have not been reported. Seven genes were found to encode proteins with AP2-EREBP domains, six of which were similar to proteins that have been implicated in ethylene and/or jasmonic acid signal transduction and defense gene regulation in Arabidopsis (Arabidopsis thaliana). We identified a gene, LjERF1, that is most similar to Arabidopsis ERF1, which is up-regulated by ethylene and jasmonic acid and activates downstream defense genes. LjERF1 showed the same pattern of up-regulation in roots as Arabidopsis ERF1. The nodulation phenotype of roots that overexpressed LjERF1 or inhibited LjERF1 expression using an RNA interference construct indicated that this gene functions as a positive regulator of nodulation. We propose that LjERF1 functions as a key regulator of successful infection of L. japonicus by Mesorhizobium loti.
Plants are sessile organisms and cannot respond to environmental insults, such as heat, drought, flooding, or lack of light, by moving their location. They are also constantly exposed to potentially damaging organisms, including bacteria, fungi, and insects. Plants have evolved highly developed systems to adjust to their surrounding conditions. These so-called stress responses and resistance programs are initially regulated at the level of gene transcription by a host of specific transcription factors.
The sequencing of the genome of the model legume Lotus japonicus is nearly complete (Sato et al., 2008). Primary annotation has assigned 30,000 genes in the draft genomic sequence, including approximately 1,500 genes that encode putative transcription factors. This number represents approximately 5% of all annotated genes in the L. japonicus genome.
Leguminous plants have developed a unique mechanism of nitrogen fixation that involves a symbiotic relationship with soil bacteria known as rhizobia. The ability to sustain this symbiotic relationship is restricted to leguminous species, with the exception of the nonlegume Parasponia genus. Successful bacterial infection is established between host plant and rhizobia through a complex set of signals, resulting in the formation of symbiotic nodules in the root of the host plant in which nitrogen fixation is carried out by the rhizobium.
To date, a small number of host transcription factors have been characterized in terms of their function in the nodulation process. NIN was originally identified in an analysis of a transposon-tagged mutant of L. japonicus that is unable to form nodules (Schauser et al., 1999). The expression of NIN is strictly dependent on infection by Mesorhizobium loti or exogenous purified nodulation (Nod) factors, and it has been postulated that NIN is required for rhizobial invasion through root hairs. GRAS family proteins that are essential for Nod factor-mediated signal transduction have been isolated from both Medicago truncatula and L. japonicus (Kaló et al., 2005; Murakami et al., 2006). M. truncatula MtHAP2-1 is a CCAAT-binding transcription factor whose expression is regulated by microRNA169. It was recently identified as a key regulator of symbiotic nodule development (Combier et al., 2006). Recently, an AP2-EREBP transcription factor, MtERN1 (for ERF required for nodulation), was found to be essential for Nod factor-mediated signal transduction (Middleton et al., 2007). Two additional AP2-EREBP genes, MtERN2 and MtERN3, were identified as trans-acting factors that regulate the expression of an early nodulin gene, ENOD11, through a Nod factor-responsive cis-element in the promoter region (Andriankaja et al., 2007).
Transcriptome analysis of M. truncatula using cDNA microarrays has identified a number of transcription factors whose expression changes early in the nodulation process (El Yahyaoui et al., 2004; Lohar et al., 2006). Previously, we used two approaches, cDNA macroarray analysis and serial analysis of gene expression, to identify nodulation-related genes in L. japonicus (Kouchi et al., 2004; Asamizu et al., 2005). We identified 20 transcription factors whose expression is up-regulated during nodulation. In this study, we have characterized the majority of these putative nodulation-associated transcription factors, with particular emphasis on members of the AP2-EREBP family of transcription factors, which appear to be the most abundant proteins in this group. We also carried out a functional analysis of one of the transcription factors, LjERF1, to determine its role in the nodulation process in more detail.
RESULTS
Classification of Transcription Factor Genes
Twenty putative nodulation-associated transcription factor genes (Kouchi et al., 2004; Asamizu et al., 2005) were classified according to their DNA-binding domains (Davuluri et al., 2003; Table I). Seven of the genes encoded proteins that are members of the AP2-EREBP family of transcription factors. The remainder encoded members of the CCAAT (two genes), bZIP (two genes), C2H2 (two genes), Homeobox (one gene), NAC (one gene), WRKY (one gene), C3H (one gene), MADS (one gene), C2C2-Dof (one gene), and CPP (one gene) families of transcription factors. These results were consistent with previous transcriptome analyses of early symbiotic events in M. truncatula and Sinorhizobium meliloti, which also identified transcription factors of the AP2-EREBP, CCAAT, NAC, WRKY, and MADS families of proteins (El Yahyaoui et al., 2004; Lohar et al., 2006). M. truncatula MtHAP2-1, which encodes a CCAAT-binding transcription factor and is regulated by microRNA169, has been shown to be essential for the differentiation of nodule cells (Combier et al., 2006). The two CCAAT-binding transcription factors of L. japonicus that emerged from our analysis, CBF-A01 and CBF-A22, exhibited 76% and 70% amino acid sequence identity, respectively, with MtHAP2-1.
Table I.
Relative expression levels compared with uninfected root after M. loti infection
The expression of each gene was normalized using an internal control (AV772463; Supplemental Table S1). ND, Not determined.
| Gene Name | Accession No. | Transcription Factor Family | 3 h | 24 h | 2N | 4N | 7N | 12N |
|---|---|---|---|---|---|---|---|---|
| Group I (initial response) | ||||||||
| LjERF1 | AB378626 | AP2-EREBP | 1.8 | 1.7 | 1.2 | 0.1 | 0.4 | 0.5 |
| HDZ-M48 | AB378627 | Homeobox | 1.8 | 1 | 0.7 | 0.1 | 0.1 | 0.1 |
| CBF-A22 | AB378628 | CCAAT | 1.6 | 1.3 | 1.1 | 0.3 | 0.5 | 0.2 |
| bZIP-R91 | AB378629 | bZIP | 1.9 | 1.1 | 0.9 | 0.2 | 0.3 | 0.9 |
| LjRAP2.4 | AB378630 | AP2-EREBP | 1.9 | 1.1 | 1.4 | 0.6 | 1.6 | 0.5 |
| ZF-M39 | AB378631 | C2H2 | 1.8 | 1.1 | 0.8 | 1 | 1.2 | 0.8 |
| LjERF2 | AB378632 | AP2-EREBP | 1.7 | 1.3 | 0.6 | 0.8 | 1.4 | 1.6 |
| Group II (induced at 2N) | ||||||||
| CBF-A01 | AB378633 | CCAAT | ND | ND | 14.3 | 6.2 | 14.7 | 12.2 |
| LjERF16 | AB378634 | AP2-EREBP | ND | ND | 5.1 | 5.8 | 3.7 | 2.9 |
| ZF-G96 | AB378635 | C2H2 | ND | ND | 2.7 | 3.1 | 1.5 | 0.7 |
| LjERF18 | AB378636 | AP2-EREBP | ND | ND | 1.6 | 15.4 | 25.2 | 11 |
| LjWRKY30 | AB378640 | WRKY | ND | ND | 1.8 | 1 | 2.7 | 1.8 |
| Group III (induced at 4N) | ||||||||
| LjERF17 | AB378637 | AP2-EREBP | ND | ND | 0.4 | 9.4 | 11.5 | 4.9 |
| LjERFI9 | AB378638 | AP2-EREBP | ND | ND | 1.2 | 1.7 | 3.1 | 4.7 |
| NAM-A43 | AB378639 | NAC | ND | ND | 1.3 | 1.8 | 2 | 2.3 |
| Group IV (induced at 12N) | ||||||||
| RING-G83 | AB378641 | C3H | ND | ND | 0.4 | 0.2 | 0.9 | 9.9 |
| MADS-A18 | AB378642 | MADS | ND | ND | 0.4 | 0.1 | 0.2 | 8 |
| bZ1P-M43 | AB378643 | bZIP | ND | ND | 1.1 | 0.2 | 0.5 | 2.1 |
| Not clear | ||||||||
| Dof-M153 | AB378644 | C2C2-Dof | 1.1 | 0.8 | 0.4 | 0.1 | 0.1 | 0.3 |
| CPP-L56 | AB378645 | CPP | 1.1 | 0.7 | 0.6 | 0.3 | 0.4 | 0.4 |
Expression Analysis of Transcription Factor Genes in Response to Inoculation with M. loti
To determine the expression profiles of transcription factor genes during the nodulation process, we used quantitative-reverse transcription (RT-Q)-PCR to examine uninfected roots, infected roots at 2 and 4 d after inoculation (DAI) with M. loti, nodule primordia collected at 7 DAI, and mature nodules at 12 DAI. Genes for which induced expression was not clearly detected were examined by RT-Q-PCR at 3 and 24 h after M. loti inoculation. The transcription factor genes were classified according to a time course of induction (Table I). Seven transcription factor genes (LjERF1, HDZ-M48, CBF-A22, bZIP-R91, LjRAP2.4, ZF-M39, and LjERF2) were classified as group I and were induced as early as 3 h after inoculation. The expression of four of the early induced genes of group I was repressed at a later stage of nodulation (4 DAI). Group II consisted of five transcription factor genes (CBF-A01, LjERF16, ZF-G96, LjERF18, and LjWRKY30) that were induced at 2 DAI, although their expression pattern in earlier stages was not examined. Persistently induced expression throughout the nodulation process (2–12 DAI) was observed for CBF-A01, LjERF16, and LjERF18. Group III included three transcription factor genes (LjERF17, LjERF19, and NAM-A43) that were induced at 4 DAI. Group IV included three transcription factor genes (RING-G83, MADS-A18, and bZIP-M43) whose expression was induced at 12 DAI. We were unable to detect the induced expression of two genes, Dof-M154 and CPP-L56, at any time point examined.
Phylogenetic Analysis of Nodulation-Induced AP2-EREBP Family Genes
Members of the AP2-EREBP family of transcription factors were the most abundant in our analysis, and their expression was induced at various time points during nodulation (Table I). These results suggested that they may play important roles in the nodulation process. To date, three AP2-EREBP genes have been shown to be essential in the nodulation process in M. truncatula. ERN1 was isolated as the causative gene in a nodulation mutant and has been identified as a component of the Nod factor signal transduction pathway downstream of DMI3 (Middleton et al., 2007). Recently, two additional genes, ERN2 and ERN3, were identified as trans-acting factors that interact with the Nod factor-responsive cis-element in the promoter region of ENOD11 (Andriankaja et al., 2007). ERN1 to ERN3 encode AP2-EREBP domain proteins and are highly homologous to Arabidopsis (Arabidopsis thaliana) RAP2.11, a gene of unknown function that was classified as a group V gene in a detailed genome-wide classification of Arabidopsis and Oryza sativa AP2-EREBP domain proteins (Nakano et al., 2006).
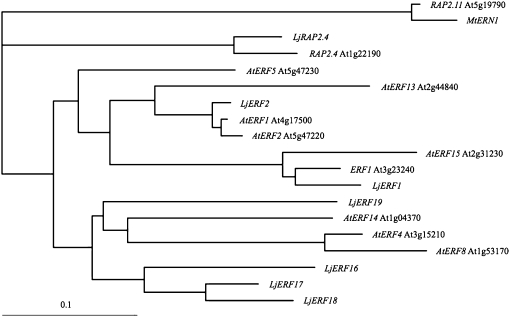
To determine the relationship of the AP2-EREBP genes of L. japonicus to their homologues in Arabidopsis, we carried out a phylogenetic analysis using the ClustalW server of the DNA Data Bank of Japan (http://clustalw.ddbj.nig.ac.jp/; Fig. 1). LjRAP2.4 showed the highest level of similarity to Arabidopsis RAP2.4, which belongs to group I in the classification scheme of Nakano et al. (2006). The remaining six L. japonicus genes were classified into groups VIII and IX. LjERF1 was closest to Arabidopsis ERF1. LjERF2 was classified in a different clade that included AtERF1 and AtERF2. The remaining four genes were relatively divergent in the legume lineage. LjERF16, LjERF17, and LjERF18 were similar to each other and classified in a clade that included AtERF4, AtERF8, and AtERF14 as well as LjERF19. These results indicated that the analyzed L. japonicus AP2-EREBP transcription factors are phylogenetically distinct from M. truncatula ERN1 to ERN3.
Figure 1.
Phylogenetic tree of nodulation-associated L. japonicus AP2-EREBP genes. Phylogenetic analysis was carried out using the sequences of the AP2-EREBP domains of the L. japonicus (Lj) genes and their Arabidopsis (At) homologues. Locus identifiers (such as At1g22190) are shown for the Arabidopsis genes. Arabidopsis RAP2.11 and M. truncatula MtERN1 were included as an outgroup.
Response to Ethylene and Jasmonic Acid of AP2-EREBP Family Genes
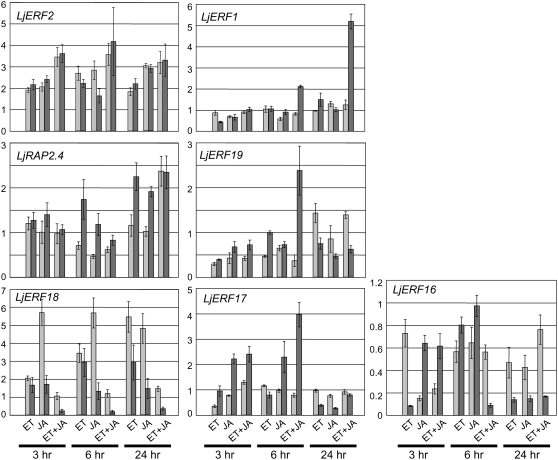
Previous studies have shown that ethylene and jasmonic acid (JA) are involved in the nodulation process (Penmetsa and Cook, 1997; Oldroyd et al., 2001; Nakagawa and Kawaguchi, 2006; Sun et al., 2006). Furthermore, Arabidopsis AP2-EREBP genes in groups VIII and IX have often been implicated in ethylene and/or JA signaling (Nakano et al., 2006). When we examined the effect of ethylene and JA on the expression of L. japonicus AP2-EREBP genes, we found that they fell into one of two groups: ubiquitously induced genes (induced in both shoots and roots) and root-specific genes (Fig. 2). One exception was LjERF16, which showed a complex pattern of down-regulation in response to the plant growth regulators. LjERF2, LjRAP2.4, and LjERF18 were induced in both shoots and root. The induction of LjERF2 and LjERF18 was observed in response to both ethylene and JA, and LjERF18 induction was higher in shoots than in root. The induction of LjRAP2.4 by ethylene and JA was observed in the root, and a synergistic effect was observed in shoots and root. A significant level of induction of LjERF1, LjERF19, and LjERF17 was observed only in the root. LjERF17 was preferentially induced by JA. A synergistic effect of ethylene and JA was observed for LjERF1 and LjERF19. LjERF1 exhibited synergistic up-regulation in response to ethylene and JA in a root-specific manner.
Figure 2.
Effects of ethylene (ET) and/or JA on the expression of AP2-EREBP genes in shoots and root. The expression levels of the indicated L. japonicus AP2-EREBP genes in shoots (light gray) and root (dark gray) are expressed relative to controls. Controls are either nontreated shoots or root. Data represent averages ± se of three independent experiments. The y axis in each bar graph indicates the relative transcript level.
Effect of the LjERF1 Overexpression on Nodulation
We chose to analyze the function of LjERF1 in early nodulation in more detail for several reasons: phylogenetic analysis indicated that LjERF1 is highly similar to Arabidopsis ERF1 (Lorenzo et al., 2003); the expression of LjERF1 was up-regulated very early (3–24 h) after M. loti inoculation (Table I); and LjERF1 exhibited synergistic induction by ethylene and JA. We first examined the effect of the overexpression of LjERF1 on the nodulation process. We transformed L. japonicus hairy roots with an LjERF1-overexpressing vector, as described in “Materials and Methods.” LjERF1 expression in transformed hairy roots was 8.9-fold higher than in control roots that were transformed with an empty vector. Roots were then inoculated with M. loti MAFF303099, and the number of nodules formed on the roots was counted at 3 weeks after infection. The average number of nodules formed on transformed, LjERF1-overexpressing hairy roots was 11.1, while that on control roots was 6.5. When we analyzed the data using Student's t test, we found that this difference was statistically significant (P < 0.01; Table II).
Table II.
Results of t test of mean numbers of nodules formed on control and LjERF1-overexpressed or RNAi hairy roots
| Plant | Control | LjERF1 Overexpression/RNAi | Statistical Significancea |
|---|---|---|---|
| Overexpressor | 6.5 ± 0.7 | 11.1 ± 1.3 | 0.01 |
| RNAi-1 | 3.3 ± 0.8 | 0.8 ± 0.5 | 0.01 |
| RNAi-2 | 3.3 ± 0.8 | 0.9 ± 0.6 | 0.02 |
Statistical significance level of observed difference in nodule number calculated by Student's t test.
Effect of Suppression of LjERF1 Expression on Nodulation
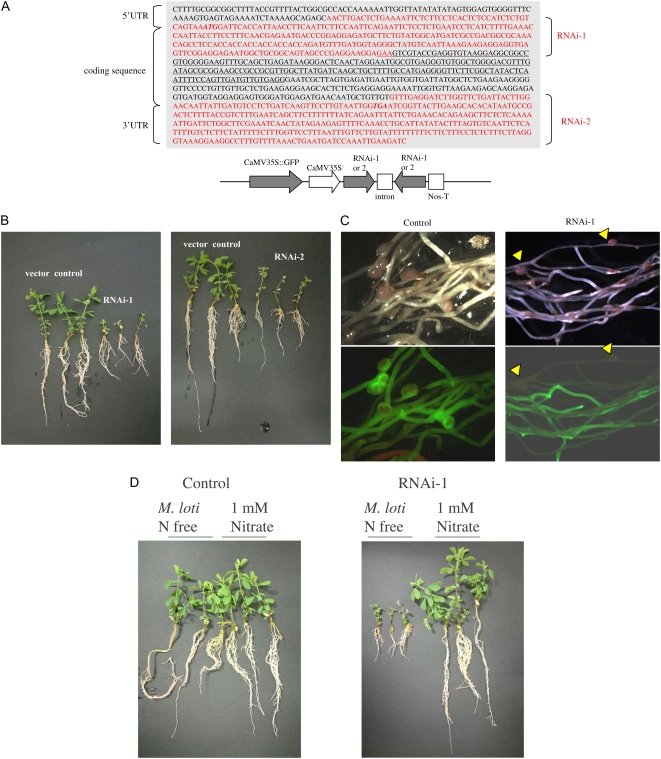
We also examined the effect of suppression of LjERF1 expression on nodulation using RNA interference (RNAi). We transformed L. japonicus hairy roots with two LjERF1-specific RNAi constructs (Fig. 3). Suppression of LjERF1 expression by RNAi was confirmed using real-time RT-PCR by amplifying a region of the LjERF1 mRNA that was not used in the RNAi constructs. LjERF1 mRNA was down-regulated approximately 60% in transformed hairy roots compared with control roots. The phenotype of transformed hairy roots was observed at 3 weeks after inoculation with M. loti MAFF303099. As shown in Figure 3, the growth of transformed plants was severely retarded compared with that of control plants. The average number of nodules formed on control hairy roots was 3.3, while that on the two RNAi-transformed plants, RNAi-1 and RNAi-2, was 0.8 and 0.9, respectively, and this difference was statistically significant (P < 0.01 and P < 0.02, respectively; Table II). To determine whether the growth suppression phenotype of RNAi-transformed plants was due to the availability of nitrogen, plants were grown in the presence of 1 mm nitrate (Fig. 3). RNAi-transformed plants grew normally when exogenous nitrogen was supplied, which indicated that the growth retardation of RNAi plants is due to nitrogen deficiency resulting from poor nodulation.
Figure 3.
Nodulation phenotype of LjERF1-specific RNAi-transformed hairy roots. A, Two constructs targeting different regions of LjERF1 (RNAi-1 and RNAi-2) were used to transform plants. B, Plants were inoculated with M. loti and grown in the absence of nitrogen. Photographs were taken 3 weeks after infection and show the growth phenotype of plants that were transformed with an empty vector (control), RNAi-1, or RNAi-2. C, Fluorescence microscopy of transformed hairy roots. The formation of nodules on control hairy roots was evident, but nodules were observed only in nontransformed hairy roots in the RNAi plants, as distinguished by GFP fluorescence. D, The growth of plants in which the expression of LjERF1 was suppressed by RNAi was similar to that of control plants in the presence of a nitrogen source.
Spatial Expression of LjERF1
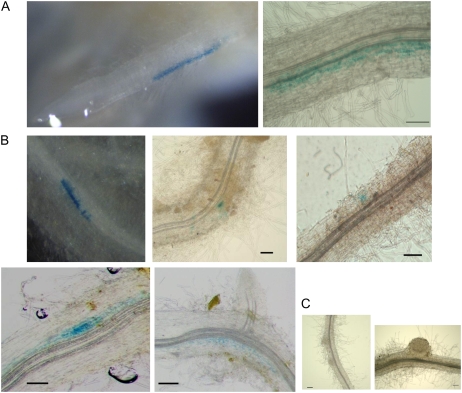
To investigate the spatial response of LjERF1 to M. loti infection, we transformed hairy roots with a chimeric gene composed of the LjERF1 promoter (1,000 bp upstream from the open reading frame) and the GUS coding sequence. Histochemical GUS staining at 3 h after the M. loti inoculation revealed GUS activity in an epidermal region 2 to 3 mm above the root tip, which corresponded to the infection zone (Fig. 4). This time point was consistent with the results of the RT-Q-PCR analysis, in which highest induction of LjERF1 was observed (Table I). No GUS-stained roots were observed at 24 h after inoculation or at later time points. We also performed a GUS assay using the entire hairy roots at 10 d after M. loti inoculation. No GUS staining was observed in the nodule primordia or mature nodules of these roots (Fig. 4), which is also consistent with the RT-Q-PCR results. GUS staining was detected in epidermal and cortical cells, where no visible morphological changes for nodule formation were observed (Fig. 4).
Figure 4.
LjERF1 promoter∷GUS assay after M. loti inoculation. A, Histochemical GUS staining was observed in an epidermal region of the LjERF1 promoter∷GUS transgenic hairy root at 3 h after M. loti inoculation. B and C, GUS staining at 10 d after M. loti inoculation indicated that LjERF1 is induced in epidermal and cortical regions where no morphological changes for nodulation were observed (B) but not in the nodule primordia or mature nodules (C). Bars = 0.1 mm.
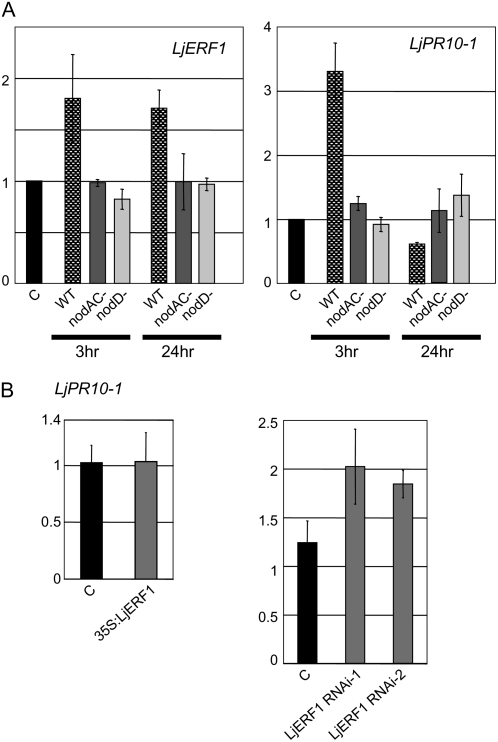
Expression of PR10-1 during Normal Nodulation and in LjERF1-Overexpressing or LjERF1 RNAi Plants
Overexpression of LjERF1 increased the number of nodules formed on hairy roots, and RNAi resulted in the inhibition of nodulation. These results were somewhat surprising, because it was previously reported that Arabidopsis ERF1 is involved in the activation of ethylene/JA-dependent pathogen defense genes (Lorenzo et al., 2003). It is generally believed that in order for legume-Rhizobium symbiosis to be established, plant resistance mechanisms must be temporarily or locally suppressed (Mithöfer, 2002). The precise mechanisms of suppression of defense responses remain to be elucidated. To determine whether LjERF1 was involved in the regulation of plant defense mechanisms, we examined the expression of LjPR10-1 in wild-type, LjERF1-overexpressing, and LjERF1 RNAi plants. M. truncatula MtPR10-1 expression is induced by pathogens (Gamas et al., 1998), and LjPR10-1 was shown to be induced during the establishment of L. japonicus-microorganism symbiosis (Kouchi et al., 2004; Deguchi et al., 2007). We first examined the expression of LjPR10-1 in response to M. loti infection in wild-type plants by RT-Q-PCR (Fig. 5). The expression of LjPR10-1 was induced at 3 h after M. loti infection and decreased after 24 h. To confirm the specificity of the response, we infected plants with two symbiosis-defective M. loti mutant strains, nodAC− and nodD−. The specificity of LjPR10-1 induction in response to a compatible rhizobium was also confirmed by examining the expression pattern of LjERF1, which does not respond to symbiosis-defective M. loti strains. These results indicated that the expression of LjPR10-1 is temporally induced only upon infection by wild-type M. loti. We then examined the expression of LjPR10-1 in nodulating hairy roots that overexpressed LjERF1 or in which LjERF1 expression was suppressed by RNAi (Fig. 5). Overexpression of LjERF1 did not affect the expression level of LjPR10-1, whereas suppression of LjERF1 expression by RNAi resulted in up to a 2-fold induction of LjPR10-1 expression.
Figure 5.
Expression of LjPR10-1 examined by RT-Q-PCR. A, Expression of LjERF1 and LjPR10-1 in response to infection by wild-type and noninfectious mutant strains of M. loti. B, Expression of LjPR10-1 in nodule-containing roots that overexpress LjERF1 (overexpressor) or nonnodulating roots in which the expression of LjERF1 was suppressed by RNAi at 3 weeks after M. loti inoculation. The y axes indicate relative expression levels compared with those in control plants.
DISCUSSION
We used a reverse genetics approach to characterize 20 transcription factor genes of L. japonicus that were identified in previous transcriptome studies as genes whose expression is induced during nodulation (Kouchi et al., 2004; Asamizu et al., 2005). One of the transcription factor genes identified in M. truncatula transcriptome studies (El Yahyaoui et al., 2004; Lohar et al., 2006), M. truncatula MtHAP2-1, encodes a CCAAT-binding transcription factor and is a key regulator of nodule development (Combier et al., 2006). In this study, we identified two L. japonicus CCAAT-binding proteins. The proteins shared 84% sequence identity with each other and exhibited distinct patterns of induction during the nodulation process (Table I). In contrast to M. truncatula, L. japonicus forms a determinate type of nodule. We speculate that the two CCAAT-binding proteins identified in this study may play a coordinated role in meristem development in the determinate type of nodule. Future studies in our laboratory will focus on the involvement of these proteins in L. japonicus nodule organogenesis.
We also identified a MADS box gene, MADS-A18, that may play a role in nodule organogenesis. The gene was induced at 12 DAI in mature nodules, in which symbiotic nitrogen fixation by resident M. loti is established. It has been suggested that nodule MADS box proteins in M. sativa are involved in defining or maintaining the differentiated state of the nodule organ (Heard et al., 1997; Zucchero et al., 2001). In situ hybridization demonstrated that the genes encoding these proteins are expressed in infected cells, and it has been suggested that two MADS box proteins, Nmh7 and Ngl9, form heterodimers, since homo/heterodimerization of MADS box proteins is a prerequisite for DNA binding (Zucchero et al., 2001). In this study, we identified one MADS box gene that was induced during the L. japonicus nodulation process. It will be interesting to identify potential binding partners of this MADS box protein in L. japonicus using protein-protein interaction screens and to determine whether it forms homodimers or heterodimers.
We identified several transcription factor genes that may be involved in the pathogen infection response, including genes that encoded WRKY and AP2-EREBP family proteins. In the establishment of legume-rhizobium symbiosis, the resistance mechanisms of the plant must be suppressed at the site of infection (Mithöfer, 2002). In the later stages of nodulation, when the supply of nitrogen is sufficient, the defense system must be switched on to prevent excessive nodule formation, which can result in the loss of photoassimilates that are required to maintain the nitrogen fixation system. None of the genes that regulate either system have been characterized to date. We identified a WRKY transcription factor, LjWRKY30, whose induction level was highest at 7 DAI, when the nodule primordium is developed. WRKY proteins are known to have regulatory functions in response to pathogen infection (Eulgem and Somssich, 2007). Previously, cDNA array analysis demonstrated that defense-related genes are up-regulated at 7 DAI, including genes for an elicitor-inducible Dof protein and an elicitor-inducible β-1,3-glucanase (Kouchi et al., 2004). The WRKY protein identified in our study may be involved in the regulation of downstream defense genes directly or indirectly to block excess rhizobium infection under nitrogen-sufficient conditions.
We identified seven AP2-EREBP genes that were induced during nodulation in L. japonicus. This is a relatively large number, as a previous transcriptome analysis of M. truncatula described only one (El Yahyaoui et al., 2004). This result is most likely due to the increased number of time points examined in our study.
L. japonicus AP2-EREBP genes found in this study may be involved in elevated ethylene and JA sensing during M. loti infection and may function by regulating downstream genes, including defense genes. A hyperinfected mutant, skl, of M. truncatula is insensitive to ethylene, which suggests that ethylene is involved in the nodulation process (Penmetsa and Cook, 1997). It has also been suggested that ethylene functions upstream or at the point of calcium spiking to inhibit the Nod factor signal transduction pathway (Oldroyd et al., 2001). The effect of JA on nodulation also has been examined. It has been suggested that JA functions as a signaling molecule in the systemic suppression of nodulation (Nakagawa and Kawaguchi, 2006) and in the antagonistic cross talk between the pathways of ethylene signaling and calcium spiking (Sun et al., 2006).
We examined the expression of seven L. japonicus AP2-EREBP genes in response to ethylene and JA (Fig. 2). Most of the genes could be grouped according to two patterns of expression: ubiquitous expression (induced in both shoots and root) and root-specific expression. LjERF16 was not activated by ethylene or JA, which suggests that it may be regulated by other inducers, such as salicylic acid or abscisic acid, as is the case for AtERF4 (McGrath et al., 2005). The expression patterns of the L. japonicus AP2-EREBP genes in response to ethylene and JA did not indicate a clear correlation between their phylogenetic relationship and expression during the nodulation process. These results indicate that L. japonicus AP2-EREBP genes are involved in various stages of nodulation and that the regulation of their expression is complex and likely involves a balance in the ethylene and JA levels within the cell.
We found that LjERF1, the closest homologue of Arabidopsis ERF1, is a positive regulator of the early process of nodulation. The overexpression of LjERF1 resulted in a statistically significant increase in nodule number (Table II), and RNAi resulted in a severe inhibition of nodulation (Fig. 3). LjERF1 expression was induced at 3 h after M. loti infection (Table I) and was specifically induced by compatible wild-type M. loti (Fig. 5). The expression was localized to an epidermal region at the infection zone (Fig. 4). GUS staining observed in hairy roots at 10 DAI may indicate that new infection events accompanied by LjERF1 expression are occurring continuously.
To determine whether LjERF1 affected the expression of defense genes, we examined the expression of LjPR10-1, an ortholog of M. truncatula MtPR10-1. Previous transcriptome studies of L. japonicus nodulation and arbuscular mycorrhizal fungi colonization have reported that the expression of several defense genes, including PR10-1 (LjPR10-1), is initially induced, then repressed (Kouchi et al., 2004; Deguchi et al., 2007). In this study, we showed that the expression of LjPR10-1 is initially induced, then suppressed in response to infection by compatible M. loti (Fig. 5). Taken together with previous results, our results indicate that LjPR10-1 is involved in pathogen defense in nodulating roots.
Unlike in Arabidopsis, in which the expression of ERF1, driven by the cauliflower mosaic virus (CAMV) 35S promoter, resulted in the activation of defense genes (Lorenzo et al., 2003), LjERF1 overexpression did not induce the expression of LjPR10-1 (Fig. 5B). However, we did not observe suppression of LjPR10-1, as expected based on the down-regulation of LjPR10-1 at 24 h after infection with wild-type M. loti (Fig. 5A). This is likely due to the fact that we used nodulating roots at 3 weeks after M. loti inoculation to examine the effect of LjERF1 overexpression. In these samples, the basal level of LjPR10-1 may have offset the effect of constitutive LjERF1 expression, even if it suppressed the expression of LjPR10-1 locally. We also observed a slight induction of LjPR10-1 expression in LjERF1 RNAi plants. The observed induction of LjPR10-1 was relatively small, and it seems likely that LjERF1 does not activate LjPR10-1 in nodulating roots. Since LjERF1 was induced in the early stages of infection (3–24 h; Table I), it is possible that it is involved in the suppression of defense genes for the establishment of rhizobium infection.
The mechanism by which the host plant recognizes a compatible rhizobium as “self” is an unsolved question. Our results provide evidence that LjERF1 functions as a key regulator of this process. However, additional studies, aimed at identifying the direct targets of this transcription factor, are needed to clarify its role in plant defense systems.
MATERIALS AND METHODS
RT-Q-PCR
RNA was extracted using the RNeasy Kit (QIAGEN) from the uninfected roots of soil-grown Lotus japonicus ‘Gifu B-129’ and Mesorhizobium loti Tono-infected samples at 3 and 24 h after inoculation and 2, 4, 7, and 12 DAI (N2, N4, N7, and N12, respectively). In the course of RNA extraction, samples were treated with RNase-Free DNase Set (QIAGEN) for genomic DNA removal. The entire root was collected for the 3- and 24-h samples, the infection zone (5 mm in length, 3 mm above the root tip) was collected for the N2 and N4 samples, and visible nodule primordia and mature nodules were collected for the N7 and N12 samples (Kouchi et al., 2004). RT-Q-PCR was performed using 1 μg of total RNA as a template and the DyNAmo HS SYBR Green qPCR Kit (Finnzymes). Results were quantitated using the DNA Engine Opticon2 system (Bio-Rad). Each experiment was performed in triplicate with two biological repeats. Transcript levels were normalized to a PERK1 homolog (AV772463), since its stable expression was suggested in a previous serial analysis of gene expression (Asamizu et al., 2005). Primers were designed for each gene using the Primer3 program (Rozen and Skaletsky, 2000). The sequences of the primers used in this study are available in Supplemental Table S1.
Treatment with Ethylene and JA
The seeds of Gifu B-129 were sterilized and then germinated on half-strength Broughton and Dilworth (B&D) medium containing 1 mm nitric acid. Fourteen-day-old seedlings were transferred to the same medium containing 10−5 m 1-aminocyclopropane-l-carboxylic acid (Wako), 10−6 m methyl JA (Wako), or both. The shoots and roots of hormone-treated seedlings were collected at 3, 6, and 24 h after treatment, separated, and then frozen in liquid nitrogen.
Overexpression of LjERF1 and LjERF1-Specific RNAi
The vector for the overexpression of LjERF1 was constructed as follows. The full-length cDNA of MWM012d12 was obtained by RT-PCR using primers specific for the 5′ and 3′ untranslated regions (5′-CTTTTGCGGCGGCTTTTACC-3′ and 5′-CAAAGACGGTAAAAGAGTCG-3′). The amplified fragment was cloned into pENTR (Invitrogen) and then transferred by Gateway reaction to the binary vector pHBR (a kind gift from Dr. Y. Murakami, RIKEN). pHBR encodes GFP [sGFP (S65T); Niwa et al., 1999] driven by the CAMV 35S promoter for the selection of transformed hairy roots. The overexpression of LjERF1 was mediated by tandem duplicated CAMV 35S promoters. The expression of LjERF1 was confirmed by triplicate RT-Q-PCR analysis that resulted in a mean overexpression value of 8.9- ± 1.5-fold.
The vectors for RNAi were constructed as follows. A 300-bp fragment of LjERF1, including 46 bp of the 5′ untranslated region and 254 bp of coding sequence that did not contain the AP2 domain (RNAi-1), and a 374-bp region of the 3′ untranslated region (RNAi-2) were amplified by RT-PCR using the following primers: 5′-CTTGACTCTGAAAATTCTC-3′, containing an XhoI or BamHI site at the 5′ end, and 5′-TTCTCCTTCCTCGGGCTAC-3′, containing a KpnI or ClaI site at the 5′ end, for RNAi-1; and 5′-TGAATCGGTTACTTGAAG-3′, containing an XhoI or BamHI site at the 5′ end, and 5′-GATCTTCAATTTGGATCATTA-3′, containing a KpnI or ClaI site at the 5′ end, for RNAi-2. Amplified fragments were digested with XhoI/KpnI or BamHI/ClaI and ligated into the corresponding sites of pKANNIBAL (Wesley et al., 2001) in inverse orientations with an intron between them. The construct, consisting of the CAMV 35S promoter and the intron-containing hairpin RNA, was transferred to the binary vector pHKN29 using NotI. pHKN29 is a modified version of pCAMBIA1300 in which the kanamycin resistance gene is replaced by sGFP (S65T) (Kumagai and Kouchi, 2003). The suppression of LjERF1 was confirmed by triplicate RT-Q-PCR analysis that resulted in a mean down-regulation value of 60% ± 4.2%.
The overexpression and RNAi binary vectors were transferred into Agrobacterium rhizogenes LBA1334 (Offringa et al., 1986) by electroporation.
Hairy Root Transformation
Induction and transformation of L. japonicus hairy root using A. rhizogenes LBA1334 were performed as described previously (Kumagai and Kouchi, 2003). Briefly, seeds were sterilized and germinated on filter paper submerged in water for 4 d in the dark at 23°C to obtain elongated hypocotyls, followed by 2 d in a photocycle of 16 h of light/8 h of dark in a growth chamber. The seedlings were cut just above the base of the hypocotyls and put into a suspension of A. rhizogenes in a petri dish for a several minutes. The seedlings were transferred onto agar plates containing Jensen (N+) medium (Díaz et al., 1989) and cocultivated for 5 d in a growth chamber. Plants were then transferred onto Schenk and Hildebrandt medium containing 100 μg mL−1 cefotaxime and grown for 10 d until the hairy roots were developed from the section of hypocotyls. Emerged hairy roots were assayed for GFP using a fluorescence microscope. Plants harboring transformed hairy roots were transferred to pots filled with vermiculite and supplied with half-strength B&D medium containing 1 mm nitric acid and grown in a growth chamber in a 16-h photocycle at 23°C. After 5 to 7 d, plants were inoculated with M. loti MAFF303099 and allowed to continue growing with the same medium without the nitrogen source.
LjERF1 Promoter∷GUS Construction
The 1,000-bp upstream promoter region of LjERF1 was PCR amplified by the primers 5′-CACCGTATAGGGTTCGAACCCTAGGGAGCAC-3′ and 5′-CATTTACTGACAGAGATGGAGAGTGAGGAAGAG-3′. The amplified fragment was cloned into the pENTR D-TOPO vector (Invitrogen) and then transferred to the modified binary vector pCAMBIA1381Z (AF234306; Hajdukiewicz et al., 1994). pCAMBIA1381Z was converted to a Gateway destination vector by ligating a bunt-ended Gateway cassette into the SmaI site in the multicloning site. In addition, for the selection of transformed hairy roots, the CaMV 35S promoter and hygromycin resistance gene of pCAMBIA1381Z were replaced by the GFP gene sGFP (S65T) (Niwa et al., 1999) driven by the nopaline synthase promoter. The LjERF1 promoter fragment was placed in front of the GUS gene by the LR recombinase reaction.
Histochemical GUS Staining
Hairy roots transformed with the LjERF1 promoter∷GUS construct were placed on nitrogen-free half-strength B&D medium for 5 d after GFP selection (see “Hairy Root Transformation” above), and the hairy roots were inoculated with M. loti MAFF303099 by placing the roots between two sheets of filter paper that were immersed in a M. loti cell suspension. After 3 and 24 h, hairy roots were immersed in a GUS staining solution (2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, and 100 mm sodium phosphate, pH 7.0) and placed under vacuum for a few minutes, followed by incubation overnight at 37°C in the dark. Otherwise, plants were transferred to pots filled with vermiculite and supplied with nitrogen-free half-strength B&D medium and grown in a growth chamber with a 16-h photocycle at 23°C. Ten days after inoculation with M. loti, hairy roots were immersed in a GUS staining solution. The stained materials were observed with a light microscope.
Noninfectious Mutant Strains of M. loti
Nod gene disruptants of M. loti MAFF303099 were generated by replacing the coding regions of each gene with a gentamicin resistance cassette (aacC1) from pMS246 (Becker et al., 1995). M. loti M1101 has a deletion of nodA and nodC genes and thus cannot produce Nod factors (Takeda et al., 2005). This strain is therefore designated nod AC−. Another strain has a deletion of the nodD gene, and this strain is also infection defective (Y. Shimoda, unpublished data). This strain is therefore called nodD−.
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession numbers AB378626 to AB378645.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of primers used in RT-Q-PCR.
Supplementary Material
Acknowledgments
We thank A. Watanabe for excellent technical assistance. We also thank Dr. H. Mitsui at Tohoku University, who kindly provided the M. loti mutant strain nodAC−. L. japonicus Gifu B-129 seeds were provided by the National BioResource Project.
This work was supported by the Kazusa DNA Research Institute Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Erika Asamizu (asamizu@gene.tsukuba.ac.jp).
The online version of this article contains Web-only data.
References
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2005) Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol Plant Microbe Interact 18 487–498 [DOI] [PubMed] [Google Scholar]
- Becker A, Schmidt M, Jager W, Puhler A (1995) New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162 37–39 [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi Y, Banba M, Shimoda Y, Chechetka SA, Suzuri R, Okusako Y, Ooki Y, Toyokura K, Suzuki A, Uchiumi T, et al (2007) Transcriptome profiling of Lotus japonicus roots during arbuscular mycorrhiza development and comparison with that of nodulation. DNA Res 14 117–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz CL, Melchers LS, Hooykaas PJJ, Lugtenberg BJJ, Kijne JW (1989) Root lectin as a determinant of host-plant specificity in the Rhizobium-legume symbiosis. Nature 338 579–581 [Google Scholar]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10 366–371 [DOI] [PubMed] [Google Scholar]
- Gamas P, de Billy F, Truchet G (1998) Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol Plant Microbe Interact 11 393–403 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Heard J, Caspi M, Dunn K (1997) Evolutionary diversity of symbiotically induced nodule MADS box genes: characterization of nmhC5, a member of a novel subfamily. Mol Plant Microbe Interact 10 665–676 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11 263–274 [DOI] [PubMed] [Google Scholar]
- Kumagai H, Kouchi H (2003) Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol Plant Microbe Interact 16 663–668 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Peñuela S, Samac D, Town C, Silverstein KA, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A (2002) Suppression of plant defence in rhizobia-legume symbiosis. Trends Plant Sci 7 440–444 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S (2006) Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13 255–265 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kawaguchi M (2006) Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol 47 176–180 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18 455–463 [DOI] [PubMed] [Google Scholar]
- Offringa IA, Melchers LS, Regensburg-Tuink AJ, Costantino P, Schilperoort RA, Hooykaas PJ (1986) Complementation of Agrobacterium tumefaciens tumor-inducing aux mutants by genes from the T(R)-region of the Ri plasmid of Agrobacterium rhizogenes. Proc Natl Acad Sci USA 83 6935–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Engstrom EM, Long SR (2001) Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275 527–530 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al (2008) Genome structure of the legume, Lotus japonicus. DNA Res (in press) [DOI] [PMC free article] [PubMed]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Sun J, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM (2006) Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J 46 961–970 [DOI] [PubMed] [Google Scholar]
- Takeda N, Okamoto S, Hayashi M, Murooka Y (2005) Expression of LjENOD40 genes in response to symbiotic and non-symbiotic signals: LjENOD40-1 and LjENOD40-2 are differentially regulated in Lotus japonicus. Plant Cell Physiol 46 1291–1298 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Zucchero JC, Caspi M, Dunn K (2001) ngl9: a third MADS box gene expressed in alfalfa root nodules. Mol Plant Microbe Interact 14 1463–1467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.