Abstract
We have isolated two dominant mutants from screening approximately 50,000 RIKEN activation-tagging lines that have short inflorescence internodes. The activation T-DNAs were inserted near a putative basic helix-loop-helix (bHLH) gene and expression of this gene was increased in the mutant lines. Overexpression of this bHLH gene produced the original mutant phenotype, indicating it was responsible for the mutants. Specific expression was observed during seed development. The loss-of-function mutation of the RETARDED GROWTH OF EMBRYO1 (RGE1) gene caused small and shriveled seeds. The embryo of the loss-of-function mutant showed retarded growth after the heart stage although abnormal morphogenesis and pattern formation of the embryo and endosperm was not observed. We named this bHLH gene RGE1. RGE1 expression was determined in endosperm cells using the β-glucuronidase reporter gene and reverse transcription-polymerase chain reaction. Microarray and real-time reverse transcription-polymerase chain reaction analysis showed specific down-regulation of putative GDSL motif lipase genes in the rge1-1 mutant, indicating possible involvement of these genes in seed morphology. These data suggest that RGE1 expression in the endosperm at the heart stage of embryo development plays an important role in controlling embryo growth.
In flowering plants seed development progresses through a series of complex processes. It begins as the egg cell, the female gamete, and the central cell in the ovule are each fertilized by one of the two male gametes, the sperm cells that are delivered to the site of fertilization by the pollen tube. The diploid zygote resulting from the union of one sperm cell with the egg cell develops into the embryo of the progeny plant. The fertilization product of the homodiploid central cell and the second sperm cell develops as the triploid endosperm (Faure et al., 2002). Endosperm is important for seed development and, in some species, for seedling development after germination because it nurtures the embryo and the seedling. After fertilization in eudicots, such as Arabidopsis (Arabidopsis thaliana), the triploid nuclei of the endosperm divide repeatedly without cell wall formation, resulting in the formation of a large multinucleate cell, the syncytium (Boisnard-Lorig et al., 2001). After the eighth round of syncytial mitoses the syncytium is partitioned by a specific type of cytokinesis called cellularization initially in the region surrounding the embryo, proceeding toward the chalazal region (Sorensen et al., 2002). The cellular endosperm is gradually consumed by the embryo during seed development, leaving only the peripheral aleurone-like cell layer in mature seeds (Olsen, 2004).
The development of the integument of the maternal organ and the new generation embryo and endosperm complete the formation of a viable seed. Some mutations that impose abnormal integument development have been isolated and genes that control embryo development have been identified through gene disruption (Busch et al., 1996; Torres-Ruiz et al., 1996; Takada et al., 2001). A number of genes that regulate endosperm cellularization and/or embryo cytokinesis have been reported (Sorensen et al., 2002). Some mutations, such as titan1 and titan2 that cause abnormal microtubule formation in the embryo, also affect endosperm development (McElver et al., 2000; Steinborn et al., 2002; Tzafrir et al., 2002). These results indicate that endosperm cellularization and embryo cytokinesis involve components of the same basic machinery. FIE, FIS2, MEA, and MSI1 have been described as genes of endosperm development regulation, because mutations in them cause autonomous endosperm development in the absence of fertilization (Chaudhury et al., 1997; Luo et al., 1999; Kohler et al., 2003). Based on the phenotype and on similarity to the polycomb group proteins in Drosophila and mammals, it has been proposed that the proteins coded by these genes form a chromatin-associated polycomb complex that represses genes involved in endosperm development before double fertilization (Grossniklaus et al., 1998; Luo et al., 2000; Spillane et al., 2000; Guitton et al., 2004).
In Arabidopsis, a dicot, and maize (Zea mays), a monocot, the endosperm has an important role in the control of seed size (Olsen, 2004). It is known that in both monocots and dicots an imbalance in maternal/paternal dosage affects the endosperm and seed size (Scott et al., 1998), indicating that control of endosperm development is by differential expression of genes that are dependent on their parent of origin, probably through imprinting. One example of imprinting is the gametophytic-limited expression of the fis class of genes on alleles in the maternal genome (Luo et al., 2000). Introduction of a maintenance DNA methyltransferase 1 antisense construct via transgenic pollen into a wild-type ovule causes precocious endosperm cellularization and a reduction in seed size (Adams et al., 2000; Luo et al., 2000). Hence these results indicate that genomic imprinting by methylation controls the critical genes for endosperm development.
Final seed size is mainly attained during growth of the endosperm (Boisnard-Lorig et al., 2001). The haiku1 (iku1) and iku2 mutations, which are sporophytic recessive, prevent an increase in the size of the syncytial endosperm by precocious cellularization, leading to reduced embryo proliferation and decreased seed size (Garcia et al., 2003). Luo and coworkers have identified MINI3, which encodes one of the WRKY family, as a growth regulator of endosperm, and have shown that the successive action of iku1, iku2, and mini3 in the same signal pathway plays an important role in the control of seed size (Luo et al., 2005). The mutant transparent testa glabra2 (ttg2) shows a reduction in integument cell elongation and is defective for proanthocyanidin synthesis and mucilage deposition in the seed coat. Crosses of ttg2 with iku2 result in a greater reduction in seed size than that of each single mutation (Garcia et al., 2005). The regulation of seed size, therefore, is coupled to the growth of endosperm and of the integument.
We have identified two independent lines that show a compact phenotype with reduced internode length from the RIKEN Arabidopsis activation-tagging lines. These two lines have T-DNA insertions close to a basic helix-loop-helix (bHLH) gene. The loss-of-function mutation results in the production of small and shriveled seeds. Our work indicates that this gene, which we have designated as RETARDED GROWTH OF EMBRYO1 (RGE1), expressed in endosperm controls embryo growth after the heart stage.
RESULTS
Z029732 and Z068035 Are Dominant Mutants That Have Short Internodes
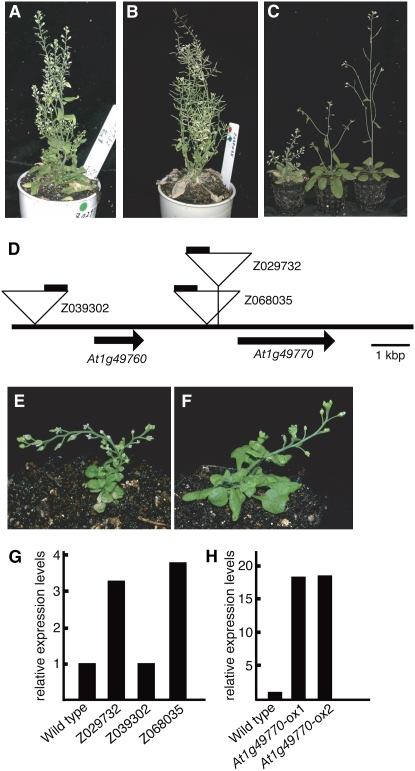
We have generated around 50,000 Arabidopsis activation-tagging lines and have observed and reported 1,262 visibly identifiable mutants during the generation of these lines (Ichikawa et al., 2003). They have been categorized according to growth rate, plant leaf color, fertility, and flowering time and morphology. Among the morphological mutants we noticed ones with short inflorescence internodes. These mutants can also be classified into several subcategories according to internode length. Two dominant mutants Z029732 and Z068035 have short internodes, small leaves, and short petioles (Fig. 1, A and B).
Figure 1.
Z029732 and Z068035 show short inflorescence internodes. A and B, Adult phenotypes of Z029732 (A) and Z068305 (B) mutants at 34 d under long-day conditions in the T1 generation. C, Comparison of adult phenotypes of Z029732 mutants at 30 d in the T2 generation. Plants from left to right are homozygous and heterozygous for the activation tag and wild type (ecotype Col-0). D, The activation-tagging T-DNA insertion sites in mutants. The triangles indicate the activation-tagging T-DNA insertion sites in Z029732, Z068305, and Z039302. The black bars on the triangles indicate the positions of the four copies of the CaMV 35S enhancer near the right border. E and F, Adult phenotype of overexpressing plants is similar, but more severe than Z029732 and Z068305. G and H, Real-time PCR analysis showing expression of At1g49770 in the wild type, Z029732, Z039302, and Z068305 (G), and the wild type and At1g49770-overexpressing plants (At1g49770-ox1 and At1g49770-ox2; H). Expression levels of genes were normalized with ACT2 expression. Relative expression levels: expression levels of the At1g49770 genes in Z029732, Z039302, Z068305, At1g49770-ox1, and At1g49770-ox2 relative to the wild type.
The homozygous mutants had a much more severe phenotype compared to the heterozygotes and had very short internodes (Fig. 1C). Both mutants showed a dominant phenotype that cosegregated with the antibiotic resistance gene on the activation T-DNA. This result suggested that these mutants were caused by the activation T-DNA that has a transcriptional enhancer at the right border.
We determined the T-DNA insertion sites of Z029732 and Z068035. Both T-DNAs were inserted in chromosome 1 in a region between At1g49760 and At1g49770 (Fig. 1D). The distances between the cauliflower mosaic virus (CaMV) 35S enhancer on the T-DNA and the predicted translation start site of At1g49760 are 6.8 kb for Z029732 and 5.8 kb for Z068035. Also the distances between the CaMV 35S enhancer and At1g49770 are 5.7 kb for Z029732 and 6.8 kb for Z068035 (Fig. 1D). From a database search of T-DNA insertion sites we found one activation-tagged line Z039302 that has a T-DNA insertion proximal to At1g49760. Although the CaMV 35S enhancer is close to At1g49760, this line did not show the morphological alterations of Z029732 and Z068035. These results suggest that At1g49760 is not responsible for these mutants and that At1g49770 is the corresponding gene for the characteristic phenotypes of Z029732 and Z068035. The expression level of At1g49770 determined by quantitative PCR was enhanced in Z029732 and Z068035 but not in Z039302 (Fig. 1G).
We overexpressed At1g49770 under the control of the CaMV 35S promoter and generated around 20 independent transgenic lines. These transgenic lines showed the characteristic short internodes and some showed a more severe phenotype than Z029732 and Z068035 (Fig. 1, E and F). We confirmed the expression level of At1g49770 was enhanced in these more severely mutant transgenic lines (Fig. 1H). From these results we confirmed that At1g49770 is the corresponding gene for the mutant phenotype of Z029732 and Z068035.
The At1g49770 Product Is a Member of the bHLH Transcription Factor Family
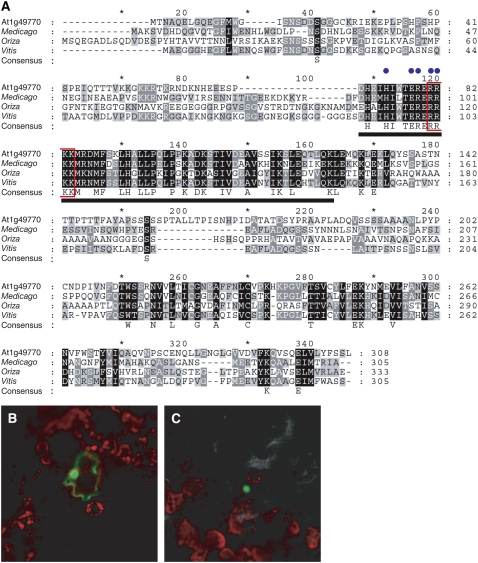
At1g49770 encodes for a protein containing a putative bHLH domain. It has been reported that bHLH proteins form a family of more than 100 members in Arabidopsis (Heim et al., 2003; Toledo-Ortiz et al., 2003). Almost all the residues in a bHLH domain that are known to be important for DNA binding are conserved (blue dots in Fig. 2A; Heim et al., 2003). The At1g49770 product has homologous sequence in other organisms. The closest homologous protein, found in Vitis vinifera, has 43% identity and 57% similarity at the amino acid level (Fig. 2A). As we found a potential nuclear localization signal in the bHLH domain of the At1g49770 product we fused GFP to the C-terminal region to establish its intracellular localization. By transient analysis using tobacco (Nicotiana tabacum) leaves and Agrobacterium infiltration, we observed that this protein localizes to the nucleus (Fig. 2, B and C).
Figure 2.
The At1g49770 product is a putative transcription factor that has a bHLH domain. A, Alignment of the At1g49770 product and its homologs. The full-length amino acid sequence of the At1g49770 product was aligned with those of Medicago, Oriza, and Vitis. All alignments were performed using ClustalW software. The bHLH domain, important amino acids for DNA binding, and putative nuclear localization signal are indicated by the bold underline, blue dots, and red box, respectively. B and C, Localization of At1g49770 product:GFP fusion protein. The fluorescence of only GFP (B) and At1g49770 product:GFP (C) are indicated. These images are merged with those of red chlorophyll fluorescence.
At1g49770 Expresses during Seed Development
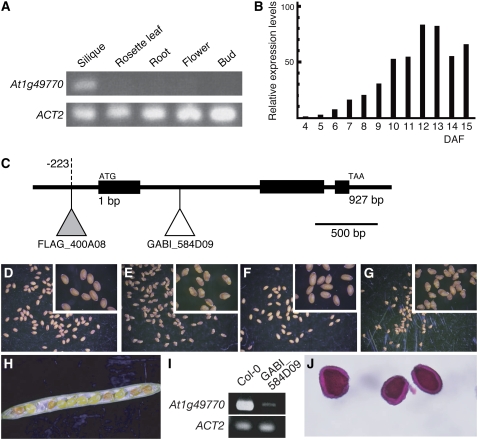
To investigate the tissue specificity of At1g49770 expression, we examined transcription levels in various organs using reverse transcription (RT)-PCR. We could not detect At1g49770 expression in seedlings (data not shown), roots, rosette leaves, flower buds, and mature flowers. However, it was strongly expressed in siliques (Fig. 3A).
Figure 3.
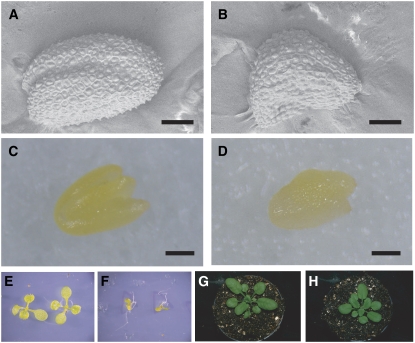
Loss-of-function At1g49770 mutants. A, RT-PCR showing At1g49770 mRNA levels in different organs (rosette leaves, buds, flowers, siliques, and roots). B, Real-time PCR analysis showing At1g49770 mRNA levels in developing siliques of wild type. Relative expression levels: expression levels in each developmental stage relative to 4 DAF. C, The T-DNA insertion sites in loss-of-function mutants. The white and gray triangles indicate the T-DNA insertion sites in GABI_584D09 (ecotype Col-0) and FLAG_400A08 (ecotype Ws), respectively. D to G, The seed phenotypes of Col-0 (D), GABI_584D09 (E), Ws (F), and FLAG_400A08 (G). A magnified image is inset. H, The phenotype seen in opened siliques from crosses of GABI_584D09 and wild type. I, RT-PCR showing At1g49770 mRNA levels in siliques of wild type and GABI_584D09. J, Ruthenium red-stained seed coat mucilage phenotype of GABI_584D09 seeds.
It is reported that seed development is controlled by the regulation of expression of many genes at several seed developmental stages. We investigated the expression level of At1g49770 at these stages by real-time RT-PCR (Fig. 3B). We observed that the expression level of At1g49770 gradually increased from 7 d after flowering (DAF), when the embryo is at the early heart stage (Fig. 5B).
Figure 5.
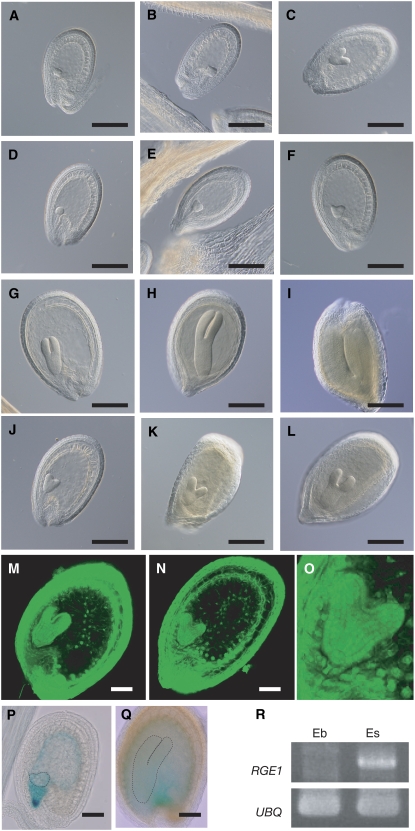
Overall embryo growth is retarded during seed development of GABI_584D09. A to L, Embryo development in wild type (A, B, C, G, H, and I) and GABI_584D09 (D, E, F, J, K, and L) at 6 (A and D), 7 (B and E), 9 (C and F), 10 (G and J), 12 (H and K), and 13 (I and L) DAF. M and N, Confocal sections of Feulgen-stained seeds from wild type (M) and GABI_584D09 (N) at 10 DAF. O, Magnified image of a GABI_584D09 embryo at 10 DAF. P and Q, At1g49770∷GUS developing seeds showing GUS activity 8 (P) and 12 (Q) DAF. Embryos are indicated with dotted lines. R, RT-PCR analysis showing expression of At1g49770 in embryos (Eb) and endosperms and integuments (Es). Expression levels of UBQ2 were analyzed as an internal control.
The Loss-of-Function Mutants of At1g49770 Have Shriveled Seeds
Since At1g49770 expresses during seed development, it is reasonable to speculate this gene has a specific function during seed formation. To understand its function we examined T-DNA insertion mutants of the At1g49770 gene. We obtained two independent T-DNA insertion mutants from GABI (Rosso et al., 2003) and INRA (Samson et al., 2002; Fig. 3C). T-DNAs were inserted in the first intron (GABI_584D09, GABI line) and 223 bp upstream of the translation start site (FLAG_400A08, INRA line). These T-DNA insertion lines were self-pollinated to obtain homozygous lines. Both GABI_584D09 and FLAG_400A08 homozygous lines had shriveled seeds that were smaller than the corresponding wild types (Columbia-0 [Col-0] for GABI_584D09 and Wassilewskija [Ws] for FLAG_400A08; Table I; Figs. 3, D–G [inset], and 4, A and B). These homozygous lines were backcrossed with the corresponding wild type to generate F1 seeds and these were self-pollinated to generate F2 seeds. Of the F2 seeds of both GABI_584D09 and FLAG_400A08 a quarter, 223:72 (χ2: 0.033, P > 0.5) and 654:175 (χ2: 6.465, P > 0.1), respectively, had shriveled seeds and all plants derived from shriveled seeds were homozygous for the T-DNA insertion, indicating that this phenotype was recessive and cosegregated with the T-DNAs (Fig. 3H; Supplemental Fig. S1). We investigated At1g49770 expression levels using RT-PCR analysis in GABI_584D09 homozygous lines that had the T-DNA inserted in the first intron. As expected, At1g49770 expression was strongly reduced in GABI_584D09 (Fig. 3I).
Table I.
Dry seed size of Col-0, Ws, GABI_584D09, and FLAG_400A08
At least 70 seeds were measured. Student's t test: P > 0.01 (*) versus each wild type, Col-0 and Ws, against each mutant, GABI_584D09 and FLAG_400A08, respectively.
| Genotype | Dimension | Length | Width |
|---|---|---|---|
| mm2 (±sd) | mm2 (±sd) | mm2 (±sd) | |
| Col-0 | 13.992 ± 1.648 | 0.470 ± 03030 | 0.309 ± 0.025 |
| GABI_584D09 | 12.110 ± 1.519* | 0.468 ± 0.043 | 0.296 ± 0.030* |
| Ws | 15.588 ± 2.431 | 0.501 ± 0.038 | 0.325 ± 0.031 |
| FLAG_400A08 | 10.025 ± 1.225* | 0.410 ± 0.033* | 0.271 ± 0.018* |
Figure 4.
Phenotypes of GABI_584D09 (A and B). A and B, Dry seed morphology of wild type (A) and GABI_584D09 (B). C and D, Mature embryos of wild type (C) and GABI_584D09 (D). E and F, Appearance of 10-d-old seedlings of wild type (E) and GABI_584D09 (F) on medium including Suc. G and H, Adult phenotypes of wild type (G) and GABI_584D09 (H) grown for 22 d under long-day conditions.
Mutation of the At1g49770 Locus Causes Retardation in Embryonic Growth
The seed integuments are formed from maternal tissue and the phenotype caused by abnormal integuments deviates from the expected ratio predicted from Mendelian rules. The mutant ttg2 is defective in mucilage deposition and seed size is also reduced in ttg2/ttg2 plants (Penfield et al., 2001; Western et al., 2001; Garcia et al., 2005). Therefore we examined the nature of the seed coat of our mutants by staining with ruthenium red that colors the mucilage released from seed integuments. We observed red staining of the seed coat, suggesting that it is the abnormal embryo and/or endosperm during seed development rather than the seed integument that causes the seed phenotype of GABI_584D09 (Fig. 3J). We examined the development of seeds in further detail using GABI_584D09.
The mature embryos in seeds of the GABI_584D09 homozygote were smaller than those of wild type (Fig. 4, C and D). The GABI_584D09 seeds did not germinate on Murashige and Skoog plates without Suc but they were able to germinate and grow on plates containing 1% Suc. At the beginning of germination, GABI_584D09 seedlings are much smaller than those of wild type (Fig. 4, E and F). During seedling development, they grow to almost the same size as wild type. The number of rosette leaves before bolting and the overall adult morphology of GABI_584D09 are also almost the same as wild type (Fig. 4, G and H).
We observed the development of the seed at several stages. Both GABI_584D09 and wild type grew without any differences until early in the heart stage (Fig. 5, B and E). After the heart stage embryogenesis of GABI_584D09 was gradually retarded and the mature embryo was smaller than wild type (Fig. 5, A–L and O). First, we examined the process of endosperm formation. Some embryo mutants with late growth show abnormal endosperm development caused by disordered cellularization after the syncytial mitosis (Sorensen et al., 2002; Luo et al., 2005). We therefore observed endosperm cellularization in GABI_584D09 using Feulgen staining that dyes intact plant tissues and nuclei. Cellularization occurs normally (Fig. 5, M and N) and, interestingly, we could not observe any defects in the morphogenesis or pattern formation of the GABI_584D09 embryo (protoderm, root, and shoot embryonic meristem) in spite of retardation in embryo growth (Fig. 5O). We therefore named At1g49770 as RGE1. We also designated GABI_584D09 and FLAG_400A08 as rge1-1 and rge1-2, respectively.
RGE1 Expresses in the Endosperm
To investigate RGE1 expression in seeds further we made transgenic plants by Agrobacterium in planta transformation that contained the RGE1 promoter region fused to the GUS gene. This pRGE1∷GUS construct contained the 2-kb region upstream from the RGE1 start codon and the first 10 codons of the RGE1 protein fused in frame to the GUS gene. We investigated more than five independent transgenic lines. GUS reporter activity was observed in the endosperm from the early heart developmental stage (Fig. 5, P and Q). RGE1 transcripts were detected only in RNA isolated from the endosperm and seed coat by RT-PCR analysis (Fig. 5R). These results indicate that RGE1 is expressed in the endosperm during seed development.
RGE1 Might Function as a Positive Regulator in the Endosperm during Embryo Growth
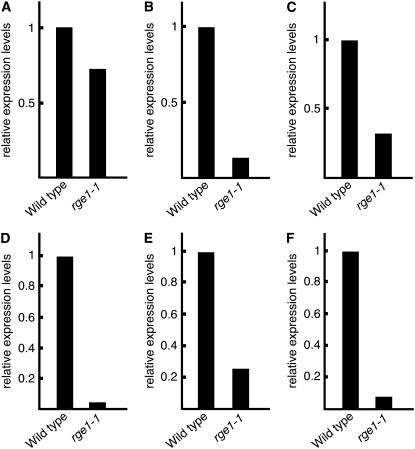
Since RGE1 is a bHLH protein that has similarity to transcription factors it was worthwhile to examine the genes that coexpress with RGE1 to understand transcription targets. We investigated the differences in gene expression profiles in siliques between rge1-1 and wild type at 12 DAF by oligomicroarray, because the expression level of RGE1 was most abundant at 12 and 13 DAF (Fig. 3B). From the microarray analysis, six genes were shown to have decreased expression levels at 12 DAF (Table II). We could not find any genes that showed increased expression at the same times. By using real-time RT-PCR, we checked expression levels of the six genes identified at 12 DAF in siliques of rge1-1 and wild type. The expression levels of these genes were expected to decrease in the rge1-1 mutant (Fig. 6). Interestingly, the expression levels of most of these genes were reported to be induced strongly during the heart stage (AtGenExpress expression atlas provided by Schimd et al., 2005; Table II). This suggests that RGE1 functions as a positive regulator for the induction of expression of these genes during seed development especially during the heart stage and that the genes that are decreased in rge1-1 might be the cause of the shriveled seed morphology of the rge1-1 mutant.
Table II.
Genes with decreased expression levels in 12 DAF siliques of rge1-1
We referred to a previous article for the column labeled Signal Levels in Col-0 (Schmid et al., 2005). Globular, Seed stage 3 (Sample ID, ATGE_76); Heart, seed stage 4 (Sample ID, ATGE_77); Late Heart, seed stage 5 (Sample ID, ATGE_78).
| 12 DAF
|
Signal Levels in Col-0
|
||||
|---|---|---|---|---|---|
| AGI Code | Annotation | Fold Change | Globular | Heart | Late Heart |
| At3g06890 | Hypothetical protein | 0.3815 | 75.34 | 74.08 | 62.06 |
| At4g38000 | Dof zinc-finger protein | 0.1175 | 19.58 | 89.45 | 105.55 |
| At1g71250 | Putative GDSL motif lipase | 0.07715 | 6.33 | 675.85 | 703.61 |
| At3g52970 | Cytochrome P450 family | 0.04475 | 13.68 | 110.08 | 128.86 |
| At5g03820 | Putative GDSL motif lipase | 0.03425 | 25.56 | 178.06 | 184.70 |
| At4g33600 | Hypothetical protein | 0.01875 | 3.21 | 239.68 | 273.06 |
Figure 6.
Real-time PCR analysis showing mRNA levels of genes whose expression levels from the results of microarray analysis were decreased in siliques of rge1-1 and wild type at 12 DAF. Relative expression levels are the expression levels in siliques of rge1-1 relative to wild type. A to F, Expression levels of At3g06890 (A), At4g38000 (B), At1g71250 (C), At3g52970 (D), At5g03820 (E), and At4g33600 (F) in siliques of rge1-1 and wild type at 12 DAF. Expression levels of all genes were normalized with ACT2 expression.
DISCUSSION
RGE1 Is a Member of a Novel bHLH Gene Family That Localizes in the Nucleus
RGE1 (At1g49770) is a member of the bHLH gene family, some of which are known to function as transcription factors. RGE1 belongs to a subgroup, the Ib bHLH subfamily (Heim et al., 2003).
The bHLH domain of RGE1 is categorized as a G-box-binding type and some important residues for DNA binding in this family are also conserved in RGE1 (Toledo-Ortiz et al., 2003). From the microarray analysis we found that six genes are down-regulated in the siliques of the rge1-1 mutant at 12 DAF. We confirmed the down-regulation of the six genes by using real-time RT-PCR analysis (Fig. 6). From the six genes we found a G-box motif (5′-CACGTG-3′) only in the promoter region of At4g38000 encoding a putative Dof zinc-finger protein (Table II). But we found stretches of 5′-CATGTG-3′, found in M-box elements, in more large promoter regions of At1g71250, At3g52970, and At5g03820. The M-box element is well conserved in the E box (5′-CANNTG-3′) as a binding site of the bHLH-Leu zipper transcription factor in mammalian cells (Lowings et al., 1992; Zeller et al., 2003). We therefore performed a gel-shift assay using RGE1 protein produced by in vitro translation and labeled promoter fragments of At5g03820 (a lipase gene) that included an M-box element rather than a G box. However, direct interaction was not observed under our assay conditions (data not shown). RGE1 has a putative nuclear localization signal in the bHLH domain. The RGE1-GFP fusion protein localized to the nucleus of tobacco cells after Agrobacterium infiltration (Fig. 2, B and C). These results might indicate that RGE1 indirectly regulates down-regulated genes or that some other factors that are expressed at the heart embryonic stage are required for specific binding to these promoters including M-box elements.
RGE1 Controls Embryo Growth from the Endosperm
We found that RGE1 is strongly expressed in seeds and that expression levels are gradually induced from the early heart stage (9 and 10 DAF). The loss-of-function mutants, rge1-1 and rge1-2, showed shriveled seeds that were smaller than wild type (Table I). These results indicate that RGE1 has a function in seed development. When rge1-1 was pollinated with wild-type pollen, seed shape and size was restored within the expected Mendelian segregation. This suggests that RGE1 is not likely to be imprinted, because a function imprinted during seed development would be expected to show deviations from the ratio predicted from Mendelian rules. The seeds restored by pollination with wild type appeared to have a wild-type phenotype even when the maternal plant was homozygous (Fig. 3G). These results indicate that there is no maternal effect on the rge1-1 seed phenotypes and that the shriveled seeds are caused by abnormal embryo and/or endosperm development. This is supported by the ability of the shriveled seed integuments, which come from maternal tissue, to release mucilage, because ttg1 and ttg2 that have defective seed integuments cannot (Penfield et al., 2001; Western et al., 2001).
The mature embryo of rge1-1 is obviously smaller than that of wild type (Fig. 4, A–D). The seedling is small in the beginning but grows to the equivalent size as wild type during seedling development and to almost the same size and shape as wild type at the adult stage (Fig. 4, E–H). This is a typical phenotype of a mutant for abnormal endosperm development (Sorensen et al., 2002; Luo et al., 2005). RGE1 expresses during seed development and more specifically in the endosperm (Fig. 5, P–R). These results indicate that RGE1 functions in the endosperm. This speculation is supported by the fact that the two genes (At1g71250 and At5g03820) down-regulated by RGE1 (Table II) strongly express in the endosperm, although germinated seeds, and not developing seeds, were used to investigate the differences in gene expression profiles between the embryo and the endosperm using microarray analysis (Penfield et al., 2006). It is known that seed and embryo size are mainly dependent on endosperm growth during seed development (Boisnard-Lorig et al., 2001), and several endosperm mutants produce abnormal endosperm caused by defective or inefficient cellularization (Sorensen et al., 2001, 2002). At the early heart stage when RGE1 expression appears (7 DAF), endosperm cellularization also starts. We therefore checked whether the endosperm of rge1-1 cellularizes normally, and observed no irregularities. To our knowledge, it has not been reported that mutation in a gene expressed specifically in endosperm causes retarded embryo growth, resulting in smaller seeds without abnormal endosperm development. Consequently, rge1-1 and rge1-2 should be novel mutants affecting seed development. These results suggest that RGE1 does not function for endosperm development, but rather controls embryo development through the endosperm.
Genes, Including Lipases and P450, Are Specifically Repressed in the rge1-1 Mutant
By microarray analysis we could not find genes known to be related with embryo and endosperm development like titan and mini3. But we found six genes that showed decreased expression in rge1-1 compared to wild type. Among these are two genes that encode for different putative GDSL motif lipases. The lipase family containing the GDSL motif has been described as a novel class of lipases that is only present in bacteria and plants and some members have been confirmed or presumed to have lipolytic activity (Brick et al., 1995; Oh et al., 2005). The bacterial lipase has been extensively studied, but few plant lipases have been characterized and there is very little knowledge about their possible biological functions. Many lipases have been implicated in signal transduction in the biosynthesis of hormones such as Brassionosteroid and auxin (Fujioka and Yokota, 2003; Woodward and Bartel, 2005). We also found a gene encoding a cytochrome P450 (CYP76G1) whose function is unknown. It is known that many P450s are concerned with plant hormone biosynthesis (Fujioka and Yokota, 2003; Woodward and Bartel, 2005). One possibility is that down-regulation of these genes, GDSL motif lipases and/or CYP76G1, causes disordered hormone flux in endosperm. This might indicate that the supply of some hormones from the endosperm to the embryo is necessary for embryo growth and decreased hormone levels in the endosperm by the absence of these lipases and/or CYP76G1 cause retardation of embryo growth.
The endosperm of many dicots is not persistent in the dry seeds, and is assumed to be used for supporting embryo morphogenesis and early maturation during seed development (Lopes and Larkins, 1993). Therefore, it is an attractive hypothesis that these GDSL motif lipases are related to the supply of the carbon source to the embryo through the breakdown of oil/lipid in the endosperm during seed development. This hypothesis is supported by the fact that a defective Suc transporter expressed specifically in endosperm causes perturbed fatty acid composition in early developing seeds and a little delay in embryo development (Baud et al., 2005). This is also supported by the fact that loss-of-function mutants of WRINKLED1 (WRI1), which encodes an APETALA2/ethylene-responsive element-binding domain protein, and of plastidic pyruvate kinase (PKP1), which plays a role in oil accumulation in maturing embryos of Arabidopsis, show similar phenotypes to rge1-1 and rge1-2, including small shriveled seeds and retardation of embryo growth (Cernac et al., 2006; Andre et al., 2007; Baud et al., 2007). In addition wri1 and pkp1 show seedling lethality on plates without Suc, as do rge1-1 seedlings (Cernac et al., 2006; Andre et al., 2007). The inability to break down lipid in the endosperm of rge1-1 and rge1-2 may cause a decreased supply of the carbon source to the embryo and depletion of storage lipids, triacylglycerols, as observed in wri1 and pkp1 seeds.
RGE1 protein can control gene expression that is specific at the embryo heart stage. We found a strong reduction in the expression of six genes including putative GDSL motif lipases and CYP76G1 in the rge1-1 mutant and the coexpression profile of these genes with RGE1 from the heart stage of embryo development (Table II; Fig. 3B). We checked seed phenotypes of loss-of-function mutants of these genes, but did not observe any characteristic phenotypes (data not shown). These genes might redundantly function in seed development. They suggest that further analysis of the relationship between RGE1 and these genes using double mutants is required.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col-0, Ws, and the transformed lines) were grown at 22°C in a cultivation container system (ARACON) in long-day conditions (16 h light and 8 h dark) under white fluorescent tubes (FL40SW; Sanyo). Arabidopsis transformation was performed as previously described (Nakazawa et al., 2003).
The GABI and INRA lines, GABI_584D09 and FLAG_400A08, which have T-DNA insertions in the gene At1g49770 (RGE1), were obtained from the Max Planck Institute in Germany and Institute Jean-Pierre Bourgin in France, respectively.
Seed Size Measurement
At least 70 seeds were put on adhesive PCR films (ABgene) and sealed with another film. We used a flatbed scanner at 3,200 dpi resolution to scan the seeds sealed between the films for the creation of TIFF files. The scanned TIFF files were analyzed using WINSEEDLE software (Regent Instrument Inc.). The customized parameters for Arabidopsis seeds were used for the measurement of dimension, length, and width of seeds. The customized parameters were as follows; seeds were identified as being more than 400 and less than 3,000 pixels in area.
Construction of Vectors for Generation of Transgenic Plants
To generate a RGE1 overexpression construct, the PCR primers RGE1-ATG-GW (GGGGACAAGTTTGTACAAAAAAGCAGGCTATGACTAATGCTCAAGAGTTG) and RGE1-STOP-GW (GGGGACCACTTTGTACAAGAAAGCTGGGTTATAGAGATGAAAAATATAACAC) were used for amplification from cDNA prepared from siliques. This amplified fragment was cloned into pBIDAVL-GWR1 using the GATEWAY cloning system (Invitrogen Corp.) as previously described (Nakazawa et al., 2003). For the promoter-GUS constructs, RGE1-Pro-F (5′-CCCAAGCTTAATTAAGGGTCATAATTACAAC-3′) and RGE1-Pro-R (5′-CGGGATCCGGAATCATCAGAATTGGATATG-3′) were used to amplify 2.2 kb upstream from the start codon and the first 10 codons of RGE1. This amplified fragment was digested with BamHI and HindIII, and cloned into the pBI101.1 vector. To construct the 35S-RGE1:GFP construct, a full-length RGE1 cDNA was cloned into the binary vector pBE2113GFP-GW, made by Dr. Kuroda (RIKEN), in frame with sGFP (S65T) using the GATEWAY cloning system (Niwa, 2003). pBE2113GFP-GW was made by insertion of the reading frame B cassette into the XbaI site of pBEGFP (Suzuki et al., 2006), provided from Dr. Takahashi (Tohoku University) and Dr. Niwa (University of Shizuoka). These constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation for transformation of Arabidopsis plants.
RNA Analysis
RNA was isolated using an RNAqueous kit (Ambion) and cDNA was synthesized using QuantiTect Reverse Transcription for RT-PCR and real-time PCR analysis (QIAGEN GmbH) according to the instructions.
Real-time PCR analysis was performed using the MX3000P Multiplex Quantitative PCR system (Promega Corp.) according to the manufacturer's instructions. SYBR Green I was used as the dye for detection of the amplified fragment and part of each gene was employed as reference DNA. The primers for amplification of the reference DNA were: RGE1, 5′-TGAAGAAGAATCACCTGATC-3′ and 5′-CTGTTGCGGTGGCGTCTATG-3′; At3g06890, 5′-ATCATACTCGCCGTCGTTGTTGC-3′ and 5′-GAACAAGATCAGTACGAGGAAGAG-3′; At4g38000, 5′-CAACCTTGCCTCGTCTTCTATCG-3′ and 5′-ATCATCATTATCTTCATGATTG-3′; At1g71250, 5′-GCGATCACCTGGAGCCATCTATG-3′ and 5′-GTCTGAGTAGGATGGAATGCATC-3′; At3g52970, 5′-GCCGGAGAGTTCATCAGAGAACG-3′ and 5′-TGGTAGGTCTTCTTCTTGGAGC-3′; At5g03820, 5′-TCAATACCAAACTCAACAACACG-3′ and 5′-GTAATTCGTAGCATTCGAACATG-3′; At4g33600, 5′-TCTCGTATAATCATTACGATTAC-3′ and 5′-TATCGATCACGGCTGACTCATTC-3; and ACT2, 5′-GTATCGCTGACCGTATGAGC-3′ and 5′-GATCTTGAGAGCTTAGAAAC-3′.
The gene-specific primers for real-time PCR were: RGE1, 5′-TTTGCTTCCCCAACTTCCTC-3′ and 5′-GCTTCTCAAGCTTTTGCATTTC-3′; At3g06890, 5′-CATCGGAGACAGTGGTGAGG-3′ and 5′-AGAAAATGGCCGACACGAAG-3′; At4g38000, 5′-CGTCACTTCCATGTTTCTCCA-3′ and 5′-CACTTGTTGCACCTCCTCCA-3′; At1g71250, 5′-AGCGATCACCTGGAGCCATC-3′ and 5′-TCCTTGGTTCCTCCCAATCC-3′; At3g52970, 5′-GAGCGACGAGAAGACGAAAGA-3′ and 5′-ATCCGTTCCAGCCGTAAACA-3′; At5g03820, 5′-TTCCCGGTCTGAAATTGGTC-3′ and 5′-CGGTTCCACAACATGCTCTTC-3′; At4g33600, 5′-GGTTCAAGATGGGGAATTGG-3′ and 5′-ACAATAGTGTCTGGCTTTGCATC-3; and ACT2, 5′-CTGGATCGGTGGTTCCATTC-3′ and 5′-CCTGGACCTGCCTCATCATAC-3′. Expression levels were normalized with ACT2 expression. Values are the means of two replicates.
RT-PCR analysis was performed using cDNA from each organ as the template. The primers for amplification of RGE1 and ACT2 were used as gene-specific primers for real-time PCR for RGE1 and ACT2, respectively. The primers for amplification of UBQ were UBQ-F (5′-CAGCTCTTGG GTGAAGACGA-3′) and UBQ-R (5′-GATGGCCGTACTTTGGCTGA-3′). To determine RGE1 expression levels in rge1-1, we used primers RGE1-ATG-long (5′-ATGACTAATGCTCAAGAGTTGGGGCAAGAG-3′) and RGE1-STOP-long (5′-TAGAGATGAAAAATATAACACCAGTTCTTG-3′).
Microarray Analysis
We used the Agilent Arabidopsis 2 Oligo Microarray for 22 K Microarray analysis (Agilent Technologies). Each RNA sample for Cy5- and Cy3-labeled cDNA probes was isolated from siliques of rge1-1 and wild type at 12 DAF. The hybridized and washed material on each glass slide was scanned by an Agilent DNA microarray scanner (model G2565BA; Agilent Technologies). We used Feature Extraction and Image Analysis software (Agilent Technologies) for establishing the location and delineation of every spot in the array. For integration of each intensity, we used filtration and normalization and for calculating the expression ratio and P value of each spot we used default parameters. Identification of genes with reliably altered levels was achieved using a false discover rate procedure from our experimental data set (Storey and Tibshirani, 2003). The calculation was done using the statistical analysis software R that includes a module for performing the q-value calculation. All default parameters in the q-value module were used. The genes were selected as having altered expression levels if the q values for differential expression were commonly below 0.005 in two experiments. Genespring GX (Agilent Technologies) was used for all gene-clustering analysis.
Intracellular Protein Localization
Individual Agrobacterium colonies that were transformed with the 35S-RGE1:GFP construct were grown for 20 h in 5-mL cultures (Luria broth, 50 μg/mL kanamaycin) at 30°C. Using a 2.5-mL syringe, the Agrobacterium solution was injected into the abaxial surface of the leaves. After 24 h we observed the injected leaves using confocal microscopy (LSM510; Carl Zeiss, Inc.).
Microscopic Observation
We carried out GUS staining of seeds of plants transformed with the RGE1-promoter construct as described previously (DeBlock and DeBrouwer, 1992). Developing seeds and GUS-stained seeds were cleared as described previously (Boisnard-Lorig et al., 2001). Specimens were examined with an Olympus microscope using differential interference contrast optics. For observations of mature seeds we used low vacuum scanning electron microscopy (JSM-5600LV; JEOL). Feulgen staining was carried out and stained seeds were observed by confocal microscopy as described previously (Sorensen et al., 2002).
Microarray data from this article have been deposited with the Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSM294800 and GSM294801.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic PCR experiment showing all plants derived from shriveled seeds of rge1-1 were homozygous.
Supplemental Table S1. The results of oligomicroarray analysis for the investigation of differences in gene expression profiles in siliques between rge1-1 and wild type at 12 DAF are shown.
Supplementary Material
Acknowledgments
We thank H. Kuroda (Plant Functional Genomics Research Group, RIKEN, Japan) for providing pBE2113GFP-GW and tobacco leaves, S. Takahashi (Tohoku University, Japan) and Y. Niwa (University of Shizuoka, Japan) for providing pBE2113GFP, M. Okamoto, A. Matsui, F. Myoga, and T. Kuromori for helpful discussions, and A. Enju for sequencing (Plant Functional Genomics Research Group, RIKEN, Japan). We also thank the Max Planck Institute and Institute Jean-Pierre Bourgin for providing the T-DNA insertion mutant lines. This work was performed during functional analysis of activation-tagging lines. These lines were generated as material for functional genomics using Arabidopsis at the RIKEN Genomic Sciences Center.
This work was supported by KAKENHI from the Ministry of Education, Culture, Sports, Science and Technology of Japan (19710055).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Minami Matsui (minami@riken.jp).
The online version of this article contains Web-only data.
References
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ (2000) Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127 2493–2502 [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43 824–836 [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick DJ, Brumlik MJ, Buckley JT, Cao JX, Davies PC, Misra S, Tranbarger TJ, Upton C (1995) A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett 377 475–480 [DOI] [PubMed] [Google Scholar]
- Busch M, Mayer U, Jurgens G (1996) Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol Gen Genet 250 681–691 [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlock M, DeBrouwer D (1992) In-situ enzyme histochemistry on plastic-embedded plant material: the development of an artefact-free β-glucuronidase assay. Plant J 2 261–266 [Google Scholar]
- Faure JE, Rotman N, Fortune P, Dumas C (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J 30 481–488 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54 137–164 [DOI] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Saingery V, Chambrier P, Mayer U, Jurgens G, Berger F (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450 [DOI] [PubMed] [Google Scholar]
- Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F (2004) Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 2971–2981 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 735–747 [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Muto S, Gohda K, Suzuki K, Ishikawa A, Kobayashi H, Yoshizumi T, Tsumoto Y, et al (2003) Sequence database of 1172 T-DNA insertion sites in Arabidopsis activation-tagging lines that showed phenotypes in T1 generation. Plant J 36 421–429 [DOI] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W (2003) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA (1993) Endosperm origin, development, and function. Plant Cell 5 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowings P, Yavuzer U, Goding CR (1992) Positive and negative elements regulate a melanocyte-specific promoter. Mol Cell Biol 12 3653–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 102 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C, Yang LJ, Meinke D (2000) The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, Suzuki K, Muto S, Matsui M (2003) Activation tagging, a novel tool to dissect the functions of a gene family. Plant J 34 741–750 [DOI] [PubMed] [Google Scholar]
- Niwa Y (2003) A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol 20 1–11 [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell (Suppl) 16 S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341 [DOI] [PubMed] [Google Scholar]
- Sorensen MB, Chaudhury AM, Robert H, Bancharel E, Berger F (2001) Polycomb group genes control pattern formation in plant seed. Curr Biol 11 277–281 [DOI] [PubMed] [Google Scholar]
- Sorensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jurgens G, Somerville C, Lepiniec L, Berger F (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129 5567–5576 [DOI] [PubMed] [Google Scholar]
- Spillane C, MacDougall C, Stock C, Kohler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J (2000) Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol 10 1535–1538 [DOI] [PubMed] [Google Scholar]
- Steinborn K, Maulbetsch C, Priester B, Trautmann S, Pacher T, Geiges B, Kuttner F, Lepiniec L, Stierhof YD, Schwarz H, et al (2002) The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev 16 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol 224 149–157 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Takahashi S, Watanabe R, Fukushima Y, Fujita N, Noguchi A, Yokoyama R, Nishitani K, Nishino T, Nakayama T (2006) An isoflavone conjugate-hydrolyzing beta-glucosidase from the roots of soybean (Glycine max) seedlings: purification, gene cloning, phylogenetics, and cellular localization. J Biol Chem 281 30251–30259 [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Lohner A, Jurgens G (1996) The GURKE gene is required for normal organization of the apical region in the Arabidopsis embryo. Plant J 10 1005–1016 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, McElver JA, Liu Cm CM, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW (2002) Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol 128 38–51 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV (2003) An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4: R69 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.