Abstract
The circadian RNA-binding protein CHLAMY1 from the green alga Chlamydomonas reinhardtii consists of two subunits named C1 and C3. Changes in the C1 level cause arrhythmicity of the phototaxis rhythm, while alterations in the level of C3 lead to acrophase shifts. Thus, CHLAMY1 is involved in maintaining period and phase of the circadian clock. Here, we analyzed the roles of the two subunits in the integration of temperature information, the basis for other key properties of circadian clocks, including entrainment by temperature cycles and temperature compensation. Applied temperatures (18°C and 28°C) were in the physiological range of C. reinhardtii. While C1 is hyperphosphorylated at low temperature, the C3 expression level is up-regulated at 18°C. An inhibitor experiment showed that this up-regulation occurs at the transcriptional level. Promoter analysis studies along with single promoter element mutations revealed that individual replacement of two DREB1A-boxes lowered the amplitude of c3 up-regulation at 18°C, while replacement of an E-box abolished it completely. Replacement of the E-box also caused arrhythmicity of circadian-controlled c3 expression. Thus, the E-box has a dual function for temperature-dependent up-regulation of c3 as well as for its circadian expression. We also found that the temperature-dependent regulation of C1 and C3 as well as temperature entrainment are altered in the clock mutant per1, indicating that a temperature-controlled network of C1, C3, and PER1 exists.
Circadian rhythms are biological rhythms that persist with a period of about 24 h under constant conditions of light and temperature. The physiological properties of circadian rhythms are well conserved among different organisms, including prokaryotes (Kondo et al., 1993). They can be entrained by light or temperature cycles. Single pulses of light, darkness, or chemicals can shift their phase. Depending on the circadian time the pulse is given, phase advances or delays can occur. Under constant conditions, their period length is almost unchanged at different temperatures (Q10 [increase in the rate of a process produced by raising temperature by 10°C] ≈ 0.8–1.3), a clock property named temperature compensation (Rensing and Ruoff, 2002).
For control of entrainment by temperature cycles and for temperature compensation, certain components of the circadian clock should be able to integrate temperature. A few key components of the circadian clock have been shown to be involved in these processes. For example, in Neurospora crassa, it was shown that FREQUENCY is thermally regulated at the translational level and in addition involves thermosensitive splicing (Liu et al., 1997; Diernfellner et al., 2005; Dunlap, 2006). Moreover, the PAS/LOV protein VIVID controls temperature compensation of the circadian clock phase in N. crassa (Hunt et al., 2007). Also, in Arabidopsis (Arabidopsis thaliana), some key clock components have been shown to be involved in temperature sensing (Edwards et al., 2005; Gould et al., 2006).
In the green alga Chlamydomonas reinhardtii, several processes are under the control of the circadian clock, including phototaxis, chemotaxis, stickiness to glass surfaces, UV sensitivity, and the cell cycle (for review, see Mittag et al., 2005). The rhythm of phototaxis, measured with a photoaccumulation assay, was already demonstrated more than 35 years ago by Bruce (1970). As biflagellate unicell, C. reinhardtii is able to orientate itself toward the light. Therefore, maximum accumulation toward a supplied light source occurs during day phase, allowing the cells to optimize their light perception for photosynthesis. Mutants with aberrant circadian rhythms of phototaxis were selected many years ago (Bruce, 1972). The genetic analysis of recombinants revealed that three long-period mutations named per1, per2, and per4 were situated at different genetic loci (Bruce, 1974). None of the per genes have been identified so far in C. reinhardtii. Although they have the same nomenclature as the per genes in Drosophila melanogaster and mammals, it is rather unlikely that the same proteins are defective, since there is no evidence for a PER homolog in C. reinhardtii (Mittag et al., 2005).
In recent years, the first components that are involved in the oscillatory machinery of C. reinhardtii have been characterized, including CASEIN KINASE1 (CK1; Schmidt et al., 2006) and the RNA-binding protein CHLAMY1 (Iliev et al., 2006b). Very recently, an insertional mutagenesis approach revealed several key components that are relevant for the normal circadian rhythmicity of a chloroplast bioluminescence reporter in C. reinhardtii (Brunner and Merrow, 2008; Matsuo et al., 2008).
The RNA-binding protein CHLAMY1 recognizes specifically UG≥7-repeat sequences that are present in the 3′-untranslated regions (UTRs) of several mRNAs of C. reinhardtii, and its binding activity is controlled by the circadian clock (Mittag, 1996; Waltenberger et al., 2001). Introduction of such UG-repeat regions within the 3′-UTR of a reporter gene showed that they could trigger circadian oscillations (Kiaulehn et al., 2007). CHLAMY1 represents a novel type of heteromeric RNA-binding protein that consists of two subunits comprising three RNA recognition motifs (C3 subunit) and three Lys homology motifs (KH; C1 subunit), respectively (Zhao et al., 2004). Their interaction is necessary to recognize their binding motif, the UG-repeat region. cDNA macroarray analysis showed that c3 mRNA fluctuates over a circadian cycle with a peak during night phase, while c1 mRNA is constantly present (Kucho et al., 2005). However, at the protein level, both subunits of CHLAMY1 are present in nearly equal amounts over the circadian cycle (Zhao et al., 2004). While the C1-C3 complex can be found during subjective day and night, an additional ≥680-kD complex is present during day phase that contains C1 but not C3 in any significant amount. This complex is absent during early subjective night, when the binding activity of CHLAMY1 is high. It was postulated that this large complex is involved in removing CHLAMY1 from the UG-repeat region during early subjective day, thus allowing translation of its mRNA (Iliev et al., 2006a).
Recently, the role of CHLAMY1 within the circadian system was characterized by independent up- or down-regulation of its two subunits. The transgenic strains were used to measure either the activity rhythm of NITRITE REDUCTASE1, whose mRNA bears a UG-repeat region, or the phototaxis rhythm. Changes in the level of the C3 or the C1 subunit resulted in disturbances of the circadian clock, either affecting its acrophase (changes in the C3 level) or causing arrhythmicity (changes in the C1 level). Thus, CHLAMY1 is involved in the maintenance of phase and period of the circadian clock (Iliev et al., 2006b). Interestingly, changes in the expression of the C1 subunit altered in parallel the level of the C3 subunit, suggesting that C1 can coregulate C3 expression (Iliev et al., 2006b). In contrast, up- and down-regulation of C3 did not significantly influence the expression of C1.
Here, we analyzed whether the two subunits of CHLAMY1 may be also involved in temperature integration. We show that there are temperature-dependent changes in the phosphorylation level of C1. At the same time, the expression of the C3 subunit is up-regulated at low temperature (18°C) and down-regulated at high temperature (28°C). We provide evidence that this temperature-dependent regulation of C3 occurs at the transcriptional level and involves predominantly an E-box situated in the c3 promoter region. Remarkably, this E-box is also involved in circadian c3 expression. We further show that temperature-dependent modulation of C1 and expression of C3 are different in the clock per1 mutant. Temperature-entrained per1 cells behave differently with regard to phase and period in comparison with per1 cells entrained by a light/dark cycle. These data indicate that temperature entrainment is altered in per1.
RESULTS
While the Level of C1 Stays Constant at Different Temperatures, the Level of C3 Is Significantly Higher at Low Temperature
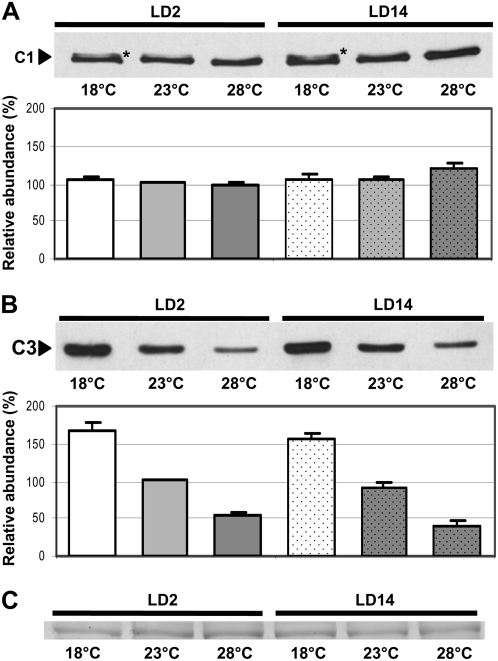
We wanted to analyze whether the C1-C3 RNA-binding protein complex CHLAMY1 could be involved in temperature integration, a prerequisite for entrainment by temperature cycles or temperature compensation. Therefore, we grew cells at different temperatures and analyzed their expression levels. Besides the usual cultivation temperature of 23°C, we selected a lower temperature (18°C) as well as a higher temperature (28°C), both being still in the physiological range of C. reinhardtii (Harris, 1989), although the difference in the lowest and highest temperatures is 10°C. Cells were harvested during early day (LD2), when the binding activity of CHLAMY1 is low, and at early night (LD14), when it is high (Mittag, 1996). Protein crude extracts were prepared, and equal amounts of protein per lane were separated by SDS-PAGE and immunoblotted with anti-C1 antibodies (see “Materials and Methods”; Zhao et al., 2004). At both time points, there was no significant change visible in the level of C1 at the different temperatures (Fig. 1A). However, at a closer look, an additional band was observed at 18°C, pointing to a possible posttranslational modification, which was analyzed further (see below). In contrast to C1, the level of C3 was significantly changed at the different temperatures, as could be seen in immunoblots with anti-C3 antibodies using the same protein extracts as used for the C1 detection and densitometry analysis (Fig. 1B). The highest amount of C3 was present at the low temperature and the lowest amount appeared at the high temperature. This was visible at both selected time points. The change in amplitude was approximately 3-fold. For comparative immunoblots, equal amounts of proteins per lane were always visually checked by Ponceau staining of the membrane (see “Materials and Methods”). In addition, we examined a duplicate gel that was stained with Coomassie blue. It corroborates that similar amounts of proteins were loaded (Fig. 1C).
Figure 1.
Immunodetection of C1 and C3 levels in wild-type cells grown at different temperatures shows differences in the case of C3. A, Cells were grown at 18°C, 23°C, and 28°C and harvested during early day (LD2) and early night (LD14). Crude extracts were prepared, and proteins (100 μg per lane) were separated by 9% SDS-PAGE along with a molecular mass standard and immunoblotted with anti-C1 antibodies (see “Materials and Methods”). Asterisks indicate a possible modified form of C1. Densitometry quantifications were done with three independent experiments (see “Materials and Methods”). Therefore, the amount of C1 detected in cells grown at 23°C and harvested at LD2 was set to 100% and used as reference. B, The same procedure described for A was carried out, but immunoblotting was done with anti-C3 antibodies (see “Materials and Methods”). C, A random nonspecific Coomassie blue-stained band from a duplicate gel shows that similar amounts of proteins were loaded.
C1 Is Hyperphosphorylated at Low Temperature
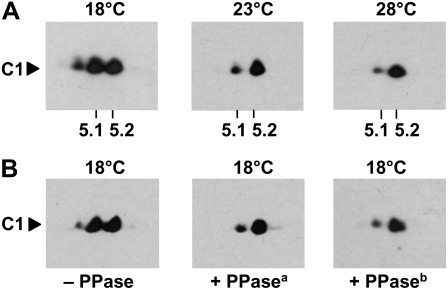
To see whether C1 might indeed be present in different posttranslationally modified forms, we separated proteins from a crude extract by a standardized two-dimensional electrophoresis (2-DE) procedure (see “Materials and Methods”) and immunoblotted the proteins with anti-C1 antibodies (Zhao et al., 2004). At 28°C, C1 appeared mostly as a single spot on 2-DE at a pH of about 5.2 (Fig. 2A). That is in accordance with its theoretical pI (5.17). A minor additional spot was present toward the lower pH, indicating that a small part of C1 is present in a posttranslationally modified form. At 23°C and especially at 18°C, this modified state of C1 was significantly increased and even a third spot was visible. Thus, C1 gets posttranslationally modified especially at low temperature. Since the modified forms of C1 did not show a significant change in molecular mass but did show a change toward the acidic pH, we postulated that phosphorylation could represent the modification. To check for this, we grew the cells again at the low temperature (18°C), at which the state of the posttranslational modification is highest, and treated the crude extract either without (control) or with λ protein phosphatase (PPase) for 30 min at 30°C. This procedure is known to remove phosphate groups from proteins. Separation of the proteins by 2-DE and immunoblotting with the anti-C1 antibodies showed that the modified forms of C1 were either missing (third spot) or significantly reduced (second spot) in the phosphatase-treated cell extracts (Fig. 2B, PPasea). These data suggest that C1 is hyperphosphorylated at the low temperature. This was corroborated when the amount of λ PPase was five times increased for the incubation (Fig. 2B, PPaseb). However, there was still a small amount of a modified C1 present. This could have different reasons, as will be discussed later. Densitometry analysis showed that the phosphorylated degree of C1 increased from approximately 28% at 28°C to approximately 51% at 18°C (Table I).
Figure 2.
C1 is hyperphosphorylated at the low temperature. A, C1 immunodetection from proteins of crude extracts separated by 2-DE. Cells were grown at 18°C, 23°C, and 28°C and harvested during early day (LD2). Crude extracts were prepared, and proteins (300 μg per assay) were separated by standardized 2-DE (see “Materials and Methods”). For the first dimension, an immobilized pH gradient strip of pH 3 to 10 was taken, and in the second dimension, 10% SDS-PAGE was used along with a molecular mass standard. The proteins were then immunoblotted with anti-C1 antibodies. The positions of pH 5.1 and 5.2 that are close to the theoretical pI of C1 (5.17) are indicated. B, The same procedure described for A was undertaken with cells grown at 18°C (− PPase). In one case (+ PPasea), the extract was treated for 30 min with λ PPase (New England Biolabs) at 30°C according to the protocol of the supplier. In another case (+ PPaseb), the amount of PPase used was increased five times.
Table I.
The temperature-dependent phosphorylation degree of C1 in the wild type and the per1 clock mutant
Percentage indicates the amount of phosphorylated C1 (see “Materials and Methods”). n = 2 unless indicated otherwise.
| Strain | Temperature | Percentage | se |
|---|---|---|---|
| Wild type | 18°C | 50.7a | 1.2 |
| Wild type | 23°C | 36.5 | 2.9 |
| Wild type | 28°C | 27.8a | 2.9 |
| per1 | 18°C | 25.3 | 3.3 |
| per1 | 28°C | 24.0 | 1.1 |
n = 3.
The c3 Promoter/5′-UTR But Not the c3 3′-UTR Is Involved in the Up-Regulation of c3 at the Low Temperature
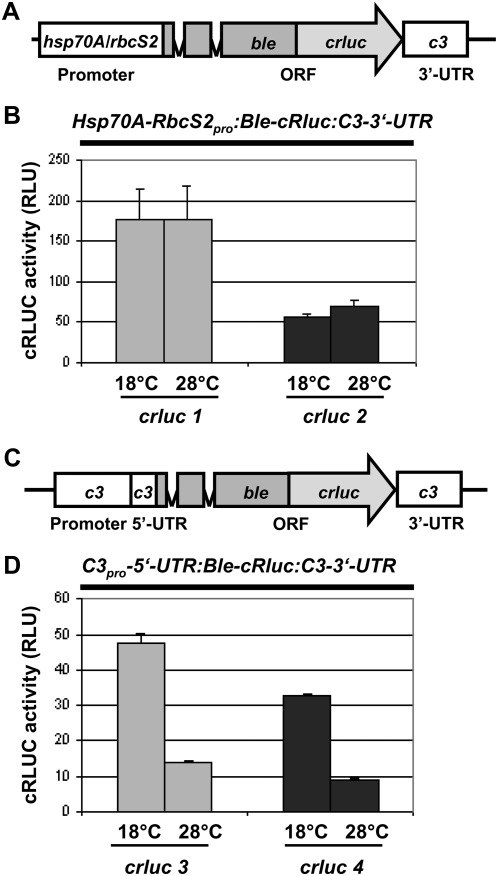
The increased amount of C3 at the low temperature could simply be due to an enhanced protein stability of C3 at 18°C or a decreased stability at the higher temperature. An alternative would be that the c3 gene is expressed in a temperature-dependent way at the transcriptional or translational level. To check for this, we carried out a reporter gene assay. As reporter, we used a fusion consisting of the zeocin resistance gene ble and the gene of Renilla reniformis luciferase that had been adapted to the nuclear codon usage of C. reinhardtii (crluc). The ble gene contained two insertions of the rbcS2 intron 1, which act as enhancers of transcription (Lumbreras et al., 1998). The chimeric gene was put under the control of the strong hsp70A-rbcS2 tandem promoter and along with the rbcS2 3′-UTR (Fuhrmann et al., 2004), resulting in a nearly equal expression over the circadian cycle, with basic levels of cRLUC activities of approximately 50 to 200 relative light units (RLU) depending on the transgenic strain (Kiaulehn et al., 2007).
Regulatory elements for transcriptional control are usually situated within the promoter region of a given gene, but they can reach into the 5′-UTR. Translational activation often involves the UTRs of an mRNA. Therefore, the 5′-UTR as well as the 3′-UTR can function as cis-acting elements. CHLAMY1 is known to bind to UG-repeat regions situated in the 3′-UTRs of several mRNAs of C. reinhardtii and can trigger their circadian expression (Kiaulehn et al., 2007). We checked first whether the c3 3′-UTR could be involved in the up-regulation of c3 at the low temperature. Therefore, we replaced the rbcS2 3′-UTR of the above-mentioned reporter gene that is under the control of the hsp70A-rbcS2 tandem promoter with the c3 3′-UTR (Fig. 3A) and transformed C. reinhardtii wild-type strains with this construct. For selection, we used the zeocin resistance marker. In a next step, we put the chimeric reporter gene construct under the control of the potential c3 promoter region along with its 5′-UTR (Fig. 3C). Again, this chimeric gene was transformed into C. reinhardtii under resistance of zeocin. We analyzed for each construct two different transgenic strains in detail. At first, we examined the expression of the BLE-cRLUC of the correct size (52 kD) by immunoblotting proteins from the transgenic lines with anti-RLUC antibodies (Supplemental Fig. S1A). We then grew these transgenic strains at different temperatures (18°C and 28°C) and measured the cRLUC activity during early day (LD2). Measurement of cRLUC activity reflects the amount of expression of cRLUC but is more sensitive and can be quantified more accurately compared with the immunological detection of cRLUC (Fuhrmann et al., 2004). No significant difference in activity was visible at 18°C and 28°C in two transgenic lines, crluc1 and crluc2, expressing the Hsp70A-RbcS2pro:Ble-cRluc:C3-3′-UTR construct (Fig. 3B). Therefore, the c3 3′-UTR does not mediate the temperature-dependent expression of c3.
Figure 3.
Up-regulation of c3 at low temperature involves its promoter/5′-UTR. A, The chimeric construct Hsp70A-RbcS2pro:Ble-cRluc:C3-3′-UTR of pSK27 (see “Materials and Methods”) that was used for transformation resulting in transgenic lines crluc1 and crluc2 is shown schematically. ORF, Open reading frame. B, Measurements of cRLUC activities in cells (crluc1 and crluc2) grown at low and high temperature. Transgenic lines crluc1 and crluc2 that express crluc along with the 3′-UTR of c3 were grown at either 18°C or 28°C and used at LD2 to measure cRLUC activities (n = 3) according to Kiaulehn et al. (2007). C, The chimeric construct C3pro-5′-UTR:Ble-cRluc:C3-3′-UTR of pSK36 (see “Materials and Methods”) that was used for transformation resulting in transgenic lines crluc3 and crluc4 is schematically shown. D, Measurements of cRLUC activities in cells (crluc3 and crluc4) grown at low and high temperature. Transgenic lines crluc3 and crluc4 that express crluc under the control of the c3 promoter/5′-UTR along with the 3′-UTR of c3 were grown at either 18°C or 28°C and used at LD2 to measure cRLUC activity according to Kiaulehn et al. (2007).
In contrast, analysis of two transgenic lines, crluc3 and crluc4, bearing the C3pro-5′-UTR:Ble-cRluc:C3-3′-UTR construct, showed significant differences in cRLUC activity at the different temperatures, with 3-fold changes in amplitude (Fig. 3D). At the low temperature, the activity was significantly increased. These results suggest that temperature-dependent regulation of c3 is controlled at the level of c3 gene expression involving its promoter and/or 5′-UTR.
The Differential Regulation of c3 at Different Temperatures Occurs at the Transcriptional Level
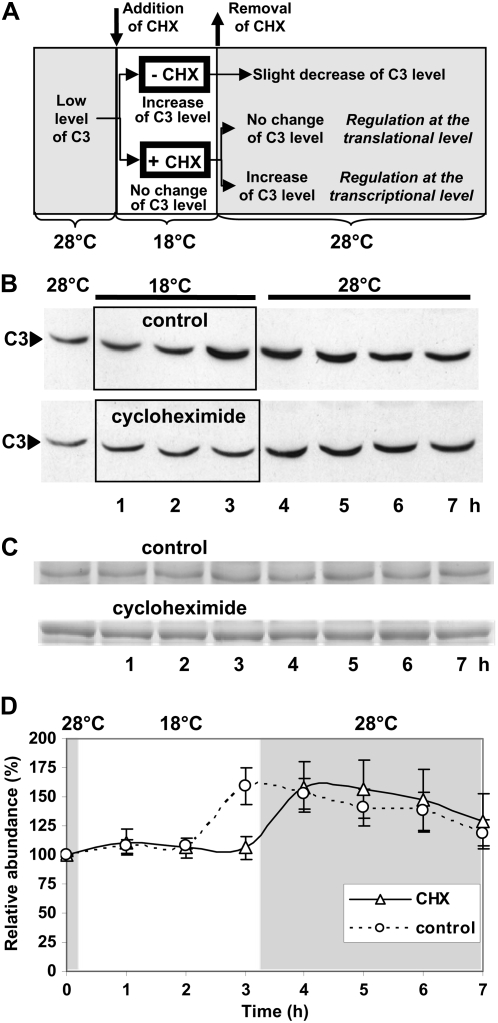
To find out whether the differential regulation of c3 occurs at the transcriptional or translational level, we decided to work out an inhibitor protocol instead of using solely the c3 promoter or its 5′-UTR along with the reporter gene. As mentioned before, it has been shown in some cases that promoter elements can reach into the 5′-UTR (Hayama et al., 2007); thus, an unambiguous result may not be achieved to decide which control is exerted. For the inhibitor experiment (Fig. 4A), we grew the cells first at the high temperature, at which the level of C3 is low. We then added cycloheximide, an inhibitor of translation, for a 3-h period, and at the time of addition we put the cells at 18°C. In parallel, a control culture without cycloheximide was put to the low temperature. If c3 up-regulation were controlled at the transcriptional level, we expected the cells to induce (with or without cycloheximide) and accumulate (in the presence of cycloheximide) c3 mRNA during this time. In the case of the control, the produced mRNA should be immediately translated and an increase of C3 protein should occur. This was found after 3 h at 18°C with the control (Fig. 4, B and D). In the presence of cycloheximide, this increase was not visible, as expected. After the 3 h at 18°C, we removed the inhibitor by washing the cells in TAP medium (see “Materials and Methods”) and transferred the cells back to 28°C, at which the induction of c3 expression will be significantly reduced, and kept them there for an additional 4 h (Fig. 4A). In the control, the level of C3 does not change after the switch to the high temperature. In the case of the culture that had been treated with cycloheximide, an increase in the level of protein was visible within the first hour after the switch to the higher temperature (Fig. 4, B and D). Equal amounts of proteins per lane were checked in two duplicate gels that were stained with Coomassie blue (Fig. 4C).
Figure 4.
Up-regulation of c3 occurs at the transcriptional level. A, Scheme of the experimental procedure with and without cycloheximide (CHX) treatment. Wild-type cells were grown at 28°C under a light/dark cycle and transferred at LD4 to 18°C by centrifuging them and dissolving them in 18°C precooled TAP medium. In one case, CHX was added (see “Materials and Methods”). After 3 h at 18°C, the two different cell cultures were centrifuged again, washed with TAP medium, and then dissolved in 28°C prewarmed TAP medium without the inhibitor. Cells were grown for an additional 4 h at 28°C. In the presence of CHX at 18°C, no translation of c3 mRNA can occur, but c3 mRNA may accumulate if the up-regulation of c3 at 18°C occurs at the transcriptional level, and thus an increase of C3 protein after the removal of CHX can be expected. B, Immunodetection of C3 levels. Cells from the control (−CHX) and cycloheximide-treated cells were harvested before, in the middle of, and after treatment as described for A at the indicated time points, and crude extracts were prepared. Proteins (50 μg per lane) were separated by 9% SDS-PAGE along with a molecular mass standard and immunoblotted with anti-C3 antibodies (see “Materials and Methods”). C, A random nonspecific Coomassie blue-stained band from duplicate gels shows that similar amounts of proteins were loaded. D, Densitometry quantifications were done with two independent experiments (see “Materials and Methods”). Therefore, the amount of C3 at time point 0 (28°C) was set to 100% and used as reference.
We conclude that c3 mRNA accumulated during the cycloheximide treatment but could not be translated due to the inhibitor. Once the stimulating low-temperature conditions were switched back to 28°C and the inhibitor was removed from the cells, accumulated c3 mRNA was translated and caused the increase in C3. This would not occur if c3 were regulated at the translational level, because during the inhibitor treatment at the low temperature no synthesis of c3 mRNA would be induced and thus none could accumulate. Therefore, the up-regulation of c3 at the low temperature occurs at the transcriptional level.
Differential Regulation of c3 Involves Predominantly an E-Box and Two DREB1A-Boxes
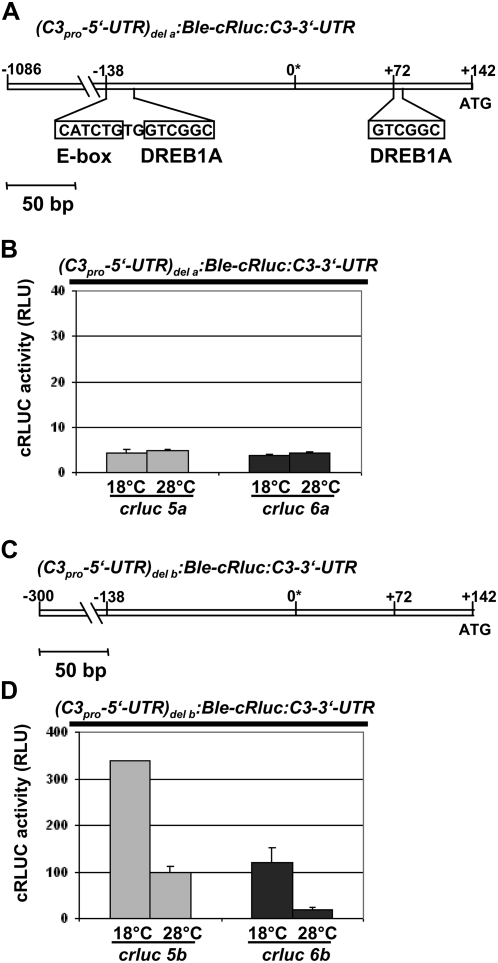
We analyzed the potential promoter region of c3 from −1,086 to +142, including the c3 5′-UTR, for potential regulatory elements that could be involved in the up- or down-regulation of c3 (see “Materials and Methods” for details). Therefore, we found three DREB1A-boxes, which can be recognized by stress-induced (e.g. cold temperature) transcription factors (dehydration-responsive element-binding proteins [DREBs]) in higher plants (Agarwal et al., 2006), either in the sense (position −573) or antisense (positions −130 and +72) direction. Furthermore, an E-box that can be bound by clock-relevant transcription factors (Yu et al., 2006) was present in the sense direction (position −138). In a first attempt to characterize any relevant cis-acting element(s) that might be involved in the temperature-dependent regulation of c3, we deleted two of the three DREB1A-boxes (at positions −130 and +72) as well as the closely situated E-box (position −138) by a splicing-overlap-extension (SOE)-PCR approach (see “Materials and Methods”; Fig. 5A). Thus, we created a (C3pro-5′-UTR)del a:Ble-cRluc:C3-3′-UTR construct that lacks two of the three DREB1A-boxes and the E-box in the c3 promoter. This construct was transformed into C. reinhardtii wild type under the selection of zeocin. Again, we checked two transgenic strains in detail and analyzed first whether they express the BLE-cRLUC construct with the correct size of 52 kD (Supplemental Fig. S1B). Then, we measured cRLUC activity in cells grown at either the low or high temperature at LD2 (Fig. 5B). In both transgenic strains (crluc5a and crluc6a), no up-regulation of cRLUC activity at the low temperature was visible. cRLUC activity stayed at a low level when cells were grown at 18°C or 28°C. These data indicate that one or two of the DREB1A-boxes and/or the E-box might be involved in up-regulation of c3 at low temperature.
Figure 5.
Up-regulation of c3 involves an E-box and/or two of the three DREB1A-boxes. A, The potential c3 promoter region including its 5′-UTR is shown from position −1,086 to +142 as it is present in pSK36. The asterisk defines the first nucleotide of the c3 EST clone (AV641734) and should be close to the +1 position of the c3 mRNA. Indicated bars include the E-box as well as the two DREB1A-boxes that have been deleted (see “Materials and Methods”) along with the two nucleotides situated between the E-box and the first DREB1A-box, resulting in pDI24. B, Measurements of cRLUC activities in cells (crluc5a and crluc6a) grown at low and high temperature that have been transformed with the chimeric construct (C3pro-5′-UTR)del a:Ble-cRluc:C3-3′-UTR of pDI24 (A). The transgenic lines were used at LD2 to measure cRLUC activities (n = 3) according to Kiaulehn et al. (2007). C, The c3 promoter region was shortened to −300 to +142 (pDI23; see “Materials and Methods”). D, Measurements of cRLUC activities (see B) in cells (crluc5b and crluc 6b) grown at low and high temperature that have been transformed with the chimeric construct (C3pro-5′-UTR)del b:Ble-cRluc:C3-3′-UTR having the shortened c3 promoter (−300 to +142; C).
In a second step, we used a shortened c3 promoter (−300 to +142; see “Materials and Methods”; Fig. 5C), in which the DREB1A-box at position −573 is missing, to examine whether it would be sufficient to mediate the temperature-dependent up-regulation of c3. In the two strains crluc5b and crluc6b, which showed correct expression of the BLE-cRLUC construct (Supplemental Fig. S1B), up-regulation of cRLUC activity at 18°C with an amplitude of 3 or greater was observed (Fig. 5D), showing that the shortened promoter is sufficient for efficient up-regulation of c3 and that the DREB1A-box at position −573 is not needed.
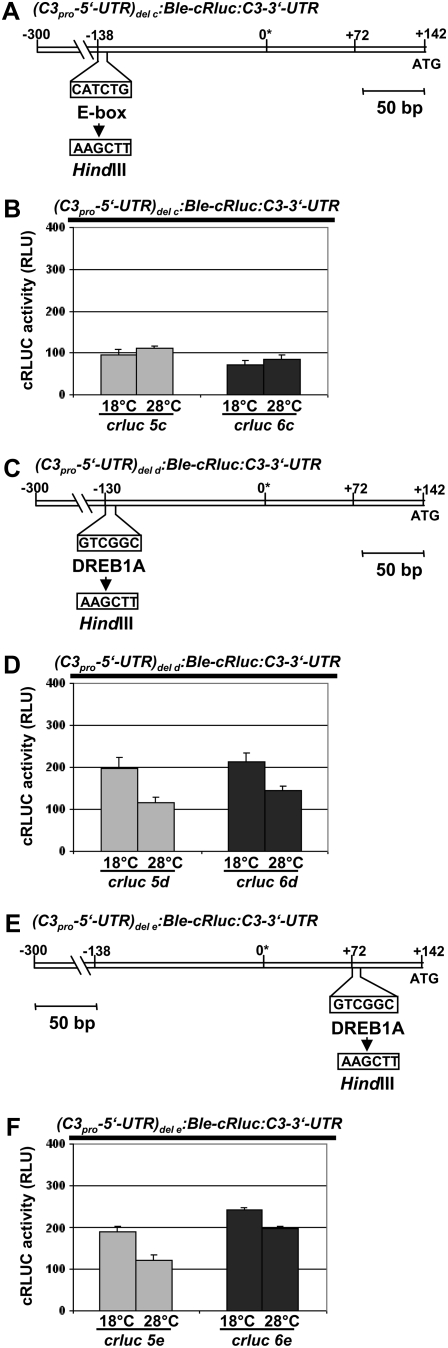
To find out whether the E-box or either of the DREB1A-boxes at −130 and +72 is relevant for the up-regulation of c3, we started single promoter element mutation assays. Therefore, each of the boxes was replaced by other nucleotides to avoid potential site-specific effects. Therefore, the shortened c3 promoter construct was used. The six-nucleotide-long E-box and either of the DREB1A-boxes at −130 and +72 were replaced in separate assays by AAGCTT, representing a HindIII restriction site (Fig. 6, A, C, and E). For each construct, two transgenic lines were checked for correct expression of the BLE-cRLUC construct with a size of 52 kD (Supplemental Fig. S1B). Then, they were analyzed for up-regulation of cRLUC activity at 18°C (Fig. 6, B, D, and F). Replacement of the E-box by the HindIII site resulted in transgenic strains crluc5c and crluc6c. In both strains, no up-regulation of cRLUC activity occurred, demonstrating that the E-box is essential for the cold-dependent up-regulation of c3. When either of the DREB1A-boxes was replaced (transgenic lines crluc5d and crluc6d for the DREB1A-box at −130 and crluc5e and crluc6e for the DREB1A-box at +72), up-regulation of c3 at 18°C was still observed; however, the amplitude of up-regulation was significantly reduced. In the lines crluc5d and crluc6d, the amplitude was decreased to 1.7 and 1.5, respectively, and in the lines crluc5e and crluc6e, it was 1.6 and 1.2, respectively. Thus, both of the DREB1A-boxes contribute to some extent to the temperature-dependent up-regulation of c3.
Figure 6.
Up-regulation of c3 involves predominantly an E-box. A, Based on the shortened c3 promoter region, the E-box was replaced by six nucleotides representing a HindIII restriction site (pD26; see “Materials and Methods”). B, Measurements of cRLUC activities in cells (crluc5c and crluc6c) grown at low and high temperature as described for Figure 5B. The chimeric construct (C3pro-5′-UTR)del c:Ble-cRluc:C3-3′-UTR, which has a shortened c3 promoter and a replaced E-box (A), was transformed into wild-type cells, resulting in transgenic lines crluc5c and crluc6c. C, Based on the shortened c3 promoter region, the DREB1A-box at position −130 was replaced by six nucleotides representing a HindIII restriction site (pDI27; see “Materials and Methods”). D, Measurements of cRLUC activities in cells (crluc5d and crluc6d) grown at low and high temperature as described for Figure 5B. The chimeric construct (C3pro-5′-UTR)del d:Ble-cRluc:C3-3′-UTR, which has a shortened c3 promoter and a replaced DREB1A-box at −130 (C), was transformed into wild-type cells, resulting in transgenic lines crluc5d and crluc6d. E, Based on the shortened c3 promoter region, the DREB1A-box at position +72 was replaced by six nucleotides representing a HindIII restriction site (pDI28; see “Materials and Methods”). F, Measurements of cRLUC activities in cells (crluc5e and crluc6e) grown at low and high temperature as described for Figure 5B. The chimeric construct (C3pro-5′-UTR)del e:Ble-cRluc:C3-3′-UTR, which has a shortened c3 promoter and a replaced DREB1A-box at +72 (E), was transformed into wild-type cells, resulting in transgenic lines crluc5e and crluc6e.
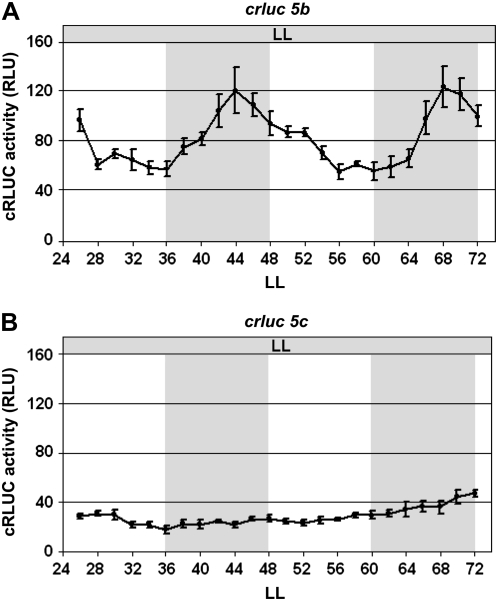
The E-Box Is Also Involved in Circadian Expression of c3
As mentioned before, c3 mRNA was shown to fluctuate over a circadian cycle by macroarray analysis (Kucho et al., 2005). We used the transgenic line crluc5b that expresses cRLUC under the control of the shortened c3 promoter to see whether this part of the c3 promoter would mediate circadian c3 expression. After entrainment by a light/dark cycle, a circadian pattern of cRLUC activity with a peak during night phase was observed under constant dim light (LL) conditions (Fig. 7A). These data show that the shortened c3 promoter contains the relevant cis-acting element for circadian c3 expression. To analyze whether the E-box could also be involved in circadian expression, we used the transgenic line crluc5c, in which it had been replaced. In this case, arrhythmicity was found (Fig. 7B). Thus, the E-box has a dual function. It is responsible for the temperature-dependent up-regulation of c3 but also for its circadian expression, linking it directly to the function of the circadian clock.
Figure 7.
Replacement of the E-box in the c3 promoter abolishes its circadian expression. The transgenic wild-type strains crluc5b (A) and crluc5c (B), expressing the chimeric crluc reporter gene under the control of the 300-bp c3 promoter in its native form (A) or under replacement of its E-box (B), were grown in a 12/12-h light/dark cycle and then released to constant dim light (LL). cRLUC activities (n = 3) were measured at the indicated time points according to Kiaulehn et al. (2007).
PER1 Is Part of a Functional Network Including C1 and C3
The per mutants of C. reinhardtii have a lengthened period of the circadian phototaxis rhythm (Bruce, 1972). We reconfirmed this for strain CC1117 (per1) by analyzing its phototaxis rhythm (Supplemental Fig. S2). So far, there are no indications that the gene/protein is defective in the per1 mutant. We wanted to know whether the functional temperature-related network of C1 and C3 is maintained or changed in the per1 mutant.
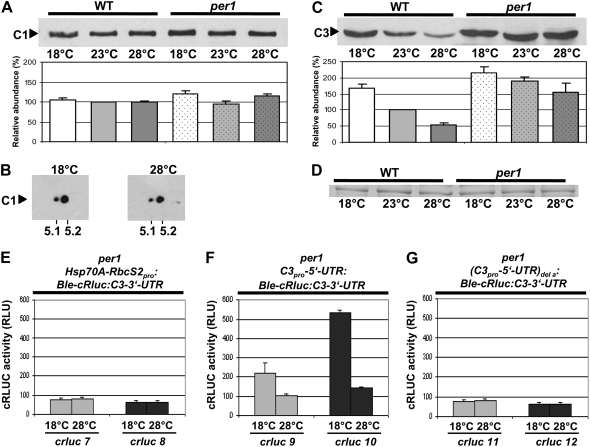
In the per1 mutant, the level of C1 stayed rather constant at the different temperatures (Fig. 8A), which is similar to what was seen in the wild type. However, the phosphorylation status of C1 changed in comparison with the wild type (Table I). C1 was relatively little phosphorylated at both low and high temperatures, thus lacking the hyperphosphorylation at 18°C (Fig. 8B) that was found in the wild type. Notably, the level of C3 was significantly increased in the per1 mutant compared with the wild type, especially at the high temperature, as judged by immunoblots (Fig. 8, C and D). We then used the chimeric reporter constructs Hsp70A-RbcS2pro:Ble-cRluc:C3-3′-UTR and C3pro-5′-UTR:Ble-cRluc:C3-3′-UTR for transformation of the per1 mutant to see whether the influence of the c3 3′-UTR and c3 promoter/5′-UTR on c3 expression would differ in per1. Correct expression of the fusion proteins in the transgenic per1 lines with a size of 52 kD was verified on immunoblots (Supplemental Fig. S1). No temperature-dependent effects on cRLUC activities were seen in the transgenic lines crluc7 and crluc8 bearing the c3 3′-UTR constructs (Fig. 8E). In contrast, the transgenic lines crluc9 and crluc10, which are under the control of the c3 promoter/5′-UTR, showed up-regulation at the low temperature, with an amplitude of about 2- and 3-fold, respectively (Fig. 8F). Therefore, it was striking that cRLUC activity was in the range of 100 RLU at 28°C in both transgenic lines, while in the corresponding transgenic wild-type lines cRLUC activity was only about 10 RLU. At the low temperature, cRLUC activities increased to approximately 200 and 500 RLU in crluc9 and crluc10. Both immunoblot and cRLUC activity measurements underline the point that C3 is expressed in general at a significantly higher rate in the per1 mutant.
Figure 8.
In the per1 mutant, the phosphorylation degree of C1 is relatively low at both different temperatures, and the expression level of C3 is increased in comparison with that in the wild type depending on the E-box and/or the DREB1A-boxes. A, Immunodetection of C1 in per1. Cells of the per1 mutant were grown at 18°C, 23°C, and 28°C and harvested during early day (LD2). Crude extracts were prepared, and proteins (50 μg per lane) were separated by 9% SDS-PAGE along with a molecular mass standard and immunoblotted with anti-C1 antibodies (see “Materials and Methods”). Densitometry quantifications were done with three independent experiments (see “Materials and Methods”). Therefore, the amount of C1 detected in wild-type cells grown at 23°C was set to 100%. B, Immunodetection of C1 from proteins separated by 2-DE. Cells of the per1 mutant were grown at either 18°C or 28°C and harvested as described for A. Crude extracts were prepared, and proteins (300 μg per assay) were separated by standardized 2-DE (see “Materials and Methods”). For the first dimension, an IPG strip of pH 3 to 10 was taken, and in the second dimension, 10% SDS-PAGE was used along with a molecular mass standard. The proteins were then immunoblotted with anti-C1 antibodies. The positions of pH 5.1 and 5.2 that are close to the theoretical pI of C1 (5.17) are indicated. C, Immunodetection of C3 in per1. The same procedure described for A was carried out, but immunoblotting was done with anti-C3 antibodies (see “Materials and Methods”). D, A random nonspecific Coomassie blue-stained band from a duplicate gel shows that similar amounts of proteins were loaded. E to G, Measurements of cRLUC activities in different transgenic per1 cells. Lines were grown at either 18°C or 28°C and used at LD2 to measure cRLUC activities (n = 3) according to Kiaulehn et al. (2007). In the first case (E), the chimeric construct Hsp70A-RbcS2pro:Ble-cRluc:C3-3′-UTR, which is under the control of the strong Hsp70A-RbcS2 promoter and bears the c3 3′-UTR, was used for transformation of the per1 mutant, resulting in transgenic lines crluc7 and crluc8. In the second case (F), the chimeric construct C3pro-5′-UTR:Ble-cRluc:C3-3′-UTR, which is under the control of the c3 promoter region, was used for transformation of the per1 mutant, resulting in transgenic lines crluc9 and crluc10. In the third case (G), the chimeric construct (C3pro-5′-UTR)del a:Ble-cRluc:C3-3′-UTR, in which the E-box and two DREB1A-boxes had been deleted from the c3 promoter region, was transformed into the per1 mutant, resulting in transgenic lines crluc11 and crluc12.
We examined whether the two DREB1A- and/or E-box elements would also be relevant for up-regulation of c3 at the low temperature in per1 and therefore would transform the c3 promoter constructs lacking two of the three DREB1A-boxes and the E-box into per1. Again, expression of the BLE-cRLUC proteins of the correct sizes was verified in the two transgenic lines crluc11 and crluc12 (Supplemental Fig. S1). No significant up-regulation of c3 at the low temperature was visible (Fig. 8G). At both low and high temperatures, the cRLUC activity levels ranged at a relatively low level close to 50 RLU. Thus, the mechanism of up-regulation of c3 is also disturbed in the per1 mutant upon deletion of the DREB1A- and E-boxes, as was found in the wild type.
Temperature Entrainment Is Altered in the per1 Mutant
If the temperature-dependent changes in C1 and C3 that we found in per1 influence temperature compensation or entrainment by temperature cycles, one of these two processes should be affected in per1. We analyzed the ability for temperature entrainment in the per1 mutant in comparison with the wild type. As reporter, we took a chimeric crluc reporter gene that is under the control of the hsp70A-rbcS2 tandem promoter and has the UG-repeat region of Gln synthetase2 (gs2) in the rbcS2 3′-UTR (Hsp70A-RbcS2pro:Ble-cRluc:Gs2-RbcS2-3′-UTR; Kiaulehn et al., 2007). Thus, expression of the reporter gene is under the control of CHLAMY1. It was shown before that the insertion of the UG repeat into the reporter 3′-UTR introduces a circadian rhythm of cRLUC activity when cells are entrained by a light/dark cycle and released to constant conditions of temperature and dim light (LL). The maximum peak of activity occurred during the middle of subjective day, and a change in amplitude of about 2-fold was observed (Kiaulehn et al., 2007).
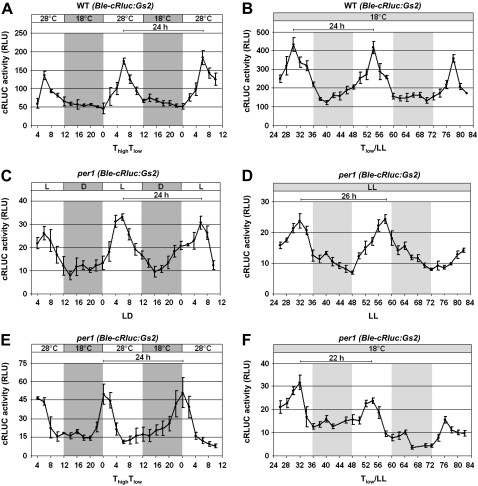
At first, we used this transgenic line, designated the wild type (Ble-cRLUC:Gs2), to test the temperature entrainment protocol. In the setup for temperature entrainment, we entrained the transgenic cells with a temperature cycle of 12 h at 28°C and 12 h at 18°C, labeled as ThighTlow. Our data show that the transgenic cells perceived the high temperature as signal for daytime and the low temperature as signal for nighttime. cRLUC activity peaked at ThighTlow6 (Fig. 9A), which is in agreement with the peak in the middle of the day phase when the cells had been entrained by a light/dark cycle (Kiaulehn et al., 2007). When the temperature-entrained cells were released under constant conditions of dim light and temperature, the rhythm continued with a period of 24 h (Fig. 9B). Thus, the C. reinhardtii clock can be entrained by a 12/12-h, 28°C/18°C cycle. The same chimeric construct, Hsp70A-RbcS2pro:Ble-cRluc:Gs2-RbcS2-3′-UTR, was then transformed into per1, resulting in transgenic line crluc13, designated per1 (Ble-cRLUC:Gs2). Correct expression of the BLE-cRLUC was analyzed (Supplemental Fig. S1). At first, we examined whether this transgenic line can be entrained by a light/dark cycle (Fig. 9C). Under the light/dark cycle, the period of the cRLUC activity rhythm was around 24 h and its maximum peak occurred at LD6. Thus, the per1 mutant can be entrained by a light/dark cycle to 24 h, and the acrophase of the maximal activity peak is maintained during the middle of the day. The transgenic per1 line that had been grown in the light/dark cycle was then released under constant conditions of dim light and temperature. Under these free-running conditions, the transgenic line had a lengthened period (approximately 26 h; Fig. 9D), as was also found with the phototaxis rhythm, in which the cells were kept under constant dark conditions (Supplemental Fig. S2). Thus, per1 behaves similarly to the clock mouse mutant (Antoch et al., 1997). Under the light/dark cycle, its period was 24 h, and under free running conditions, its period was lengthened by about 2 h. We then entrained the transgenic per1 cells with a temperature cycle (12/12 h, 28°C:18°C). A rhythmic activity of cRLUC was observed. However, the maximal activity peak occurred at ThighTlow0 on both measured day/night cycles, although a period of 24 h was maintained (Fig. 9E). These data show that the per1 clock is disturbed in its acrophase during the process of temperature entrainment.
Figure 9.
Temperature entrainment is altered in per1. A and B, The transgenic wild-type strain Hsp70A-RbcS2pro:Ble-cRluc:Gs2-RbcS2-3′-UTR, designated WT (Ble-cRluc:Gs2) (Kiaulehn et al., 2007), expressing the chimeric crluc reporter gene under the control of the hsp70A-rbcS2 tandem promoter and the gs2 UG-repeat region within the rbcS2 3′-UTR, was grown in a 12/12-h temperature cycle (ThighTlow; A) and then released under constant temperature and dim light (B). cRLUC activities (n = 3) were measured at the indicated time points according to Kiaulehn et al. (2007). C and E, The transgenic per1 strain crluc 13, which expresses the chimeric Hsp70A-RbcS2pro:Ble-cRluc:Gs2-RbcS2-3′-UTR construct, designated per1 (Ble-cRluc:Gs2), was grown either under a light/dark cycle (C) or in a temperature cycle (ThighTlow; E). cRLUC activities (n = 3) were measured at the indicated time points according to A. D and F, The transgenic line per1 (Ble-cRluc:Gs2) was released under conditions of constant temperature and dim light either after LD entrainment (D) or after treatment with a temperature cycle (F). cRLUC activities (n = 3) were measured at the indicated time points according to A.
Then transgenic per1 cells that had been exposed to a temperature cycle were released under constant dim light and low temperature (labeled as Tlow/LL) to see how their clock would free run. Under these conditions, a circadian rhythm of cRLUC activity was visible, but the period was shortened by about 4 h (22 h; Fig. 9F) in comparison with the period of 26 h that had been observed after entrainment by a light/dark cycle (Fig. 9D).
DISCUSSION
In contrast to biochemical reactions, the period length of circadian rhythms has a Q10 value that is close to 1. Thus, circadian clocks are temperature compensated (Pittendrigh, 1954; Hastings and Sweeney, 1957). Moreover, temperature cycles, in addition to light cycles, are among the main environmental cues that can synchronize circadian clocks (Rensing and Ruoff, 2002). For control of both processes, certain components of the circadian clock should be able to integrate temperature.
The two subunits of the RNA-binding protein CHLAMY1 cause arrhythmicity (C1) or shifts in the acrophase (C3) when their expression level is altered (Iliev et al., 2006b). Here, we show that the expression of these clock-relevant components is altered in different ways by temperature changes of 10°C, ranging from 18°C up to 28°C. These temperatures are still in the physiological range of C. reinhardtii (Harris, 1989); thus, these clock components can integrate temperature. For true stress-related responses (e.g. heat stress), a significantly higher temperature (40°C) should be applied (Schulz-Raffelt et al., 2007). The temperature-dependent regulation of C1 and C3 occurred at different levels. In the case of C1, posttranslational modifications were altered at the different temperatures, and with C3, a temperature-dependent change in its abundance was observed, with a maximum amount at low temperature. Studies with cycloheximide treatment suggested that the up-regulation of C3 is controlled at the transcriptional level. To study the involvement of cis-acting elements in temperature-dependent c3 up-regulation, we used chimeric ble-crluc constructs along with the c3 3′-UTR and the hsp70A-rbcS2 tandem promoter or with the c3 promoter/5′-UTR. The resistance against zeocin was used as a selection marker for positive transformation of the transgenic lines. Nuclear transformation of C. reinhardtii usually occurs via nonhomologous recombination. Depending on the place of integration in the genome, epigenetic effects can contribute to an expression level that is basically higher or lower. We always analyzed a few different transgenic lines for each chimeric construct and used two exemplary transgenic lines for detailed analysis.
Using different chimeric ble-crluc constructs, we found that the c3 promoter/5′-UTR region from −1,086 to +142 is involved in the up-regulation of c3 at low temperature. In this region, three DREB1A-boxes and an E-box were evident. Since a shortened promoter from −300 to +142 that lacks the DREB1A-box at −573 still can efficiently up-regulate c3 at 18°C, this DREB1A-box seems to be not relevant. Further analysis of the “shortened” promoter region of c3 along with its 5′-UTR showed that two DREB1A-boxes at positions −130 and +72, and especially an E-box at −138, are the relevant cis-acting elements. Replacement of the DREB1A-boxes at −130 and +72 reduced the amplitude of cold-induced c3 up-regulation, suggesting that these boxes contribute to some extent to the up-regulation. DREB-boxes are recognized by DREB proteins that are important transcription factors for abiotic stress in higher plants (Agarwal et al., 2006). Therefore, these transcription factors are dichotomized as DREB1 and DREB2. It should be noted that DREB1A, -B, and -C are also known as CBF3, -1, and -2, respectively (for C-REPEAT BINDING FACTOR). DREBs are involved in two separate signal transduction pathways with regard to low temperature or dehydration stress. They belong to the ERF (for ETHYLENE-RESPONSIVE ELEMENT-BINDING FACTORS) family of transcription factors. ERF proteins are a subfamily of the APETLA2 (AP2)/ethylene-responsive element binding protein transcription factors that is distinctive to plants. Recently, it was shown that a DREB factor (named PpDBF1) also occurs in the moss Physcomitrella patens and is involved in salt, drought, and cold stresses there (Liu et al., 2007). Additional members of the cold signaling pathway in higher plants that are in a functional network with the DREB factors include ICE1 (for INDUCER OF CBF/DREB EXPRESSION1) and its SIZ1-mediated (for SAP and MIZ) sumoylation (Miura et al., 2007). Interestingly, it has been shown in Arabidopsis that low temperature induction of CBF1, -2, and -3 is gated by the circadian clock (Fowler et al., 2005). In C. reinhardtii, several gene models (genome version 3) were found that encode proteins with ERF/AP2-like domains. The two with highest homology to the Arabidopsis DREB1A and DREB2A factors have the protein identifiers 167010 and 205920. However, the reported temperature shift activating DREB1 in higher plants is usually a switch from room temperature (about 22°C) to 4°C or even below. To our knowledge, there are no data available on whether 18°C can act as a stimulating low temperature. It seems possible that the two DREB1A-boxes cold stimulate c3 expression at a high degree, as in higher plants, at a temperature that is significantly lower than 18°C. This has not been checked in our experiments, because we focused on the physiological temperature range of C. reinhardtii, in which temperature compensation can be observed.
Replacement of the other cis-acting element, the E-box at −138, abolished completely the up-regulation of c3 at low temperature (18°C), showing that it represents the key element for temperature integration of c3. It also abolished the circadian rhythm of cRLUC driven by the shortened c3 promoter, showing that is has a dual function for temperature-dependent and circadian expression. E-boxes are known as relevant cis-acting elements for circadian regulation. For example, in D. melanogaster, they are situated in the promoter regions of per and timeless (tim). Transcriptional activation of both genes occurs via the CLOCK-CYCLE (CLK-CYC) heterodimer and is blocked when the PER/TIM complex interacts with CLK-CYC (Yu et al., 2006). But potential homologues to the heterodimer CLK-CYC that recognize the E-box have not been found in C. reinhardtii (Mittag et al., 2005). Positive transcriptional regulation by an E-box was also reported in other cases, such as with the polyserase gene, in which the E-box is situated in the 5′-UTR and is required for maximal promoter activity (Hayama et al., 2007). In the case of the c3 promoter, a special situation exists, because it might be recognized by a circadian transcription factor that is activated especially at low temperature. In this context, it will also be interesting to find out whether the cis-acting elements involved in the coregulation of c3 are identical to or different from the cis-acting elements involved in its circadian and temperature-dependent regulation. As mentioned before, strains that overexpress or underexpress C1 show in parallel an up- or down-regulation, respectively, of C3 (Iliev et al., 2006b). Preliminary experiments indicate that this coregulation is also mediated at the transcriptional level (S.B. Seitz and M. Mittag, unpublished data).
The posttranslational modifications of C1 that increased at low temperature involve phosphorylation. Treatment of extracts with λ PPase showed the disappearance of one modified C1 form and a significant reduction of the other one, both at 18°C. However, even after PPase treatment with higher amounts of enzyme, there was still a small percentage of a modified C1 form present. This could mean that C1 has some additional posttranslational modifications. But it is also possible that the phosphorylated C1 that is present in the C1-C3 complex and in addition in the ≥680-kD complex during day phase (Zhao et al., 2004) cannot be fully accessed by the PPase.
Based on these data, it seems possible that the different phosphorylation status of C1 at low and high temperature might be involved in the regulation of c3 at different temperatures in a direct or indirect way. C1 bears three KH domains that are known as RNA-binding domains and binds in combination with C3 to UG-repeat RNAs (Zhao et al., 2004). But there are reports that KH domain-containing proteins recognize RNA and DNA sequences, as was shown with poly(C)-binding proteins (Du et al., 2007). Moreover, DNA-binding protein complexes such as NF-κB, which mediate selective gene regulation, can contain KH domain subunits. In the case of the NF-κB complex, the KH domain ribosomal protein S3 is a subunit of this complex (Wan et al., 2007). These findings suggest the possibility that C1 could be directly involved in up-regulating c3, for example, as part of a DNA-binding protein complex. The finding that c3 expression is coregulated depending on the level of C1 (Iliev et al., 2006b) would be in concert with such a postulation, which has to be carefully analyzed in the future.
Another point of interest is the kinase that is involved in the phosphorylation of C1 at low temperature. Theoretical predictions of phosphorylation sites (NetPhosK 1.0 server at www.cbs.dtu.dk/services/NetPhosK/; Blom et al., 2004) of C1 highlight several protein kinases that should phosphorylate C1. Among them, CK1 was indicated. CK1 will be an interesting candidate to study with regard to phosphorylation of C1, because it is already known to be clock relevant. Its silencing causes period shortening of the circadian clock in C. reinhardtii (Schmidt et al., 2006).
As mentioned earlier, Bruce (1972) isolated per mutants that show a lengthened period decades ago. One of them, per1, was included in our studies to see whether there could be a connection between PER1 and the temperature integration of C1 and C3. In the per1 mutant, temperature-dependent hyperphosphorylation of C1 as well as c3 expression were altered, suggesting that PER1 is indeed part of the network regulating temperature sensing of C1 and C3. In the case of C1, we observed that the hyperphosphorylation at low temperature did not occur. It was little phosphorylated in the per1 mutant at both 18°C and 28°C. This could be due to a missing kinase or increased activity of a PPase. Notably, c3 expression was significantly increased in the per1 mutant, both at low and high temperature, while its up-regulation at the low temperature was still present. Deletion of the DREB1A- and E-boxes also resulted in the loss of low-temperature-mediated up-regulation of c3, showing the relevance of the elements also in per1. It can be hypothesized that a transcription factor that increases c3 expression might be activated at both 18°C and 28°C in per1. This could be achieved, for example, by changing the phosphorylation status of such a transcription factor. The involvement of a PPase or a kinase that could be altered in the per1 mutant would also explain the changes in the phosphorylation level of C1 in per1. Another possibility is that there is some positively acting mutation in the c3 promoter that causes the increased c3 expression. To check for this, we sequenced the promoter region of the c3 gene (positions −874 to +274) in the per1 mutant. However, there was no difference in comparison with the wild-type sequence depicted from the Joint Genome Institute, excluding this possibility.
The circadian clock in per1 can be entrained by a light/dark cycle to 24 h with a peak of activity at LD6, as was also found in the wild type. But it free runs with a period of about 26 h under constant conditions. As mentioned before, the clock mouse mutant shows a similar behavior (Antoch et al., 1997). In contrast, synchronization by a temperature cycle resulted in a shift in acrophase in per1 at the transition at the end of the night to day phase. Under free-running conditions, a period of 22 h was observed in per1 after entrainment by the temperature cycle. These data show that temperature entrainment is altered in per1. Although the identity of PER1 is still unknown, it is becoming evident that it is part of a functional temperature-dependent network involving C1 and C3.
MATERIALS AND METHODS
Standard molecular biology methods were used according to Sambrook and Russell (2001).
Cell Culture
Chlamydomonas reinhardtii cells (wild-type strain SAG73.72) were grown in TAP medium (Harris, 1989) under a 12-h-light/12-h-dark cycle with a light intensity of 71 μE m−2 s−1 (1 E = 1 mol of photons) at 18°C, 23°C, or 28°C, as indicated for each experiment. Cells were grown under a light/dark cycle unless indicated otherwise. The beginning of the light period is defined as time zero (LD0), and the beginning of the dark period is defined as LD12. In some cases, cells were released after the light/dark cycle into constant dim light (LL; 20 μE m−2 s−1) and temperature (23°C). For temperature entrainment, cells were grown in LL under a 12-h, 28°C/12-h, 18°C cycle (ThighTlow). For circadian conditions, cells were released in LL to a constant temperature of 18°C (Tlow/LL). In some cases, cycloheximide (Sigma-Aldrich) was added to the culture to an end concentration of 35.5 μm (Kawazoe et al., 2000).
Preparation of Crude Extracts of C. reinhardtii and Immunoblots
Cells were grown to a cell density of 1 to 5 × 106 cells mL−1, harvested by centrifugation at the indicated LD time point, and stored at −80°C after being frozen in liquid nitrogen. Protein extracts were prepared according to Zhao et al. (2004) for further use in one-dimensional electrophoresis. All extracts were prepared in the presence of Complete Proteinase Inhibitor Cocktail (Roche-Applied-Science) according to the user's manual. In some cases, 1% phosphatase inhibitors I and II (Sigma) were also added. Protein concentration was measured either with the Bio-Rad protein assay according to its manual or with the Neuhoff et al. (1979) method. Equal loadings of the proteins were in parallel and visually checked by Ponceau staining of the membrane directly after blotting of the proteins. Immunoblotting was done as described with either anti-C1 or anti-C3 antibody (Zhao et al., 2004; Iliev et al., 2006b) along with chemiluminescence detection. In some cases, monoclonal anti-RLUC antibodies (Chemicon Europe) were used with a dilution of 1:2,000. As secondary antibody, monoclonal anti-mouse IgG antibodies (Sigma) were used with a dilution of 1:20,000. Detection was again done by chemiluminescence (Iliev et al., 2006b).
Standardized 2-DE
2-DE was basically done according to Wagner et al. (2004) with some modifications. Crude extracts for 2-DE were prepared as described by Wagner et al. (2004), and proteins were then precipitated with 12% trichloroacetic acid and 0.1% dithiothreitol (DTT) in acetone and centrifuged for 40 min at 26,800g. The protein pellet was washed with 0.1% DTT in acetone, dried for 15 min at room temperature, and solubilized for 1 h at room temperature in rehydration buffer (8 m urea, 0.5% [w/v] CHAPS, 20 mm DTT, 0.2% IPG buffer [pH 3–10; Amersham Biosciences], and 10% glycerol) to a final concentration of 2 μg μL−1. The resuspended sample was then centrifuged for 5 min at 26,800g. A total of 150 μL of the supernatant was mixed with 300 μL of rehydration buffer and applied into the strip holder. After the IPG strip had been placed in the strip holder over the solution and covered with paraffin, rehydration was carried out for 12 h. Then, isoelectric focusing was started with the following IPGphor program: 200 V (1 h), 500 V (1 h), 1,000 V (1 h), linear gradient from 1,000 to 8,000 V (1 h), and 8,000 V (6 h); thereby, current was limited to 0.05 mA per IPG strip. After the end of this program, the strip was removed from the strip holder and equilibrated for 15 min in solution 1 (6 m urea, 50 mm Tris-HCl, pH 8.8, 2% [w/v] SDS, 30% [w/v] glycerol, and 1% [w/v] DTT) and for 15 min in solution 2 (6 m urea, 50 mm Tris-HCl, pH 8.8, 2% [w/v] SDS, 30% [w/v] glycerol, and 4% [w/v] iodoacetamide). Each strip was placed on a 10% polyacrylamide gel that contained 0.02% Na-thiosulfate. Strips were fixed on the gels with 0.5% agarose in electrode buffer (0.1% SDS, 25 mm Tris, and 0.192 m Gly), and electrophoresis was performed at 4°C and 2.5 W per gel overnight (approximately 12 h) using an Ettan Dalt 6 electrophoresis unit (Amersham Biosciences). After electrophoresis, immunoblotting was done along with anti-C1 antibodies. Directly after western blotting, the membrane was stained with the MemCode Reversible Protein Stain Kit (Pierce), and the biggest spots were marked and used as position markers for the comparison of different samples. Then, membranes were further used for antibody incubation.
Densitometry Analysis
Quantifications were done with the Image Master 2D Elite (version 4.01) software from Amersham Pharmacia Biotech. For 2-DE immunoblots of C1, the area of all spots was set to 100% and compared with the phosphorylated spot area.
Construction of Chimeric cRLUC-Containing Vectors
A vector (pSK27) expressing the Hsp70A-RbcS2pro:Ble-cRluc:C3-3′-UTR construct was created. For this purpose, pSK24 was constructed first. It contains the c3 3′-UTR that was obtained by PCR from pCS31 (Zhao et al., 2004) with the following primers (5′-GCCGCCGCCATGTAAATAG-3′ and 5′-CGACTCACTATAGGGCGGGTAC-3′) and blunt-end cloned into the MluI site of pCAPs (Roche). Thus, suitable restriction sites for replacing the rbcS2 3′-UTR of pSK1 (Kiaulehn et al., 2007) with the c3 3′-UTR were obtained. Therefore, pSK24 was restricted with KpnI and ZraI, and its 1,650-bp fragment bearing the c3 3′-UTR was ligated with the 3,562-bp KpnI and EcoRV fragments of pSK1, resulting in pSK27.
Another vector (pSK36) expressing the C3pro-5′-UTR:Ble-cRluc:C3-3′-UTR construct was created. It is identical to pSK27 except that it contains the potential promoter region (positions −1086 to 0) and 5′-UTR of the c3 gene instead of the hsp70A-rbcS2 tandem promoter. Therefore, pRbcBRL(HSP196) (Fuhrmann et al., 2004) was used as a basis that also contains the chimeric Hsp70A-RbcS2pro:Ble-cRluc:C3-3′-UTR construct. At first, a linker that was made by annealing the oligonucleotides 5′-CGGGGAATTCGGGGGTCACCGGGCATG-3′ and 5′-CCCGGTGACCCCCGAATTCCCCGAGCT-3′ was cloned into the SacI and SphI sites of pRbcBRL(HSP196). Thus, the linker replaced the tandem promoter. The linker introduced EcoRI and BstEII sites that were used to insert the 1,341-bp fragment containing the above-mentioned c3 promoter region along with its 5′-UTR from pAN4 cut with the same restriction enzymes (Iliev et al., 2006b). The resulting plasmid was named pSK22. In a second cloning step, pSK22 was opened with BamHI and SapI to insert the 1,628-bp c3 3′-UTR from pSK24 (see above), which was cut out with SphI and XbaI. Both fragments were treated with Klenow enzyme to create blunt ends prior to ligation.
Promoter Analysis
For the search of putative transcription factor binding sites in the c3 promoter/5′-UTR, the sequence between positions −1,086 and +142 bp was analyzed with PROMO 3.0 free online software at http://alggen.lsi.upc.es (Messeguer et al., 2002; Farré et al., 2003). Using the following settings and cutoffs, the prediction was performed. “Plant factors” and “plant sites” were selected, the “maximum matrix dissimilarity rate” was set to zero, and cutoffs for the RE equally ≤0.3 and for the RE query ≤0.6 were made. Therefore, three DREB1A-binding sites (GCCGAC) were identified. One of the sites (starting at position −573) was identified in the sense strand, and the other two (starting at positions −130 and +72) were identified in the antisense strand. Furthermore, an E-box-binding site (CATCTG; Wozniak et al., 2007) that starts at position −138 was manually found.
For all pDI vectors, PCR-amplified DNA was cloned into pSK36 (see above) that had been restricted with EcoRI and SphI. In one case, the E-box and the DREB1A-boxes at positions −130 and +72 were deleted from the c3 promoter by a SOE-PCR procedure (Horton et al., 1989). Three DNA fragments named A (1,090 bp), B (196 bp), and C (185 bp) were amplified and fused together. As a template, vector pSK36 (see above) was used. Fragment A was amplified by oligonucleotides OMM 461 (5′-CAGGGTTTTCCCAGTCACGA-3′) and OMM 462 (5′-CGCACTCGCGCTCGACTTTCACACACAAATAATTGCGAATTTGGG-3′), fragment B was amplified by OMM 463 (5′-GTCGAGCGCGAGTGCG-3′) and OMM 464 (5′-AAATTGAGAGGACGAGTGGAGC-3′), and fragment C was amplified by OMM 465 (5′-GCTCCACTCGTCCTCTCAATTTTCGAACCGTTTCCACACG-3′) and OMM 460 (5′-CCGGTTTATCAGGAGGGCA-3′). After the amplifications, fragments A, B, and C were separated on an agarose gel, sliced out, eluted with the GenElute gel extraction kit (Sigma) according to the manufacturer's protocol, mixed in equimolar quantities, and fused together. The fusion was performed in two steps. In the first step, fragments B and C were fused together, using flanking primers OMM 463 and OMM 460 and forming product BC (359 bp). In a next step, fragment BC was sliced out from the gel and fused to fragment A, using flanking primers OMM 461 and OMM 460. The final PCR product (1,433 bp) was cloned into pSK36, resulting in pDI24. All PCR amplifications were done with the GC-Rich PCR System (Roche) according to its protocol. The overlapping regions in the primers are underlined. For the amplification of fragment B, an annealing temperature of 53°C was used; for all other amplifications and fusions, an annealing temperature of 54°C was used. The extension time was 1 min kb−1 DNA, with 30 cycles per PCR. Sequencing of pDI24 was performed at Eurofins Medigenomix. It confirmed the deletion of the E- and DREB1A-boxes in the c3 promoter. Additionally, one substitution of T to C was identified at position −53.
In another case, a shortened c3 promoter was constructed by amplification with oligonucleotides OMM 459 (5′-CATTGCGAATTCGTCCAGCCGTTGCTATGG-3′) and OMM 460 (see above) and cloned into pSK36. It ranges from positions −300 to +142. The resulting vector was named pDI23. Based on this vector, the E-box as well as the DREB1A-boxes at positions −130 and +72 were replaced individually by the restriction target site for HindIII (AAGCTT). pDI26, in which the E-box was replaced by AAGCTT, was constructed by SOE-PCR using mutation primers OMM 475 (5′-GCTCGACGCCGACCAAAGCTTTTTCACACACAAATAATTGCGAATTTGGG-3′) and OMM 470 (5′-TGGTCGGCGTCGAGC-3′) and flanking primers OMM 460 and OMM 459. For construction of pDI27, in which the DREB1A-box at −130 was replaced by AAGCTT, mutation primers OMM 471 (5′-CGCACTCGCGCTCGACAAGCTTCACAGATGTTTCACACACAAATAATTGC-3′) and OMM 463 (5′-GTCGAGCGCGAGTGCG-3′) and flanking primers OMM 459 and OMM 460 were used. In the case of pDI28, in which the DREB1A-box at +72 was replaced by AAGCTT, mutation primers OMM 464 (5′-AAATTGAGAGGACGAGTGGAGC-3′) and OMM 472 (5′-GCTCCACTCGTCCTCTCAATTTAAGCTTTCGAACCGTTTCCACACG-3′) and flanking primers OMM 459 and OMM 460 were used. DNA sequencing of the resulting vectors confirmed all of these changes.
Transformations of C. reinhardtii, and cRLUC Activity Measurements
Transformations of C. reinhardtii wild-type strain SAG73.72 and of the per1 mutant CC1117 were carried out as described previously using zeocin as selection marker (Kiaulehn et al., 2007). cRLUC activity measurements were carried out as described before (Kiaulehn et al., 2007).
Automated Measurement of Circadian Phototaxis (Photoaccumulation Assay)
Circadian phototaxis measurement was done with a self-made phototaxis machine, developed by Mergenhagen (1984). Preparation of cell culture for the assay, phototaxis measurement, and data evaluation were done as described previously (Iliev et al., 2006b; Schmidt et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Immunodetection of the BLE-cRLUC fusion protein in different transgenic lines with an anti-RLUC antibody shows its correct expression with a size of 52 kD.
Supplemental Figure S2. The per1 mutant has a lengthened period in comparison with wild-type strain 137C.
Supplementary Material
Acknowledgments
We thank Elena Govorunova, Volker Wagner, and Carsten Milkowski for helpful comments on the manuscript, Markus Fuhrmann for giving us pRbcBRL(HSP196), Lib Harris at the Chlamydomonas Center at Duke University for sending us the per1 mutant, Dieter Mergenhagen for the donation of the automated phototaxis machine, and Monika Fiedler for technical assistance with the measurement of phototaxis. We also appreciate the free delivery of the EST and genome sequences from the genome projects in the United States (Joint Genome Institute) and Japan. We thank Harald Paulsen, who inspired our work with his idea that the C1 and C3 duo of RNA-binding proteins might be involved in temperature compensation.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Mi 373/6 and 7 to M.M.) and the Alexander von Humboldt Stiftung (grant no. Z:3.4–FoKoop–DEU1124590 to M.M.). S.B.S. has a PhD fellowship from the Studienstiftung des Deutschen Volkes.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Maria Mittag (m.mittag@uni-jena.de).
The online version of this article contains Web-only data.
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25 1263–1274 [DOI] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS (1997) Functional identification of the mouse circadian clock gene by transgenic BAC rescue. Cell 89 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4 1633–1649 [DOI] [PubMed] [Google Scholar]
- Bruce VG (1970) The biological clock in Chlamydomonas reinhardtii. J Protozool 17 328–334 [Google Scholar]
- Bruce VG (1972) Mutants of the biological clock in Chlamydomonas reinhardtii. Genetics 70 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VG (1974) Recombinants between clock mutants of Chlamydomonas reinhardtii. Genetics 77 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner M, Merrow M (2008) The green yeast uses its plant-like clock to regulate its animal-like tail. Genes Dev 22 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner AC, Schafmeier T, Merrow MW, Brunner M (2005) Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev 19 1968–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Lee JK, Fenn S, Tjhen R, Stroud RM, James TL (2007) X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains poly(C)-binding protein-2. RNA 13 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC (2006) Proteins in the Neurospora circadian clockworks. J Biol Chem 281 28489–28493 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ (2005) Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, Messeguer X (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31 3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Hausherr A, Ferbitz L, Schödl T, Heitzer M, Hegemann P (2004) Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol Biol 55 869–881 [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego
- Hastings JW, Sweeney BM (1957) On the mechanism of temperature independence in a biological clock. Proc Natl Acad Sci USA 43 804–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama M, Okumura Y, Takahashi E, Shimabukuro A, Tamura M, Takeda N, Kubo T, Kido H (2007) Identification and analysis of the promoter region of the type II transmembrane serine protease polyserase-1 and its transcript variants. Biol Chem 388 853–858 [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77 61–68 [DOI] [PubMed] [Google Scholar]
- Hunt SM, Elvin M, Crosthwaite SK, Heintzen C (2007) The POS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes Dev 21 1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Mittag M (2006. a) The circadian system of Chlamydomonas reinhardtii. Biol Rhythm Res 37 323–333 [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M (2006. b) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe R, Hwang S, Herrin DL (2000) Requirement for cytoplasmic protein synthesis during circadian peaks of transcription of chloroplast-encoded genes in Chlamydomonas. Plant Mol Biol 44 699–709 [DOI] [PubMed] [Google Scholar]
- Kiaulehn S, Voytsekh O, Fuhrmann M, Mittag M (2007) The presence of UG-repeat sequences in the 3′-UTRs of reporter luciferase mRNAs mediates circadian expression and determine acrophase in Chlamydomonas reinhardtii. J Biol Rhythms 22 275–277 [DOI] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH (1993) Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA 90 5672–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Okamoto K, Tabata S, Fukuzawa H, Ishiura M (2005) Identification of novel clock-controlled genes by cDNA macroarray analysis in Chlamydomonas reinhardtii. Plant Mol Biol 57 889–906 [DOI] [PubMed] [Google Scholar]
- Liu N, Zhong NQ, Wang GL, Li LJ, Liu XL, He YK, Xia GX (2007) Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta 226 827–838 [DOI] [PubMed] [Google Scholar]
- Liu Y, Garceau NY, Loros JJ, Dunlap JC (1997) Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell 89 477–486 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S (1998) Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J 14 441–447 [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergenhagen D (1984) Circadian clock: genetic characterization of a short period mutant of Chlamydomonas reinhardtii. Eur J Cell Biol 33 13–18 [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farré D, Nuñez O, Martìnez J, Albà MM (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18 333–334 [DOI] [PubMed] [Google Scholar]
- Mittag M (1996) Conserved circadian elements in phylogenetically diverse algae. Proc Natl Acad Sci USA 93 14401–14404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M, Kiaulehn S, Johnson CH (2005) The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol 137 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth E, Bressan RA, Yun D-J, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V, Philipp K, Zimmer HG, Mesecke S (1979) A simple, versatile, sensitive and volume-independent method for quantitative protein determination which is independent of other external influences. Hoppe Seylers Z Physiol Chem 360 1657–1670 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS (1954) On temperature independence in the clock system controlling emergence time in Drosophila. Proc Natl Acad Sci USA 40 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing L, Ruoff P (2002) Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol Int 19 807–864 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Woodbury, NY
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, et al (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Raffelt M, Lodha M, Schroda M (2007) Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J 52 286–295 [DOI] [PubMed] [Google Scholar]
- Wagner V, Fiedler M, Markert C, Hippler M, Mittag M (2004) Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett 559 129–135 [DOI] [PubMed] [Google Scholar]
- Waltenberger H, Schneid C, Grosch JO, Bareiß A, Mittag M (2001) Identification of target mRNAs from C. reinhardtii for the clock-controlled RNA-binding protein Chlamy1. Mol Genet Genomics 265 180–188 [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al (2007) Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131 927–939 [DOI] [PubMed] [Google Scholar]
- Wozniak RJ, Boyer ME, Grass JA, Lee Y, Bresnick EH (2007) Context-dependent GATA factor function: combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J Biol Chem 282 14665–14674 [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE (2006) PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY1 represents a novel type heteromer of RNA recognition motif and lysine homology-containing subunits. Eukaryot Cell 3 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.