Recent data on the plasma membrane (PM)-located LRR-RLKs (for Leu-rich repeat receptor-like kinases) BRI1 (for brassinosteroid insensitive 1) and the coreceptors BAK1 (for BRI1-associated kinase 1) and SERK1 (for somatic embryogenesis receptor-like kinase 1) that participate in the perception of brassinosteroids (BRs) suggest that they are organized into heterooligomeric protein complexes. Other components of this complex include members of the 14-3-3 family, and, in the case of SERK1, the kinase-associated protein phosphatase (KAPP) and the AAA ATPase cell division cycle 48A (CDC48A). CDC48 proteins interact with ubiquitinated target proteins in animal and plant cells. In this Update we describe the role of several of the nonreceptor partners of the PM receptor complex with an emphasis on the role of CDC48 proteins in translocation and ubiquitination as a proposed mode of regulation of plant PM receptors.
THE BRI1/SERK RECEPTOR COMPLEX
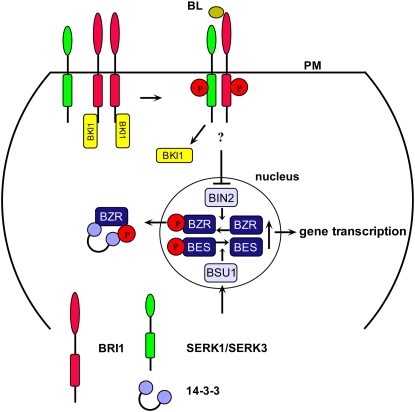
Receptors such as BRI1 and SERK1 are synthesized in the endoplasmic reticulum (ER), from where they pass through the Golgi network to be inserted into the PM. Indeed, the use of fluorescently tagged BRI1 and SERK proteins has shown clearly both plasma and internal membrane localization (Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002; Russinova et al., 2004; Albrecht et al., 2005; Kwaaitaal et al., 2005). In Arabidopsis (Arabidopsis thaliana), removal of receptors from the PM by endocytosis is documented for BRI1, BAK1, SERK1, and the flagellin-sensing receptor FLS2 (Russinova et al., 2004; Chinchilla et al., 2007; Geldner et al., 2007). In the case of FLS2, ligand-induced endocytosis was documented (Robatzek et al., 2006; Chinchilla et al., 2007) while the BRI1 and SERK receptors seem to undergo non-ligand-dependent recycling (Russinova et al., 2004; Geldner et al., 2007). The SERK1 protein is one of five members of a small family belonging to subgroup II of the LRR-RLKs in Arabidopsis. SERK1, together with SERK2, is instrumental in male fertility (Albrecht et al., 2005) while SERK3, also known as BAK1, forms a heterodimer with BRI1 (Li et al., 2002; Nam and Li, 2002). Upon binding of BR to the extracellular domain of BRI1, the BRI1 kinase inhibitor 1 (BKI1), a negative regulator of BRI1 activity, is released from the PM and increases the affinity of BRI1 for BAK1/SERK3 (Wang and Chory, 2006; Fig. 1). Oligomerization of the BRI1/BAK1 receptors and transphosphorylation of the kinase domains takes place (Wang et al., 2005; Wang and Chory, 2006). This inhibits, via an unknown pathway, the phosphorylation of BZR1 (for brassinazole resistant 1) by the BIN2 (for BR insensitive 2) kinase that, when phosphorylated, is translocated to (Ryu et al., 2008) and retained in the cytoplasm via 14-3-3 proteins (Gampala et al., 2007). Moreover, BR activates the phosphatase BSU1 (for BRI1 suppressor protein 1) that dephosphorylates BES1 (for BRI1 EMS suppressor 1). Subsequently, accumulation of dephosphorylated nuclear-localized BES1 and dephosphorylated BZR1 transcription factors induces the genetic response to brassinolide in Arabidopsis. BAK1/SERK3 alone also serves a BR-independent pathway involved in plant immunity (Heese et al., 2007; Kemmerling et al., 2007), interacts with FLS2 (Chinchilla et al., 2007), and together with SERK4 is involved in cell death (He et al., 2007). BAK1/SERK3 is not the only coreceptor of BRI1; SERK1 also interacts with BRI1 (Karlova et al., 2006), and evidence has been presented that BKK1 (for BAK1-like kinase 1), identical to SERK4, also participates in BR signaling (He et al., 2007). Thus, the classical model for ligand-induced heterooligomerization followed by auto- and transphosphorylation as well as translocation of target proteins appears to apply to receptors of the BRI1/SERK class while complexes of different composition may have different specificity. So far no ligands have been shown to interact with the SERK receptors. Where and how the receptors and interacting proteins such as BKI1 are assembled into functional units have not been described so far. Structural elements involved in sorting of animal receptors such as Yxxφ and di-Leu motifs, involved in clathrin-mediated processes (Ortiz-Zapater et al., 2006), are found in the primary sequences of all receptors mediating the BR response, suggesting complex sorting and trafficking pathways.
Figure 1.
Interacting partners in BR signaling. BKI1 binds to the BRI1 homodimer, thus preventing interaction with BAK1/SERK3. Upon binding of brassinolide (BL) to BRI1, BKI1 dissociates from the receptor and dimerization and transphosphorylation take place with SERK1/SERK3 (BAK1). Subsequently phosphorylation of BZR1 by BIN2 is inhibited, and the phosphatase BSU1 is activated. This leads to accumulation of dephosphorylated BZR1 and BES1 in the nucleus, which induces gene transcription. Furthermore, phosphorylated BZR1 translocated to the cytoplasm is retained there by 14-3-3 proteins, and only dephosphorylated BZR1 translocates back to the nucleus.
DIFFERENT SERK1 RECEPTOR COMPLEXES EXIST
The SERK1 complex was immunoprecipitated from transgenic seedlings and found to contain both the BRI1 and SERK3 proteins. In addition were identified the KAPP protein, a member of the 14-3-3 family, and the AAA ATPase CDC48A (Fig. 2), as well as two putative transcriptional regulators. Of these the KAPP, 14-3-3, and CDC48A proteins also were found previously in a yeast two-hybrid screening (Rienties et al., 2005), suggesting a direct interaction with the SERK1 receptor. In mammalian cells, the CDC48A ortholog p97/VCP is phosphorylated by the JAK-2 kinase and dephosphorylated by PTPH1 that in turn is associated with a 14-3-3 protein (Zhang et al., 1999; Lavoie et al., 2000). The interaction between KAPP and SERK1 is dependent on the phosphorylation status of the Thr residues in the SERK1 activation loop. In vitro, KAPP not only dephosphorylates SERK1 but also actively prevents autophosphorylation of the receptor, possibly maintaining the dephosphorylated state of SERK1 (Shah et al., 2002). In cells, KAPP colocalizes at the PM and in the cytoplasm in endosomal compartments with SERK1 and colocalizes with the endosomal marker Ara6 and the membrane dye FM4-64 (Aker et al., 2006). In contrast, CDC48A does not colocalize with SERK1 in endosomal compartments but colocalizes and interacts with SERK1 in ER-membrane-based regions in close proximity to the PM (Aker et al., 2006). It is still unclear if CDC48A and KAPP can interact directly, suggesting that the three plant proteins may not be a direct copy of the corresponding animal complexes.
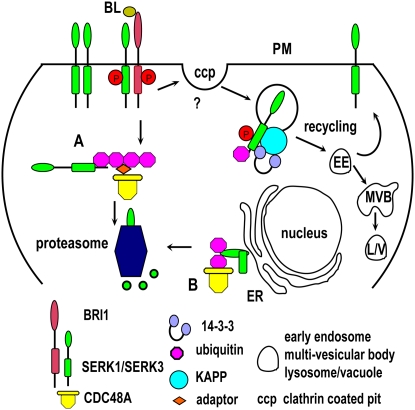
Figure 2.
Hypothetical degradation pathways of the SERK1 receptor after activation. Upon binding of the BR brassinolide, BRI1, and SERK1 (and/or SERK3), receptors dimerize and transphosphorylate. After activation, monoubiquitination is a signal for internalization (via clathrin-coated pits?), acquiring the phosphatase KAPP, to give rise to early endosomes. From these endosomes, the receptors can recycle to the PM or are targeted to MVBs and subsequently to lysosomes or vacuoles to be degraded. A, Polyubiquitination after activation of the receptors is a signal for degradation in the proteasome, acquiring adaptor proteins and CDC48A. B, Misfolded SERK1 receptors are pulled out of the ER lumen, polyubiquitinated, and targeted to the proteasome, acquiring CDC48A again.
After activation of animal receptor complexes, changes in interacting partners and/or location often occur. Members of the regulatory 14-3-3 family of proteins were shown to be instrumental in protein translocation between the cytosol and the nucleus in animal cells (Aitken, 2006). In Arabidopsis BR signaling it was shown that 14-3-3λ retains phosphorylated BZR1 in the cytoplasm, thereby preventing pBZR1 degradation and inhibiting BR-induced gene expression (Gampala et al., 2007). Likewise, in human cells, 14-3-3σ is up-regulated by the tumor suppressor gene p53 in response to DNA damage and retains the E3 ubiquitin ligase murine double minute oncogene (MDM2) in the cytoplasm, thereby preventing p53 degradation and inhibiting tumor growth (Yang et al., 2007).
SERK1 also contains a putative 14-3-3 binding domain, RPPS394QPP, and was shown to interact with 14-3-3λ and 14-3-3ν (Rienties et al., 2005; Karlova et al., 2006), but a role in translocation of the SERK1 protein has yet to be demonstrated.
To summarize, these data suggest that not all the proteins identified are part of the same SERK1-containing complex and therefore can represent different stations during the trafficking of SERK1 through the cell.
UBIQUITIN AS A SORTING SIGNAL
Turnover of receptors and other membrane proteins is dependent on endocytosis and/or ubiquitin-mediated protein degradation via the proteasome pathway. Monoubiquitination serves as a signal for endocytosis of PM proteins, sorting of proteins to the multivesicular bodies (MVBs), budding of retroviruses, DNA repair, and transcriptional activation. Polyubiquitination has been associated mainly with targeting substrates to the proteasome (for review, see Haglund et al., 2003).
Examples of trafficking of cell surface molecules mediated by ubiquitin is the recruitment of G-protein-coupled receptors (GPCRs) and other cargo into vesicles budding from either the PM or the MVB (Reggiori and Pelham, 2001) in yeast. The same mechanism is used by receptor Tyr kinases (RTKs) such as the EGF receptor (Shtiegman and Yarden, 2003). Unlike the yeast GPCRs and the RTKs, the known mammalian GPCRs appear to require ubiquitination only as a sorting signal at the MVB level, not at the PM. At the PM other ubiquitinated proteins, such as β-arrestin, bind to the tail of the GPCR to recruit the receptor after ligand stimulation to the clathrin-dependent endocytic machinery. Apparently not ubiquitination of the receptor itself but ubiquitination of β-arrestin promotes rapid endocytosis (Shenoy and Lefkowitz, 2003).
In plant receptor biology only a few cases have been described that suggest involvement of ubiquitination in membrane protein trafficking. Internalization of plant-specific auxin efflux carriers (PIN proteins) in the PM is required for the regulation of auxin efflux (Geldner et al., 2003; Paciorek et al., 2005). Recently it was shown that ubiquitin-mediated degradation contributed to the asymmetric localization of PIN2 during the gravitropic response (Abas et al., 2006). Proteasomal degradation also plays an important role in developmental responses to the phytohormone auxin that are controlled by the activity of the auxin-inducible genes. Without auxin, the AUX/IAA transcriptional repressors inhibit transcription. In response to auxin, AUX/IAA repressors are degraded, thereby inducing gene expression. The F-box protein TIR1 (conferring E3 ligase specificity), which binds to these proteins in an auxin-dependent manner, controls this process (Gray et al., 2001). Because the TIR1 protein was identified as the auxin receptor in plants (Dharmasari et al., 2005; Kepinski and Leyser, 2005), this may provide a biochemical link between hormone homeostasis and trafficking of membrane proteins (Wisniewska et al., 2006). So far, no evidence has been presented for a role of monoubiquitination in trafficking or degradation of LRR-RLKs involved in other signaling pathways, including those mediated by BRI1 and SERKs.
DIFFERENT CELLULAR PATHWAYS EMPLOY CDC48 ACTIVITY
One partner of the BRI1/SERK receptor complex possibly involved in regulation of the SERK1 receptor degradation is the AAA ATPase CDC48A.The yeast and animal homologous proteins CDC48 and p97 are involved in various activities as diverse as cell cycle regulation, transcriptional activation, membrane fusion, and ER-associated degradation (ERAD) of misfolded proteins (Woodman, 2003; Müller et al., 2005). Although the ATPases also are involved in proteasome-independent functions, many substrates of CDC48 proteins eventually are degraded in the proteasome. Polyubiquitinated substrates are delivered to the proteasome via ubiquitin receptors that bind to the ubiquitin moiety and the proteasome. Most cofactors of p97/CDC48 also bind to ubiquitin conjugates, whether they are associated with proteolytic activity or not. This suggests that ubiquitination is a common regulator in various functions of p97/CDC48 and does not result in proteolytic degradation by default (Ye, 2006).
Meyer et al. (2000) proposed that a certain number of p97-interacting proteins define a set of distinct CDC48/p97 complexes, conferring different cellular activities. This hypothesis was based on their observation that a complex of Ufd1 (ubiquitin fusion degradation) and Np14 proteins competes for binding to p97 with another cofactor, p47. p47 plays a role in the fusion of Golgi and ER membrane fragments and links p97 to its substrate, the t-SNARE syntaxin 5. Syntaxin 5 is a membrane-localized p47 receptor and a family member of the t-SNARE proteins (target soluble NSF attachment protein receptors; Rabouille et al., 1998). Syntaxin 5 proteins on two close membranes pair to form a complex, resulting in fusion of these membranes. The p97/p47 complex is suggested to act as a chaperone in this process. p47 recruits p97 via its ubiquitin regulatory X domain. The Ufd1/Np14 complex binds to p97 in an exclusive manner different from p47 and is probably involved in nuclear transport, showing that the different participants in a certain CDC48/p97 complex determine its cellular function and localization.
Similar to the situation in yeast (http://www.yeastgenomics.org/), also in Arabidopsis CDC48 proteins are found to be interacting with a large variety of different proteins, such as ubiquitin-binding proteins, SNARE proteins, including the cell plate-associated protein KNOLLE, as well as SYP21 and SYP31 (Rancour et al., 2002), suggesting that in plants they also perform a general role. This makes the AAA ATPase p97/CDC48 an intriguing membrane complex protein potentially involved in recognizing ubiquitinated membrane proteins.
WHAT MECHANISM UNDERLIES THE INTERACTION OF THE CDC48A PROTEIN WITH THE SERK1 RECEPTOR?
CDC48 proteins were shown to be involved in proteasomal targeting of proteins. Proteasome activity has been shown to mediate turnover of plant membrane proteins including PIN2. Treatment of PIN2 seedlings with the proteasome inhibitors MG132 and lactastatin resulted in elevated PIN2 levels. Furthermore, proteasome inhibition caused a dramatic increase in the amount of ubiquitinated PIN2 (Abas et al., 2006).
Proteasome-dependent turnover of SERK1 receptors might be a way to regulate its activity. CDC48A may be involved in targeting polyubiquitinated receptors for degradation in the proteasome, like it was shown for the mammalian cytokine receptors IL-2 and IL-9, the erythropoietin receptor, and the CDC48 homolog p97 (Yen et al., 2000). Another hypothesis is that the CDC48A protein mediates ERAD-like quality control of SERK1 receptors. As much as 30% of newly synthesized proteins contain peptides arising from degraded misfolded proteins, which partly are ubiquitinated (Schubert et al., 2000). In the ER, a strict quality control therefore must ensure that aberrant proteins are not exported. The conformational change of p97/CDC48 during ATP binding and subsequent release supplies the mechanical force to pull ERAD substrates from the ER membrane and translocate them to the proteasome machinery in the cytoplasm. Overexpression of dominant negative CDC48A mutants indeed resulted in accumulation of a mutant barley (Hordeum vulgare) powdery mildew resistance o receptor (mlo-1) in Arabidopsis protoplasts, showing that CDC48A is involved in the degradation of aberrant proteins (Müller et al., 2005). Mutant MLO receptors became highly polyubiquitinated upon treatment with the proteasomal inhibitor MG132. SERK1 expressed in protoplasts also was accumulating in the perinuclear ER upon MG132 treatment (Aker et al., 2006), suggesting that in protoplasts a large amount of newly synthesized protein is misfolded and must be degraded. However, no data are presently available to support the hypothesis that SERK1 interaction with hexameric CDC48A at the ER is indeed dependent on ubiquitination.
OUTLOOK
Recent evidence from animal receptor biology suggests that ubiquitination is one of the first biochemical events after receptor activation (Lowe et al., 2006; Shenoy, 2007) and therefore also could be involved in later steps in plant signaling pathways via PM LRR-RLKs. We propose that the AAA ATPase protein CDC48A interacts with the SERK receptor as a chaperone to promote the formation of correctly folded receptors in the ER and to remove misfolded proteins by degradation via the ubiquitin-proteasome pathway. In Figure 2 the possible degradation pathways of the SERK1 receptor after activation are depicted.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sacco C. de Vries (sacco.devries@wur.nl).
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wisniewska J, Moulinier-Azola J, Sieberer T, Friml J, Luschnig C (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8 249–256 [DOI] [PubMed] [Google Scholar]
- Aitken A (2006) 14-3-3 proteins: a historic overview. Semin Cancer Biol 16 162–172 [DOI] [PubMed] [Google Scholar]
- Aker J, Borst JW, Karlova R, de Vries SC (2006) The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the Somatic Embryogenesis Receptor-like Kinase 1 receptor at the plasma membrane. J Struct Biol 156 62–71 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robazek S, Kemmerling B, Nurnberger T, Jones J, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Dharmasari N, Dharmasari S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Gampala S, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron J, et al (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell 13 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport and auxin-dependent plant growth. Cell 112 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J (2007) Endosomal signalling of plant steroid receptor kinase BRI1. Genes Dev 21 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276 [DOI] [PubMed] [Google Scholar]
- Haglund K, Di Fiore P, Dikic I (2003) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci 28 598–603 [DOI] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J (2007) BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol 17 1109–1115 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann D, Gimenez-Ibanez S, Jones A, He K, Li J, Schroeder J, Peck S, Rathjen J (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriquez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Mussig C, et al (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17 1116–1122 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Kwaaitaal MA, de Vries SC, Russinova E (2005) Arabidopsis thaliana Somatic Embryogenesis Receptor Kinase 1 protein is present in sporophytic and gametophytic cells and undergoes endocytosis. Protoplasma 226 55–65 [DOI] [PubMed] [Google Scholar]
- Lavoie C, Chevet E, Roy L, Tonks N, Fazel A, Posner B, Paiement J, Bergeron J (2000) Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro. Proc Natl Acad Sci USA 97 13637–13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222 [DOI] [PubMed] [Google Scholar]
- Lowe E, Doherty T, Karahashi H, Arditi M (2006) Ubiquitination and de-ubiquitination: role in regulation of signaling by Toll-like receptors. J Endotoxin Res 12 337–345 [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G (2000) A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J 19 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Piffanelli P, Devoto A, Miklis M, Elliott C, Ortmann B, Schulze-Lefert P, Panstruga R (2005) Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212 [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Soriano-Ortega E, Marcote M, Ortiz-Masia D, Aniento F (2006) Trafficking of the human transferrin receptor in plant cells: effects of tyrphostin A23 and brefeldin A. Plant J 48 757–770 [DOI] [PubMed] [Google Scholar]
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof Y, Kleine-Vehn J, Morris DA, Emans N, Jurgens G, Geldner N, et al (2005) Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92 603–610 [DOI] [PubMed] [Google Scholar]
- Rancour DM, Dickey CE, Park S, Bednarek SY (2002) Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol 130 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Pelham H (2001) Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J 20 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienties IM, Vink J, Borst JW, Russinova E, Vries SC (2005) The Arabidopsis SERK1 protein interacts with the AAA-ATPase AtCDC48, the 14-3-3 protein GF14λ and the PP2C phosphatase KAPP. Planta 221 394–405 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin YH, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Hwang I (2008) Spatial redistribution of key transcriptional regulators in brassinosteroid signaling. Plant Signal Behav 3 278–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton L, Gibbs J, Norbury C, Yewdell J, Bennink J (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404 770–774 [DOI] [PubMed] [Google Scholar]
- Shah K, Russinova E, Theodorus WJ, Gadella J, Willemse J, DeVries SC (2002) The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev 16 1707–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S (2007) Seven-transmembrane receptors and ubiquitination. Circ Res 100 1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S, Lefkowitz R (2003) Multifaceted roles of β-arrestins in the regulation of seven-membrane-spanning receptor trafficking and signalling. Biochem J 375 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtiegman K, Yarden Y (2003) The role of ubiquitylation in signaling by growth factors: implications to cancer. Semin Cancer Biol 13 29–40 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313:1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer P, Ruzicka K, Blilou I, Rouguié D, Benkova E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312 883. [DOI] [PubMed] [Google Scholar]
- Woodman PG (2003) p97, a protein coping with multiple identities. J Cell Sci 116 4283–4290 [DOI] [PubMed] [Google Scholar]
- Yang H, Wen Y, Lin Y, Pham L, Su C, Yang H, Chen J, Lee M (2007) Roles for negative cell regulator 14-3-3sigma in control of MDM2 activities. Oncogene 26 7355–7362 [DOI] [PubMed] [Google Scholar]
- Ye Y (2006) Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J Struct Biol 156 29–40 [DOI] [PubMed] [Google Scholar]
- Yen CH, Yang YC, Ruscetti SK, Kirken RA, Dai RM, Li CC (2000) Involvement of the ubiquitin-proteasome pathway in the degradation of nontyrosine kinase-type cytokine receptors of IL-9, IL-2, and erythropoietin. J Immunol 165 6372–6380 [DOI] [PubMed] [Google Scholar]
- Zhang SH, Liu J, Kobayashi R, Tonks NK (1999) Identification of the cell cycle regulator VCP (p97/CDC48) as a substrate of the band 4.1-related protein-tyrosine phosphatase PTPH1. J Biol Chem 274 17806–17812 [DOI] [PubMed] [Google Scholar]