Abstract
Members of the Arabidopsis (Arabidopsis thaliana) DYNAMIN-RELATED PROTEIN1 (DRP1) family are required for cytokinesis and cell expansion. Two isoforms, DRP1A and DRP1C, are required for plasma membrane maintenance during stigmatic papillae expansion and pollen development, respectively. It is unknown whether the DRP1s function interchangeably or if they have distinct roles during cell division and expansion. DRP1C was previously shown to form dynamic foci in the cell cortex, which colocalize with part of the clathrin endocytic machinery in plants. DRP1A localizes to the plasma membrane, but its cortical organization and dynamics have not been determined. Using dual color labeling with live cell imaging techniques, we showed that DRP1A also forms discreet dynamic foci in the epidermal cell cortex. Although the foci overlap with those formed by DRP1C and clathrin light chain, there are clear differences in behavior and response to pharmacological inhibitors between DRP1A and DRP1C foci. Possible functional or regulatory differences between DRP1A and DRP1C were supported by the failure of DRP1C to functionally compensate for the absence of DRP1A. Our studies indicated that the DRP1 isoforms function or are regulated differently during cell expansion.
Dynamin and dynamin-related proteins (DRPs) constitute a structurally similar, yet functionally distinct, protein superfamily of GTPases found in all eukaryotes. A common feature of dynamin and DRPs is their ability to homo-oligomerize around lipid bilayers and modulate membrane structure (Praefcke and McMahon, 2004). The most well-studied protein of this family is MAMMALIAN DYNAMIN1, which functions in clathrin-mediated endocytosis (CME). During mammalian CME, adaptor complexes bind to cargo in the plasma membrane and subsequently recruit clathrin triskelia composed of clathrin heavy chain and light chain (CLC). Polymerization of clathrin triskelia into a lattice, membrane remodeling by accessory proteins, and the force generated by actin polymerization cause the plasma membrane to invaginate (Conner and Schmid, 2003). Dynamin 1 subunits are recruited to the invaginated membrane through a lipid-interacting pleckstrin homology domain and a protein-interacting Pro-rich domain (Vallis et al., 1999). Polymerization of the dynamin 1 subunits around the neck subsequently helps to sever the vesicle from the plasma membrane upon GTP hydrolysis (Damke et al., 1994; Roux et al., 2006).
The plant-specific dynamin family (DRP1) is common to many plant species, including the model systems Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and soybean (Glycine max), but has unknown molecular functions. The Arabidopsis DRP1 family is required during cytokinesis at the cell plate and during rapid cell expansion at the plasma membrane (Kang et al., 2001, 2003a, 2003b; Hong et al., 2003b; C.A. Konopka and S.Y. Bednarek, unpublished data). The Arabidopsis genome encodes five DRP1 isoforms (DRP1A through DRP1E) that have unique, yet overlapping, expression patterns based on analyses of promoter-reporter fusion constructs (Kang et al., 2003a, 2003b), native promoter-driven GFP fusion proteins (Kang et al., 2003a; C.A. Konopka and S.Y. Bednarek, unpublished data), northern-blot analysis (Kang et al., 2003b), and Arabidopsis gene expression databases (Zimmermann et al., 2004). DRP1A is expressed throughout most tissues, but its expression was not detected during pollen germination (C.A. Konopka and S.Y. Bednarek, unpublished data). Likewise, DRP1C is expressed in most tissues, including female reproductive organs and trichomes (C.A. Konopka and S.Y. Bednarek, unpublished data), but unlike DRP1A is expressed during pollen development (Kang et al., 2003b). DRP1E is expressed in cells throughout the plant, but its expression in pollen is approximately 20-fold lower than DRP1C (Zimmermann et al., 2004). DRP1B and DRP1D have the lowest expression of the DRP1s throughout the plant (Zimmermann et al., 2004).
The DRP1s share 65% to 84% amino acid sequence identity, which is most dissimilar throughout a 15- to 24-amino acid stretch between the middle domain and GTPase effector domain. Interestingly, the lipid-interacting pleckstrin homology domain of MAMMALIAN DYNAMIN1 is also positioned between the middle domain and the GTPase effector domain. By homology to known structures of dynamin (Zhang and Hinshaw, 2001) and animal DRPs (Prakash et al., 2000), the variable region in DRP1 is most likely surface exposed during oligomerization. The relevance of this variable domain remains to be determined.
drp1A, drp1C, and drp1E mutants have been isolated and characterized (Kang et al., 2001, 2003a, 2003b). The protein null drp1A-2 allele has defects in seedling development, trichome branching, fertility (Kang et al., 2001, 2003a), and venation (Sawa et al., 2005). After germination, drp1A-2 seedlings arrest unless grown on media supplemented with Suc. The direct cause of rescue is not well understood, but Suc is both a major transport metabolite in the plant and a signaling molecule. In contrast to the seedling lethality phenotype, the fertility defect of drp1A-2 is well characterized (Kang et al., 2003a). In wild-type flowers, stigmatic papillar cells undergo rapid polar expansion prior to pollen release (dehiscence), forming flask-shaped cells, which is required for pollen tube penetration and subsequent fertilization (Kandasamy et al., 1990). Stigmatic papillae from drp1A-2 flowers fail to undergo polar expansion and instead give rise to spherical, bloated cells, which correlates with low fertility (Kang et al., 2003a). In addition, the isotropically expanded papillae have an excess of plasma membrane characterized by large ingrowths and folds into the cytoplasm, indicating a requirement for DRP1A in plasma membrane maintenance during rapid polar growth. drp1E-1 mutants exhibit no obvious morphological phenotypes; however, drp1A-2/drp1E-1 double mutants are embryonic lethal and exhibit defects in cytokinesis, cell expansion, and morphology of the plasma membrane. This suggests that DRP1A and DRP1E function redundantly during embryogenesis.
In contrast, drp1C-1 mutants exhibit male gametophytic lethality. drp1C-1 pollen are small, shriveled, and do not germinate (Kang et al., 2003b). The mutant pollen also display defects in plasma membrane maintenance as drp1A-2 papillae. drp1C-1 pollen have excess plasma membrane, which forms large furrows and undulations that reach into the cytoplasm. Despite major morphological disruptions of the plasma membrane in drp1 mutants, there does not appear to be any defects in intracellular organelles, suggesting that the DRP1 family functions primarily at the plasma membrane in nondividing cells.
The Arabidopsis genome has undergone duplication events throughout its evolution, leading to gene families (Arabidopsis Genome Initiative, 2000) whose members sometimes, but not always, act redundantly. It is unclear whether the morphological defects in drp1A-2 and drp1C-1 mutants are due to differences in gene expression or whether DRP1A and DRP1C function in different pathways. DRP1C is hypothesized to be a component of the clathrin-associated endocytic machinery in Arabidopsis (C.A. Konopka and S.Y. Bednarek, unpublished data). To determine whether DRP1A is also a component of the CME machinery and whether DRP1A and DRP1C are functionally redundant, we have used live cell imaging and genetic complementation. Exogenous expression of DRP1C could not rescue drp1A-2 papillae expansion, but could compensate for the lack of DRP1A in seedlings. In addition, a DRP1A-GFP fusion protein displayed distinctive dynamics relative to DRP1C in the cell cortex of root cells, suggesting that the DRP1 isoforms act redundantly in some pathways, but also have distinct functions or regulatory mechanisms at the cell cortex during cell expansion.
RESULTS
DRP1A and DRP1C Are Conserved in Rice and Legumes
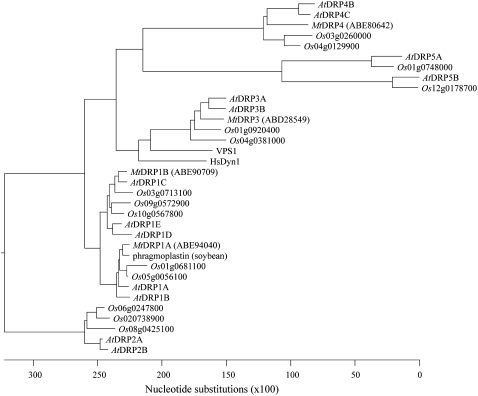
Using BLASTP database searches of the published rice and Medicago truncatula genomes, DRPs were identified in these species by the presence of the large GTPase domain (approximately 300 amino acids) conserved in all DRPs (SMART domain SM00053), and compared to the DRPs in Arabidopsis (Hong et al., 2003a). The rice genome contained 14 DRPs, five of which were closely related to the DRP1 family. We identified four DRPs from the available M. truncatula genome sequence, two of which were homologs of the Arabidopsis DRP1 family. A phylogenetic tree based on amino acid sequence including all known Arabidopsis, rice, and Medicago DRPs, as well as the soybean DRP1 homolog phragmoplastin, the yeast (Saccharomyces cerevisiae) DRP Vps1, and human dynamin 1, is shown in Figure 1. Interestingly, one MtDRP1 and two OsDRP1s formed a clade with DRP1A, while one MtDRP1 and three OsDRP1s were more similar to DRP1C than to DRP1A. This conservation of the different DRP1 isoforms in legume and nonlegume dicots and monocots suggests independent functions for the various plant-specific DRP1s.
Figure 1.
Phylogenetic tree of DRPs in Arabidopsis, rice, and Medicago. ClustalW alignment of the entire primary amino acid sequence of DRPs identified in BLAST searches from the published Arabidopsis, Oryza, and Medicago genomes and human dynamin 1, yeast Vps1, and soybean phragmoplastin were used to generate the phylogenetic tree. The scale represents to the number of nucleotide substitution events based on amino acid differences.
DRP1A-GFP Forms Discrete Foci at the Plasma Membrane
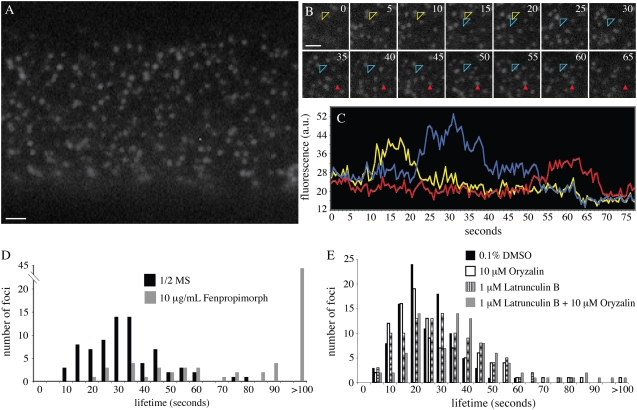
The dynamics of DRP1C at the plasma membrane and its organization into discrete mobile foci have been described previously (C.A. Konopka and S.Y. Bednarek, unpublished data). To determine the cortical localization and dynamics of DRP1A, seedling roots expressing a functional DRP1A-GFP fusion protein under the control of the DRP1A promoter (Kang et al., 2003a) were imaged using variable angle epifluorescence microscopy (VAEM; Konopka and Bednarek, 2008). In expanding root epidermal cells, DRP1A-GFP was not distributed uniformly throughout the cell cortex, but instead was organized into dynamic, discrete foci (Fig. 2A; Supplemental Video S1). DRP1A-GFP formed an average of 3.17 (±0.79) foci per micron at the cell cortex (Fig. 2A) with an average focus lifetime of 37.2 ± 20.0 s (Fig. 2, B–D).
Figure 2.
Analysis of DRP1A-GFP focus dynamics at the cell cortex. A, Epidermal root cell in the elongation zone expressing DRP1A-GFP imaged with VAEM. B, Image montage taken from time lapse sequence of the epidermal root cell shown in A. Numbers in top right corners indicate time elapsed from first image in seconds. The focus indicated by the open arrowhead changed position during its lifetime in the cell cortex (first position, yellow arrowhead; second position, blue arrowhead). The focus indicated by the solid red arrowhead did not change position. C, Intensity profiles of the foci indicated in B. The mobile focus (open arrowhead) is indicated by both yellow (first position) and blue (second position) lines. D, Lifetime distribution of DRP1A-GFP foci in cells from plants grown on one-half-strength Murashige and Skoog (1/2 MS) with no drug (black bars) and from plants grown in the presence of 10 μg/mL fenpropimorph (gray bars). E, Lifetime analysis of DRP1A-GFP from plants treated for 20 min with 10 μm oryzalin (white bars), 1 μm latB (striped bars), concurrently with 10 μm oryzalin + 1 μm latB (gray bars), or mock treated with 0.1% DMSO (black bars). Scale bars = 1 μm.
The DRP1A-GFP foci displayed various mobile behaviors both in the focal plane of the cell cortex and further within the cell. Approximately 50% of DRP1A foci (n = 175) were observed moving in the cytoplasm (not in the focal plane) for one to two frames prior to becoming immobile at the cell cortex. Approximately 23% of the foci moved within the imaging plane during their lifetime, establishing a new immobile position before disappearing from the cortex (Fig. 2, B and C, yellow and blue arrowheads). DRP1A-GFP foci movements in the imaging plane made it difficult to track some foci, so it is possible that the average foci lifetime was underestimated.
One striking difference between DRP1C-GFP and DRP1A-GFP (as well as between DRP1A-mOrange and DRP1C-mOrange, see below) was the rate of photobleaching. The estimated t0.5 of photobleaching was 20 min for DRP1C-GFP. In contrast, during the first 1.5 min of imaging DRP1A-GFP, the photobleaching t0.5 was approximately 2 min using identical optical parameters. After 1.5 min, the photobleaching rate decreased to that observed for DRP1C. The DRP1A-GFP foci that appeared in the cell cortex after 1.5 min of imaging exhibited a photobleaching t0.5 similar to DRP1C foci.
DRP1A-GFP Dynamics Are Perturbed upon Cytoskeleton, Sterol, and CME Disruption
DRP1C-GFP foci dynamics are disrupted with pharmacological inhibitors of membrane sterol composition (10 μg/mL fenpropimorph; He et al., 2003; Schrick et al., 2004), adaptin 2 (AP2)-dependent CME (50 μm typhostin A23 [tyrA23]; Crump et al., 1998; Banbury et al., 2003), microtubule dynamics (10 μm oryzalin; Baskin et al., 1994), and microtubule dynamics in conjunction with inhibition of actin dynamics (1 μm latrunculinB [latB]; Kandasamy and Meagher, 1999). To determine if DRP1A foci have a similar susceptibility to cytoskeletal, sterol, or endocytic traffic disruption, we analyzed DRP1A-GFP foci dynamics in seedlings treated with the pharmacological agents listed above.
DRP1A-GFP dynamics were not disrupted when seedlings were treated with oryzalin or latB alone under conditions that caused complete depolymerization of the microtubule or actin cytoskeleton, respectively (Fig. 2E). The average lifetime of DRP1A-GFP foci when treated with 0.1% dimethyl sulfoxide (DMSO; control) was 25.0 ± 10.4 s and increased to 28.3 ± 17.9 s upon microtubule depolymerization. In addition, the cortical lateral movements of DRP1A foci described above were unaffected after oryzalin treatment. Likewise, neither focus lifetime (31.0 ± 15.9 s) nor lateral movements within the cell cortex were significantly altered upon F-actin depolymerization (P > 0.001). When both cytoskeletal arrays were depolymerized, the average focus lifetime was nearly 1.5 times that of the control (37.0 ± 19.3 s), which was statistically significant (P < 0.001), but the percentage of laterally mobile foci was unchanged, indicating that the foci were not propelled by cytoskeletal associated forces.
The AP2 inhibitor tyrA23 causes rapid immobilization of DRP1C-GFP foci and concentration of DRP1C-GFP fluorescence in large unknown structures at the cell cortex and in the cytoplasm as previously shown (C.A. Konopka and S.Y. Bednarek, unpublished data). When seedlings expressing DRP1A-GFP were treated with tyrA23, the cytoplasmic pool of DRP1A-GFP fluorescence was unchanged and DRP1A-GFP foci did not increase in size or fluorescence intensity, like DRP1C foci. However, DRP1A-GFP foci became less dynamic at the cell cortex. After 30 min, 97% of foci did not cycle in or out of the cell cortex and an average lifetime could not be determined. As upon cytoskeletal inhibition with latB and oryzalin, the cortical lateral movements of DRP1A-GFP foci within the cell cortex were unaffected by tyrA23 when compared to DMSO-treated seedlings.
To assess the requirement for specific sterols in DRP1A-GFP dynamics, seedlings were grown on 10 μg/μL fenpropimorph, an inhibitor of the sterol biosynthetic pathway in plants. DRP1A-GFP foci in root epidermal cells from plants grown on fenpropimorph had a higher residence time than foci in seedlings grown on one-half-strength Murashige and Skoog (Fig. 2D; Supplemental Video S2). An average lifetime could not be determined, but 56% of foci analyzed remained at the cell cortex longer than 2 min. In addition, the foci did not display the characteristic movements in the cell cortex that occurred in approximately 20% of foci in untreated roots. Finally, the photobleaching that affected the DRP1A-GFP fluorophore when seedlings were grown on one-half-strength Murashige and Skoog was absent when seedlings were grown in the presence of fenpropimorph. Collectively, the response of DRP1A-GFP foci to various inhibitors differed from the response of DRP1C-GFP foci, suggesting that DRP1A was regulated differently than DRP1C at the cell cortex.
DRP1A and DRP1C Foci Colocalize in the Cell Cortex
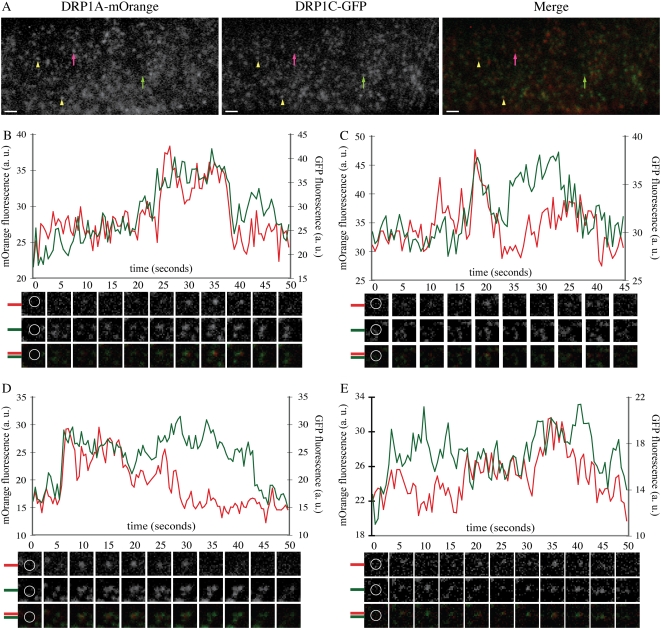
Previous studies have shown that DRP1C colocalizes with and resides on the same structures as CLC in the cell cortex (C.A. Konopka and S.Y. Bednarek, unpublished data). DRP1A-GFP and DRP1C-GFP both organize into foci with different behaviors and responses to various inhibitors. To determine whether DRP1A and DRP1C also colocalize in the cell cortex, an mOrange-tagged (Shaner et al., 2004) DRP1A cDNA fusion construct under the control of the DRP1A promoter was introduced into drp1A-2 plants. drp1A-2:DRP1A-mOrange plants did not exhibit the seedling lethality or fertility phenotypes associated with drp1A-2 (Kang et al., 2001, 2003a), indicating that the DRP1A-mOrange fusion protein was functional. DRP1A-mOrange was organized into foci in the cell cortex when imaged with VAEM (Fig. 3A), with similar dynamics to the DRP1A-GFP fusion protein (data not shown).
Figure 3.
DRP1A-mOrange foci colocalize with DRP1C-GFP foci in the cell cortex. A, Epidermal root cell in the expansion zone expressing DRP1A-mOrange and DRP1C-GFP imaged with VAEM with filter set for simultaneous GFP and mOrange fluorescence capture (see “Materials and Methods”). DRP1A-mOrange and DRP1C-GFP foci are present without the other DRP1 (DRP1A, pink arrow; DRP1C, green arrow) and also colocalize (yellow arrowheads). B to E, Intensity profiles of GFP (green) and mOrange (red) fluorescence from foci that had overlapping fluorescence of DRP1C-GFP and DRP1A-mOrange. Corresponding mOrange fluorescence (top), GFP fluorescence (middle), and merged (bottom) images are below each time (in seconds) indicated in the graph. Montage images are 1.2 × 1.2 μm. Yellow circles in the first frames indicate measured regions for fluorescence intensity. Bars = 1 μm.
To determine whether DRP1A-mOrange foci colocalized with clathrin and DRP1C at the cell cortex, drp1A-2:DRP1A-mOrange plants were crossed with drp1C-1:DRP1C-GFP and WS:CLC-GFP plants, and the F2 progeny used for analysis. Root epidermal cells from nine independent seedlings were imaged with dual color VAEM imaging (Fig. 3; Supplemental Video S3). Approximately 87% of DRP1A-mOrange foci overlapped with fluorescence from DRP1C-GFP foci. Conversely, 80% of DRP1C-GFP foci overlapped with fluorescence from DRP1A-mOrange foci (Fig. 3A). Overall, fluorescence from DRP1C-GFP and DRP1A-mOrange overlapped in 72% of foci imaged. To rule out the possibility that the fluorescence overlap was random due to the high density of both foci, the red channel images from six different cells were rotated 180 degrees with respect to the green channel, an analysis technique that has been used previously to show nonrandom colocalization (Delcroix et al., 2003; Dedek et al., 2006). The distance between each GFP focus intensity peak and the nearest mOrange focus intensity peak was calculated for the original images, and each respective mOrange rotated image. The average intensity peak distance was 4.12 pixels for the original images and 6.52 pixels for the rotated images, which was statistically different for each cell using the Student's t test (P < 0.00001), indicating that the high coincidence of DRP1A-mOrange and DRP1C-GFP foci colocalization was not random.
To examine cortical DRP1A and DRP1C dynamics, the fluorescence intensity profiles for DRP1C-GFP and DRP1A-mOrange were determined for foci in which both DRP1A and DRP1C were present. Examples of the intensity profiles are shown in Figure 3. Forty-seven percent of foci examined showed simultaneous disappearance of DRP1C-GFP and DRP1A-mOrange from the image plane (Fig. 3B), suggesting they were present on the same structure. Of these, the majority had a concurrent increase in fluorescence of both fluorophores (33% of all foci). Other foci had an initial mOrange fluorescence (3%) or GFP fluorescence (5%) increase. Another population of foci (6%) maintained a constant fluorescence of one DRP1-FFP throughout their lifetime, while fluorescence of the other DRP1-FFP fluctuated (Fig. 3C). In contrast, a majority of all foci examined did not exhibit simultaneous disappearance of both fluorophores. These either had coordinated appearance of both fluorophores (28%; Fig. 3D) or no coordination of their entrance or departure (26%; Fig. 3E). Like DRP1A-GFP, a small fraction of DRP1A-mOrange foci were also mobile in the cell cortex between periods of immobility. This population of DRP1A-mOrange foci associated with DRP1C-GFP foci when immobile, but rarely while in transit. From these colocalization and dynamics analyses, it appears that DRP1A and DRP1C can exist on the same structures, but also may function independently at the cell cortex.
DRP1A and CLC Foci Colocalize in the Cell Cortex
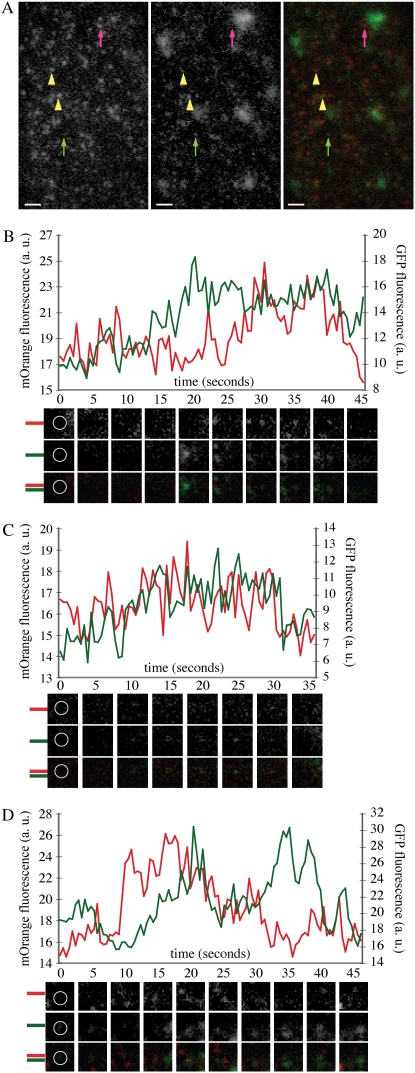
DRP1C and CLC colocalize in the cell cortex where they have coordinated dynamics (C.A. Konopka and S.Y. Bednarek, unpublished data). Based on the colocalization of DRP1A and DRP1C foci, it is expected that the foci formed by DRP1A and CLC would colocalize. Indeed, this was the case (Fig. 4A; Supplemental Video S4). A total of 80.3% of DRP1A-mOrange foci had overlapping CLC-GFP fluorescence during their lifetime (n = 600). Conversely, 72.8% of CLC-GFP foci had overlapping fluorescence from DRP1A-mOrange foci during their lifetime. In total, 60.8% of foci imaged in root epidermal cells expressing DRP1A-mOrange and CLC-GFP contained both fluorescent fusion proteins. Intensity profiles for 36 foci that contained both DRP1A-mOrange and CLC-GFP fluorescence were analyzed. Like with colocalizing DRP1A and DRP1C foci, fewer than half (39%) of the foci had simultaneous disappearance of both DRP1A-mOrange and CLC-GFP (Fig. 4B). Only 14% had simultaneous recruitment and disappearance of the DRP1-FFPs (Fig. 4C). Forty-four percent of foci displayed uncoordinated dynamics of the fluorescence intensity (Fig. 4D). In summary, a majority of DRP1A foci colocalized with DRP1C and CLC structures, but had distinct dynamics from DRP1C and CLC. This suggested that DRP1A could associate with the clathrin machinery, but may also act independently from clathrin-coated structures at the cell cortex.
Figure 4.
DRP1A-mOrange foci colocalize with CLC-GFP foci in the cell cortex. A, Epidermal root cell in the expansion zone expressing DRP1A-mOrange and CLC-GFP imaged with VAEM with filter set for simultaneous GFP and mOrange fluorescence capture (see “Materials and Methods”). DRP1A-mOrange and CLC-GFP foci are present independently (DRP1A, pink arrow; CLC, green arrow) and also colocalize (yellow arrowheads). B to D, Intensity profiles of GFP (green) and mOrange (red) fluorescence from foci that had overlapping fluorescence of CLC-GFP and DRP1A-mOrange. Corresponding mOrange fluorescence (top), GFP fluorescence (middle), and merged (bottom) images are below each time (in seconds) indicated in the graph. Montage images are 1.2 × 1.2 μm. White circles in the first frames indicate measured regions for fluorescence intensity. Bars = 1 μm.
DRP1C Can Functionally Compensate for DRP1A during Seedling Development
DRP1A and DRP1C have different dynamics at the cell cortex in expanding root epidermal cells, suggesting that the two DRP1 isoforms may have distinct roles. To determine if DRP1C and DRP1A are functionally redundant, we examined if expression of DRP1C under the control of the DRP1A promoter or constitutive expression using the viral promoter cauliflower mosaic virus 35S (35S) could complement the various phenotypes observed in drp1A-2 mutants. drp1A-2 plants are characterized by: (1) seedling lethality on soft agar (0.6% phytagar) plates, which can be rescued by supplementation with 1% Suc (Kang et al., 2001); (2) infertility due to the inability of their stigmatic papillae to undergo rapid polar expansion prior to fertilization (Kang et al., 2003a); (3) trichome branching defects (Kang et al., 2003a); and (4) defects in venation continuity in cotyledons (Sawa et al., 2005). The seedling lethality and infertility phenotypes were assayed in the complementation analysis.
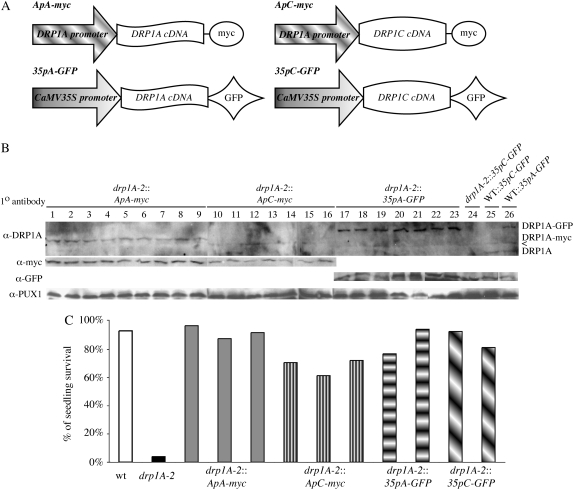
drp1A-2 plants expressing the following constructs were generated: DRP1A promoter:DRP1A cDNA C-terminal myc fusion protein (ApA-myc), DRP1A promoter:DRP1C cDNA C-terminal myc fusion protein (ApC-myc), 35S promoter:DRP1A cDNA C-terminal GFP fusion (35pA-GFP), or 35S promoter:DRP1C cDNA C-terminal GFP fusion (35pC-GFP; Fig. 5A). Protein expression of the transgenes were comparable across all lines as verified using DRP1A-specific antibodies (Kang et al., 2001), either anti-myc (Evan et al., 1985) or anti-GFP antibodies, and anti-Pux1 as a loading control (Rancour et al., 2004; Fig. 5B).
Figure 5.
Exogenous expression of DRP1C rescues drp1A-2 seedling lethality. A, Schematic of the four constructs used for complementation analysis. B, Immunoblot of total protein extracts from drp1A-2 (lanes 1–24) or wild-type (lanes 25 and 26) seedlings expressing ApA-myc (lanes 1–9), ApC-myc (lanes 10–16), 35pA-GFP (lanes 17–23, 26), or 35pC-GFP (lane 24). The top blot was blotted with anti-DRP1A specific antibodies, the middle blot with anti-myc (left) or anti-GFP (right) antibodies, and the bottom blot with anti-PUX1 antibodies (loading control). All lines express the transgene approximately equally well. The bands in the anti-DRP1A blot were DRP1A-GFP (top), DRP1A-myc (middle), and native, untagged DRP1A (bottom). A cross-reactive band is indicated by <. C, Histogram indicating the percentage of seedlings of individual lines (with the genotype indicated) that survived and developed a second set of true leaves on one-half-strength Murashige and Skoog, 0.6% agar without Suc.
Nine (ApA-myc and ApC-myc), seven (35pA-GFP), and four (35pC-GFP) independent lines were evaluated for growth on one-half-strength Murashige and Skoog + 0.6% phytagar without Suc. Wild-type plants expressing any of the constructs did not display any morphological or developmental defects (data not shown). A total of 93.3% of wild-type seedlings and 4% of drp1A-2 seedlings produced at least one pair of true leaves and survived when transferred to soil. The survival rate of seedlings from three representative lines for ApA-myc and ApC-myc and two representative lines for 35pA-GFP and 35pC-GFP is shown in Figure 5C. A total of 88.7% ± 6.8% and 59% ± 18.1% of drp1A-2 seedlings expressing ApA-myc or ApC-myc, respectively, developed normally without Suc. Although the lower survival rate of drp1A-2:ApC-myc plants was statistically significant versus the control, drp1A-2:ApA-myc, the drp1A-2:35pC-GFP lines did not have significantly lower survival rates than the control drp1A-2:35pA-GFP lines (80.3% ± 27.2% for 35p1C-GFP versus 83.5% ± 7.9% for 35pA-GFP). These data indicate that DRP1C can functionally compensate for the lack of DRP1A during seedling development, suggesting at least a partial functional redundancy of DRP1C with DRP1A.
DRP1C Cannot Functionally Compensate for DRP1A during Stigmatic Papillae Expansion
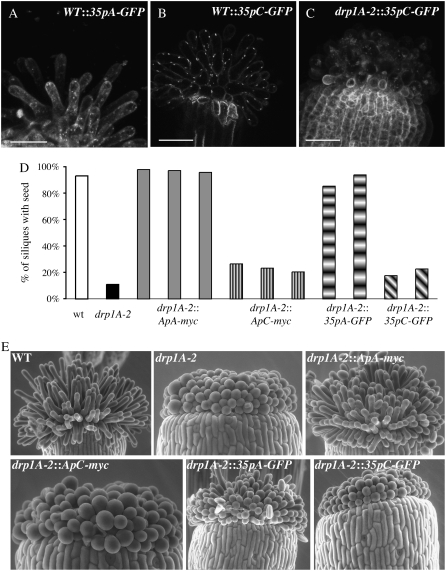
The same transgenic lines expressing ApA-myc, ApC-myc, 35pA-GFP, or 35pC-GFP described above were evaluated for fertility and stigmatic papillae expansion. drp1A-2 homozygous plants have reduced fertility, most likely due to the failure of the stigmatic papillae to expand just prior to pollination (Kang et al., 2003a). The expression of the 35pA-GFP or 35pC-GFP in papillae was confirmed with confocal microscopy (Fig. 6, A–C). Siliques were collected from four to six plants from wild type, drp1A-2, and each of the transgenic lines in the drp1A-2 background, to determine if seeds were present. In drp1A-2 homozygous plants, fewer than 12% of siliques contained at least one seed, whereas over 93% of siliques from wild-type plants contained seeds (Fig. 6D). Expression of the transgenes in wild-type plants did not affect fertility (data not shown). Unlike the seedling lethality phenotype, neither ApC-myc nor 35pC-GFP was able to rescue the fertility defect of drp1A-2. drp1A-2 plants expressing ApC-myc or 35pC-GFP averaged between 18% and 36% of siliques containing at least one seed, whereas drp1A-2 plants expressing ApA-myc or 35pA-GFP averaged 85% to 98% of seed-containing siliques (Fig. 6D).
Figure 6.
Expression of DRP1C cannot rescue the expansion defect of drp1A-2 stigmatic papillae. A to C, Maximal Z projections (A and C) or a single confocal section (B) of stigmatic papillae from first stage 13 flower from plants with genotypes indicated. C, Histogram indicating the percent of siliques in individual lines with the genotype indicated that had at least one seed. Siliques from four to six plants from each line were evaluated for seed content. D, Environmental scanning electron microscopy images of stigmatic papillae from the first open flower from plants with the genotypes indicated. Bars = 50 μm.
To confirm that the reduced fertility of drp1A-2:ApC-myc and drp1A-2:35pC-GFP plants was due to abnormal papillae expansion as in the drp1A-2 mutant, flowers from untransformed wild-type, drp1A-2, drp1A-2:ApA-myc, drp1A-2:ApC-myc, drp1A-2:35pA-GFP, and drp1A-2:35pC-GFP plants were imaged by environmental scanning electron microscopy (Fig. 6E). Stage 13 or 14 flowers were chosen to ensure that dehiscence of the pollen and papillae expansion had occurred. Papillae from wild-type flowers were elongated and flask shaped, whereas papillae from drp1A-2 flowers were small and balloon shaped, as previously described (Kang et al., 2003a). Twenty-five out of 26 stigmas from drp1A-2 plants expressing either ApA-myc or 35pA-GFP had elongated papillae (Fig. 6E). In contrast, one out of 31 stigmas from drp1A-2 plants expressing ApC-myc or 35pC-GFP plants had elongated stigmatic papillae, indicating that exogenous expression of DRP1C could not rescue the papillae expansion defect.
DISCUSSION
The plant-specific DRP1 family is essential for cytokinesis, venation, trichome development, and cell expansion (Kang et al., 2001, 2003a, 2003b; Sawa et al., 2005). Specifically, two isoforms, DRP1A and DRP1C, have been shown to be required for proper plasma membrane maintenance in expanding stigmatic papillae and pollen development, respectively (Kang et al., 2003a, 2003b). In addition, both proteins localize to the cell plate during cell division, but only DRP1A has been shown to be required for cytokinesis (Kang et al., 2003a). Here we have compared DRP1A and DRP1C, which exhibit 66% amino acid sequence identity, using genetic complementation analysis and live cell imaging at the cell surface. These studies suggest that although both DRP1A and DRP1C may be components of the CME machinery, they have different dynamics at the cell cortex, and distinct roles during cell expansion. The critical role for CME in plants has recently been demonstrated for the internalization of the auxin efflux carrier, PIN1, and several other cargos. Uptake of these factors was blocked by the AP2 inhibitor tyrA23 and by the expression of expression of a dominant negative CHC (Dhonukshe et al., 2007). The endocytic pathway for the hormone receptor BRI1 (Russinova et al., 2004), the boron transporter BOR1 (Takano et al., 2005), the plant defense receptor FLS2 (Robatzek et al., 2006), and the plant steroid receptor kinase BRI1 (Geldner et al., 2007) have not been identified.
DRP1A-GFP and DRP1C-GFP Dynamics Differ at the Cell Cortex
A common feature of dynamin and DRPs is their ability to oligomerize around lipid bilayers and deform membranes (Praefcke and McMahon, 2004). DRP1C and mammalian dynamin 1 form foci at the plasma membrane where they colocalize with CLC and exhibit similar dynamics (Merrifield et al., 2002; C.A. Konopka and S.Y. Bednarek, unpublished data). Dynamin 1 polymerizes around the necks of clathrin-coated structures (Damke et al., 1994), and although it may act similarly, the biochemical role of DRP1C at clathrin-coated structures has not been determined. Likewise, DRP1A-FFP forms foci at the plasma membrane of root epidermal cells; however, DRP1A dynamics differ from those of DRP1C. First, DRP1A foci had a greater average lifetime and a wider lifetime distribution than DRP1C (Fig. 2D). Second, DRP1A displayed different mobilities within the cell cortex than DRP1C (Fig. 3B; C.A. Konopka and S.Y. Bednarek, unpublished data). Third, the response of DRP1A-GFP foci to cytoskeletal inhibitors differed from that of DRP1C-GFP foci (Fig. 2E; C.A. Konopka and S.Y. Bednarek, unpublished data). Fourth, DRP1A-GFP had a greater dependence on plasma membrane sterol composition for its dynamics. Finally, the rate of photobleaching of GFP and mOrange fused to DRP1A and DRP1C differed, suggesting the immediate environment of the DRP1A- and DRP1C-tagged fluorophores was not equal (Murphy, 2001).
Despite these differences, DRP1C-GFP and DRP1A-GFP foci had a high coincidence of overlap, indicating that at least a subset of DRP1A functioned in the same pathway as DRP1C. The percentage of colocalization and coincidental dynamics of DRP1A with either DRP1C or CLC was lower than that of DRP1C with CLC (Figs. 3 and 4; C.A. Konopka and S.Y. Bednarek, unpublished data). This suggests that DRP1A, DRP1C, and CLC are part of the same clathrin machinery in the approximately 25% of foci that exhibited coordinated dynamics of the three proteins, whereas in the other 75% of foci, DRP1A was acting independently of DRP1C and CLC. A putative role for DRP1A or DRP1C in CME correlates with the phenotype of drp1A-2 and drp1C-1 mutants. Defects in dynamin-dependent endocytic pathways have been previously shown to cause large plasma membrane invaginations in flies (Kessell et al., 1989) and mice (Ferguson et al., 2007). Whether the DRP1 and CLC proteins facilitate endocytosis at the plasma membrane in expanding and nonexpanding plant cells remains to be clarified.
DRP1A Formed Distinct Populations at the Cell Cortex
Distinct populations of cortical-associated DRP1A were observed. As described above, one population of DRP1A foci exhibited similar dynamics to DRP1C. However, 35% of DRP1A-GFP foci have longer lifetimes than 48 s, which is the longest recorded lifetime of DRP1C foci in untreated cells (Fig. 2; C.A. Konopka and S.Y. Bednarek, unpublished data). In addition, 25% of DRP1A-GFP foci move laterally within the cell cortex at least once before disappearing entirely, which was not observed with DRP1C. It is not clear whether or not these two populations (longer lifetime and mobile) represent the same population, because the difference in residence time between mobile and nonmobile populations was not significant.
A majority of DRP1A-FFP foci that were present at the start of imaging were photobleached within the first 2 min. Subsequently, new foci appeared in the cell cortex after 2 min that did not photobleach. It is possible that the DRP1A population that was not vulnerable to photobleaching was localized in a chemically distinct environment. Interestingly, this rapid photobleaching was not apparent when plants were grown in the presence of the sterol synthesis inhibitor, fenpropimorph. Plants grown on fenpropimorph have a modified sterol profile (Schrick et al., 2004). The major plant sterols stigmasterol, sitosterol, and campesterol are virtually absent and are replaced with biosynthetic pathway intermediates (Schrick et al., 2004). One possibility is that the photobleaching-sensitive population was either absent when grown in the presence of fenpropimorph or the entire population of DRP1A-GFP was protected from photobleaching by a different sterol environment.
Sterols Are Required for DRP1A Dynamics
Disrupting sterol synthesis in plants causes defects in cytokinesis, cell expansion, cell polarity, and cell wall formation (He et al., 2003; Schrick et al., 2004), all processes that require membrane trafficking for proper maintenance. Plant sterols are required for polar localization of auxin efflux carriers, a major determinant of plant polarity (Willemsen et al., 2003), and are a component of endocytic vesicles that accumulate in ARA6-positive endosomes (Grebe et al., 2003). In addition to the change in photobleaching rate of DRP1A-GFP upon sterol disruption, fenpropimorph also caused a 5-fold increase in foci residence time and reduced mobility in the cell cortex. Neither DRP1C nor DRP1A have a canonical lipid-binding domain, thus it is not clear how sterol composition affected DRP1A dynamics. By homology to mammalian dynamin 1, the variable domain (15–24 least identical in amino acid sequence among DRP1s) may interact with membrane components when DRP1A and DRP1C oligomerize. It is plausible that these amino acids are important for conferring the different susceptibility of DRP1A and DRP1C to changes in plasma membrane sterol composition upon treatment with fenpropimorph. Live cell imaging of GFP-tagged DRP1 mutants altered in these residues may be key to understanding DRP1A's dependence on sterols.
DRP1A and DRP1C Functional Redundancy
The gametophytic lethality of the drp1C-1 mutant has prohibited the generation of double or triple DRP1 mutants as a means to determine functional redundancy with DRP1C. To bypass this, we expressed DRP1A and DRP1C under the control of the native DRP1A promoter or constitutive 35S promoter and assayed their ability to complement drp1A-2 phenotypes. The drp1A-2 mutant has a well-characterized defect in papillae expansion (Kang et al., 2003a) as well as defects in seedling development in the absence of Suc (Kang et al., 2001). The plasma membrane of the defective papillae is highly elaborated and undulated, which is similar to the plasma membrane observed in drp1C-1 mutant pollen that fail to germinate (Kang et al., 2003b). The resemblance of the plasma membrane defects suggests that DRP1A and DRP1C have a common function in the different cell types. However, DRP1C failed to complement the papillae expansion defect (Fig. 6, D and E) when expressed in these cells (Fig. 6, A–C), indicating that DRP1C functions differently in stigmatic papillae. The inability of DRP1C to complement drp1A-2 could be due to a lack of DRP1C recruitment to the proper plasma membrane domain or a lack of activation or inactivation of DRP1C by DRP1A-specific regulators. In contrast to the papillae defect, exogenously expressed DRP1C was able to bypass the need for Suc in drp1A-2 seedlings (Fig. 5B). Overall, complementation revealed that DRP1C can compensate for the absence of DRP1A during seedling growth, but that an element of specificity exists in the expanding papillae cells.
CONCLUSION
Live cell imaging and genetic complementation have demonstrated that although exogenously expressed DRP1C can compensate for the absence of DRP1A during seedling development, the DRP1 isoforms are not completely functionally redundant and display distinct dynamics. Whether these dissimilarities in dynamics account for the inability of DRP1C to complement the stigmatic papillae expansion defect of drp1A-2 mutants still needs to be elucidated. Further research will help determine the molecular and biochemical bases of the differences in DRP1A and DRP1C dynamics. Live cell imaging and genetic complementation have demonstrated that although exogenously expressed DRP1C can compensate for the absence of DRP1A during seedling development, the DRP1 isoforms are not completely functionally redundant and display distinct dynamics. Whether these dissimilarities in dynamics account for the inability of DRP1C to complement the stigmatic papillae expansion defect of drp1A-2 mutants remains to be elucidated. These differences may represent specificity in the endocytic pathways of various cargos. Further research will help determine the molecular and biochemical bases of the differences in DRP1A and DRP1C dynamics and their role in CME.
MATERIALS AND METHODS
Identification and Phylogeny of Rice and Medicago DRPs
AtDRP1A and human dynamin 1 amino acid sequences were used as queries in BlastP searches of the published rice (Oryza sativa) and Medicago truncatula sequences to identify DRPs in these organisms. The putative DRPs were verified as belonging to the dynamin superfamily if they contained the Dynamin GTPase domain in the SMART database (http://smart.embl-heidelberg.de). The amino acid sequences were aligned using the ClustalW method. The phylogenetic tree was created using MegAlign in the DNASTAR Lasergene software suite.
Plant Transformation Vector Construction
For live cell imaging of DRP1A dynamics: The coding sequence for mOrange (Shaner et al., 2004) was PCR amplified from pRSET-B mOrange (a gift from R. Tsien, University of San Diego) using primers 5′-GGATCCGATGGTGAGCAAGGGCGAG-3′ and 5′-GGTACCTTACTTGTACAGCTCGTCCATG-3′, subcloned into pPZP221 (Hajdukiewicz et al., 1994) containing the nopaline synthase (NOS) terminator from pBL121 (CLONTECH) using BamHI and KpnI sites, resulting in pPZP221-mOrange-NOS. A DRP1A promoter and cDNA fusion construct (Kang et al., 2003a) was subcloned into pPZP221-mOrange-NOS as a HindIII/XhoI fragment to generate the DRP1A-mOrange translational fusion expression vector.
For drp1A-2 complementation analysis: 2.0 kb upstream sequence of DRP1A promoter (Kang et al., 2003) was subcloned into pPZP211B containing the NOS terminator (Kang et al., 2001) to generate pPZP211B-Ap. Coding sequence for the c-myc tag (five tandem myc epitopes) was amplified from pJR1265 (Ziman et al., 1996) using primers 5′-GAGCTCATGGAGCAAAAGCTCATTTC-3′ and 5′-GAGCTCTCACAAGTCCTCTTCAGAAATGAGC-3′ and subcloned into pPZP211B-Ap using SacI sites. Full-length DRP1A and DRP1C cDNAs were amplified from total RNA isolated from 7-d-old Arabidopsis (Arabidopsis thaliana) seedlings as described (Kang et al., 2001) using primers 5′-CCGGGATATCGGAAAATCTGATCTCTCTG-3′ and 5′-GATATCAACTTGGACCAAGCAACAGCATCG-3′ (DRP1A) or 5′-TAGTCCCGGGTAAAAGTTTGATAGGTCTG-3′ and 5′-GAGCTCCTTCCAGCCACTGAATCGATG-3′ (DRP1C) and subcloned into pPZP221B-Ap-myc as an EcoRV/SacI fragment (DRP1A) or an XmaI/SacI fragment (DRP1C) to generate the constructs ApA-myc and ApC-myc, respectively. DRP1A and DRP1C cDNAs were also amplified using primers 5′-CTCGAGATGGAAAATCTGATCTCTCTGGTTAC-3′ and 5′-AAGCTTCTTGGACCAAGCAACAGCATCG-3′ (DRP1A) or 5′-CTCGAGGATGAAAGTTGATAGGTCTGATAAAC-3′ and 5′-GAATTCGCTTCCAAGCCACTGCATCGATGTCG-3′ (DRP1C) and subcloned into pEZT-NL (D. Ehrhardt, Carnegie Institution of Washington) downstream of the cauliflower mosaic virus 35S promoter (35S) as an XhoI/HindIII fragment (DRP1A) or an XhoI/EcoRI fragment (DRP1C) to generate 35pA-GFP and 35pC-GFP, respectively.
DRP1A/drp1A-2 plants (ecotype Wassilewskija) were transformed with the constructs encoding DRP1A-mOrange, ApA-myc, ApC-myc, 35pA-GFP, or 35pC-GFP using the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Transgenic plants were selected either on solid medium (0.6% phytagar, one-half-strength Murashige and Skoog; Murashige and Skoog, 1962; Caisson Labs) containing 75 μg/mL gentamycin sulfate (Amresco Inc.; DRP1A-mOrange) or on soil, sprayed once with 20 μg/mL ammonium glusofinate (Liberty; ApA-myc, ApC-myc, 35pA-GFP, and 35pC-GFP).
Plant Growth Conditions
For visualization of epidermal cells, conditions were as reported (Konopka and Bednarek, 2008). For analysis of drp1A-2 complementation seedlings were grown horizontally on one-half-strength Murashige and Skoog, 0.6% phytagar ± 1% Suc. Seedlings that did not grow in the absence of Suc within 10 d were transferred under sterile conditions to media containing 1% Suc and allowed to grow for another 10 d before transfer to soil.
VAEM
DRP1A-FFP and DRP1C-GFP foci dynamics were captured using VAEM as described (Konopka and Bednarek, 2008). Briefly, seedlings were transferred from vertically growing plates to a glass slide with 150 μL of one-half-strength Murashige and Skoog and covered with a coverslip. Plants were imaged with a Nikon Eclipse TE2000-U fitted with the Nikon T-FL-TIRF attachment, Nikon 100×/N.A. 1.45 CFI Plan Apo TIRF objective, and 1.5× intermediate magnification. For single fluorophore imaging, GFP was excited with 488-nm argon laser and filtered through a 535/30 filter (Chroma Technology). For double fluorophore imaging, GFP and mOrange were excited with 488 and 543 nm laser, respectively, and the emission spectra were separated with a 540LP dichroic mirror and filtered through a 515/30 (GFP) or 585/65 (mOrange) filter in a Dual View filter system (Photometrics). The incident angle was varied to give the highest signal-to-noise ratio. Fluorescence emission light was captured using a CoolSnapES cooled CCD camera (Roper Scientific) using Metamorph Imaging system version 6.2r6 (Molecular Devices) with 500-ms exposure times.
Inhibitor Studies
TyrA23 and latB were purchased from EMD Biosciences, oryzalin was purchased from Restek, and fenpropimorph was purchased from Sigma-Aldrich. Fenpropimorph was dissolved in water and all other inhibitors were dissolved in 100% DMSO for stock solutions. Inhibitors were diluted in one-half-strength Murashige and Skoog for VAEM imaging of root epidermal cells. The [DMSO] was 0.1% or less in all working solutions. Five- to 7-d-old vertically grown seedlings were transferred from 1% agar plates to a well of a 12-well culture plate containing 4 mL of final working concentration in one-half-strength Murashige and Skoog. After the indicated time, seedlings were transferred to a glass slide with 150 μL of inhibitor solution, covered with a glass coverslip, the excess liquid wicked away and imaged as above. For fenpropimorph studies, seedlings were grown vertically on one-half-strength Murashige and Skoog, 1% agar plates with or without 10 μg/mL fenpropimorph for 10 to 12 d prior to imaging in one-half-strength Murashige and Skoog media.
Immunoblot Analysis
To determine expression level of DRP1A-myc, DRP1C-myc, DRP1A-GFP, and DRP1C-GFP, total protein extracts were prepared from drp1A-2:ApA-myc, drp1A-2:ApC-myc, drp1A-2:35pA-GFP, drp1A-2:35pC-GFP, WT:35pA-GFP, and WT:35pC-GFP (WT, wild type) seedlings grown horizontally on one-half-strength Murashige and Skoog + 0.6% phytagar without Suc for 10 d. Seedlings with at least two pairs of leaves were ground in 15 μL of SDS-PAGE sample buffer (Laemmli, 1970) per seedling and incubated at 65°C for 15 min. Insoluble debris was cleared by centrifugation at 16,000g for 10 min at room temperature. Fifteen microliters of supernatant was separated on a 12.5% (w/v) SDS-PAGE and analyzed by immunoblotting as described (Kang et al., 2001) using anti-DRP1A (Kang et al., 2001), anti-myc (Evan et al., 1985), biotin-conjugated anti-GFP (Rockland Immunochemicals Inc.), and anti-Pux1 (Rancour et al., 2004) antibodies. HRP-conjugated anti-rabbit secondary antibodies (GE Healthcare) and HRP-conjugated streptavidin (Rockland Immunochemicals Inc.) were used to detect the primary antibodies anti-DRP1A, anti-myc, or anti-Pux1 and anti-GFP, respectively.
Environmental Scanning Electron Microscopy
Stage 13 flowers from wild-type plants and drp1A-2 plants expressing no transgene, ApA-myc, ApC-myc, 35SpA-GFP, or 35SpC-GFP were excised and imaged using a Quanta 200 environmental scanning electron microscope (FEI) at 3.78 Torr and 4°C using a 20.0-kV electron beam. Electron emission was detected with the gaseous secondary electron detector.
Confocal Microscopy
To image DRP1A-GFP and DRP1C-GFP in papillar cells, stage 13 flowers from plants expressing 35SpA-GFP and 35SpC-GFP in wild-type or drp1A-2 background were excised, flattened, placed on a glass slide with one-half-strength Murashige and Skoog media, and covered with a coverslip. Papillae were imaged using a Nikon TE2000-U inverted laser scanning confocal microscope (Nikon Instruments Inc.) fitted with a 60× (numerical aperture 1.4) PlanApo VC objective lens and excited with 488 nm light (Melles Griot). Z stacks were captured using the EZ-C1 software (Nikon Corporation) and images were recombined using the maximum projection command in Image J (National Institutes of Health).
Image Analysis
A focus was defined as a local increase in intensity above a designated threshold assigned to each time lapse image and that was present for at least 2 s. The intensity profiles in Figures 2 to 4 were generated using Image J's ROI Multi Measure Plugin. Circular ROIs with a 6-pixel diameter that included all pixels of the focus was created and a mean intensity for the ROI was recorded. All images for figures were processed in Adobe Photoshop CS2 (Adobe Systems). All statistical figures are listed as averages ± sd.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Video S1. DRP1A-GFP forms dynamic foci at the cell cortex.
Supplemental Video S2. DRP1A-GFP focus dynamics are disrupted upon sterol synthesis inhibition.
Supplemental Video S3. DRP1A and DRP1C colocalize but display different dynamics at the cell cortex.
Supplemental Video S4. DRP1A and CLC colocalize at the cell cortex.
Supplementary Material
Acknowledgments
We thank T. Martin and members of his lab for help and extensive use of their epifluorescence-TIRF microscope. Scanning electron microscopy was performed at the Plant Imaging Facility at the University of Wisconsin, Madison. We thank members of our lab, especially S. Backues, D. Rancour, S. Park, and C. McMichael, for critical reading of the manuscript and helpful discussions.
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (project no. 2004–03411 to S.Y.B.), a Howard Hughes Medical Institute Predoctoral Fellowship (to C.A.K.), a National Institutes of Health National Research Service Award (award no. T32 GM07215 to C.A.K.) from the National Institute of General Medical Sciences, and the National Science Foundation (grant no. DBI–0421266).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sebastian Y. Bednarek (sybednar@wisc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Banbury DN, Oakley JD, Sessions RB, Banting G (2003) Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J Biol Chem 278 12022–12028 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE (1994) Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol 35 935–942 [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL (2003) Regulated portals of entry into the cell. Nature 422 37–44 [DOI] [PubMed] [Google Scholar]
- Crump CM, Williams JL, Stephens DJ, Banting G (1998) Inhibition of the interaction between tyrosine-based motifs and the medium chain subunit of the AP-2 adaptor complex by specific tyrphostins. J Biol Chem 273 28073–28077 [DOI] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL (1994) Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol 127 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedek K, Schultz K, Pieper M, Dirks P, Maxeiner S, Willecke K, Weiler R, Janssen-Bienhold U (2006) Localization of heterotypic gap junctions composed of connexin45 and connexin36 in the rod pathway of the mouse retina. Eur J Neurosci 24 1675–1686 [DOI] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC (2003) NGF signaling in sensory neurons evidence that early endosomes carry NGF retrograde signals. Neuron 39 69–84 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J (2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17 520–527 [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 5 3610–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, Collesi C, Giovedi S, Raimondi A, Gong LW, Ariel P, Paradise S, et al (2007) A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science 316 570–574 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M, Xu J, Mobius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol 13 1378–1387 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- He JX, Fujioka S, Li TC, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang JC (2003) Sterols regulate development and gene expression in Arabidopsis. Plant Physiol 131 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Bednarek SY, Blumwald E, Hwang I, Jurgens G, Menzel D, Osteryoung KW, Raikhel NV, Shinozaki K, Tsutsumi N, et al (2003. a) A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol Biol 53 261–265 [DOI] [PubMed] [Google Scholar]
- Hong Z, Geisler-Lee CJ, Zhang Z, Verma DP (2003. b) Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol Biol 53 297–312 [DOI] [PubMed] [Google Scholar]
- Kandasamy M, Nasrallah J, Mikhail N (1990) Pollen-pistil interaction and developmental regulation of pollen tube growth in Arabidopsis. Development 120 3405–3418 [Google Scholar]
- Kandasamy MK, Meagher RB (1999) Actin-organelle interaction: association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton 44 110–118 [DOI] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY (2003. a) Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY (2001) The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol 126 47–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Rancour DM, Bednarek SY (2003. b) The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J 35 1–15 [DOI] [PubMed] [Google Scholar]
- Kessell I, Holst BD, Roth TF (1989) Membranous intermediates in endocytosis are labile, as shown in a temperature-sensitive mutant. Proc Natl Acad Sci USA 86 4968–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY (2008) Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J 53 186–196 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W (2002) Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol 4 691–698 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Murphy DB (2001) Fundamentals of Light Microscopy and Electronic Imaging, Ed 1. Wiley-Liss, New York
- Praefcke GJ, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5 133–147 [DOI] [PubMed] [Google Scholar]
- Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403 567–571 [DOI] [PubMed] [Google Scholar]
- Rancour DM, Park S, Knight SD, Bednarek SY (2004) Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of arabidopsis CDC48. J Biol Chem 279 54264–54274 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P (2006) GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 441 528–531 [DOI] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Koizumi K, Naramoto S, Demura T, Ueda T, Nakano A, Fukuda H (2005) DRP1A is responsible for vascular continuity synergistically working with VAN3 in Arabidopsis. Plant Physiol 138 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Fujioka S, Takatsuto S, Stierhof YD, Stransky H, Yoshida S, Jurgens G (2004) A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J 38 227–243 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22 1567–1572 [DOI] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan L, von Wiren N, Fujiwara T (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT (1999) Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol 9 257–260 [DOI] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Hinshaw JE (2001) Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol 3 922–926 [DOI] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Schekman RW (1996) Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol Biol Cell 7 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.