Abstract
The regulation of abscisic acid (ABA) biosynthesis is essential for plant responses to drought stress. In this study, we examined the tissue-specific localization of ABA biosynthetic enzymes in turgid and dehydrated Arabidopsis (Arabidopsis thaliana) plants using specific antibodies against 9-cis-epoxycarotenoid dioxygenase 3 (AtNCED3), AtABA2, and Arabidopsis aldehyde oxidase 3 (AAO3). Immunohistochemical analysis revealed that in turgid plants, AtABA2 and AAO3 proteins were localized in vascular parenchyma cells most abundantly at the boundary between xylem and phloem bundles, but the AtNCED3 protein was undetectable in these tissues. In water-stressed plants, AtNCED3 was detected exclusively in the vascular parenchyma cells together with AtABA2 and AAO3. In situ hybridization using the antisense probe for AtNCED3 showed that the drought-induced expression of AtNCED3 was also restricted to the vascular tissues. Expression analysis of laser-microdissected cells revealed that, among nine drought-inducible genes examined, the early induction of most genes was spatially restricted to vascular cells at 1 h and then some spread to mesophyll cells at 3 h. The spatial constraint of AtNCED3 expression in vascular tissues provides a novel insight into plant systemic response to drought stresses.
The phytohormone abscisic acid (ABA) plays a central role in responses to abiotic and biotic stresses, such as drought, salinity, low temperature, and pathogen attack (Zeevaart and Creelman, 1988; Zhu, 2002; de Torres-Zabala et al., 2007). Plants accumulate ABA when they are subjected to drought stress, and these changes in cellular ABA levels trigger the activation of numerous stress-responsive genes and the closure of stomata to restrict transpiration (Schroeder et al., 2001; Shinozaki and Yamaguchi-Shinozaki, 2007).
The details of de novo ABA biosynthesis in higher plants have been worked out in the last decade (Nambara and Marion-Poll, 2005). Molecular genetic studies of ABA-deficient mutants from various plant species contributed to the identification of genes involved in the ABA biosynthetic pathway (Seo and Koshiba, 2002; Schwartz et al., 2003; Xiong and Zhu, 2003). Based on these studies, it has become clear that ABA is synthesized from zeaxanthin, a C40 carotenoid. The conversion of zeaxanthin to xanthoxin, which is the C15 intermediate, is catalyzed in plastids by possibly four distinct enzymes: zeaxanthin epoxidase (Marin et al., 1996; Agrawal et al., 2001; Xiong et al., 2002), neoxanthin synthase (North et al., 2007), an unidentified epoxycarotenoid isomerase, and 9-cis-epoxycarotenoid dioxygenase (NCED; Schwartz et al., 1997; Tan et al., 1997; Qin and Zeevaart, 1999; Iuchi et al., 2000, 2001). Xanthoxin is then converted to ABA via abscisic aldehyde in the cytosol (Sindhu and Walton, 1987). The oxidation of xanthoxin to produce abscisic aldehyde is catalyzed by AtABA2, a short-chain dehydrogenase/reductase in Arabidopsis (Arabidopsis thaliana; Cheng et al., 2002; Gonzalez-Guzman et al., 2002). In turn, the conversion of abscisic aldehyde to ABA is catalyzed by Arabidopsis aldehyde oxidase 3 (AAO3; Seo et al., 2000b).
A variety of studies have indicated that the carotenoid cleavage reaction catalyzed by NCEDs is a key regulatory step in ABA biosynthesis (Qin and Zeevaart, 1999; Thompson et al., 2000; Iuchi et al., 2001). In several plant species, it has been shown that transgenic plants constitutively expressing the NCED gene accumulated higher amounts of ABA in their leaves and seeds compared with the wild type (Thompson et al., 2000; Iuchi et al., 2001; Qin and Zeevaart, 2002). Among the nine Arabidopsis genes encoding carotenoid cleavage dioxygenase, five (AtNCED2, -3, -5, -6, and -9) are implicated in ABA biosynthesis (Iuchi et al., 2001; Toh et al., 2008). Several features make the AtNCED3 gene particularly interesting with respect to its role in stress responses. First, the transcript levels of AtNCED3 have been shown to increase rapidly in response to dehydration, while those of other AtNCED genes showed almost no response to drought stress (Iuchi et al., 2001; Tan et al., 2003). Furthermore, plants with a knocked-out (or knocked-down) AtNCED3 have been shown to exhibit enhanced transpiration in turgid conditions and higher sensitivity to dehydration. In contrast, transgenic plants overexpressing AtNCED3 have enhanced stress tolerance (Iuchi et al., 2001). However, despite its apparent importance in stress physiology, the regulatory mechanisms of AtNCED3 gene expression in response to drought remain elusive.
ABA and its catabolites are mobile, possibly through the phloem and xylem flow (Zeevaart and Boyer, 1984; Wilkinson and Davies, 1997; Sauter et al., 2002). Grafting experiments have indicated that the shoot genotype is more important than that of the root to supply the active ABA pools in whole plants (Fambrini et al., 1995; Holbrook et al., 2002). In this respect, Christmann et al. (2005) utilized transgenic plants expressing an ABA-inducible reporter gene construct to monitor the active ABA pools in whole plants. The induction of the reporter gene was observed primarily in vascular tissues and guard cells in shoots when the root was subjected to osmotic stresses. In the short term, the osmotic stress in roots stimulated the expression of the reporter gene in vascular tissues of cotyledons, and ultimately expression spread throughout the cotyledons. After a longer period of stress, intense reporter gene expression was observed in guard cells. These results suggest that osmotic stress-induced ABA biosynthesis is activated by an unknown mobile signal(s) emanating from the root and that stress-induced ABA also moves quickly throughout the plants (Christmann et al., 2005, 2007).
Drought stress triggers several stress responses. Multiple drought stress signals, including ABA, are thought to mediate ABA-dependent and ABA-independent pathways to regulate the expression of various drought-inducible genes (Shinozaki and Yamaguchi-Shinozaki, 2007). One important upstream node of stress signaling is drought-induced ABA biosynthesis, and drought-induced AtNCED3 expression is the committed step of the following downstream ABA-dependent stress responses. Despite its importance, the regulation of AtNCED3 expression remains unknown. Elucidation of the site of stress-induced ABA biosynthesis is of particular importance to understanding ABA-dependent stress signaling, because the sites of ABA biosynthesis and action might be different due to the mobile nature of ABA. Moreover, the determination of stress-induced ABA biosynthesis sites is key to envisioning the molecular mechanisms of ABA movement.
Considerable effort has been undertaken to elucidate the regulatory mechanisms of ABA biosynthesis; however, our understanding of the spatial localization of ABA biosynthetic enzymes is still fragmentary (Cheng et al., 2002; Tan et al., 2003; Koiwai et al., 2004). To determine the spatial expression patterns of ABA biosynthetic enzymes during dehydration in Arabidopsis, we conducted immunological analyses using specific antibodies against AtNCED3, AtABA2, and AAO3. Our results showed that an increase in AtNCED3 was restricted to vascular parenchyma cells, in which the AtABA2 and AAO3 were localized. Furthermore, the increase in AtNCED3 mRNA in response to drought stress was found in vascular tissues. These findings provide new insights into the systemic regulation of drought stress responses.
RESULTS
Production of Specific Antibodies against ABA Biosynthetic Enzymes
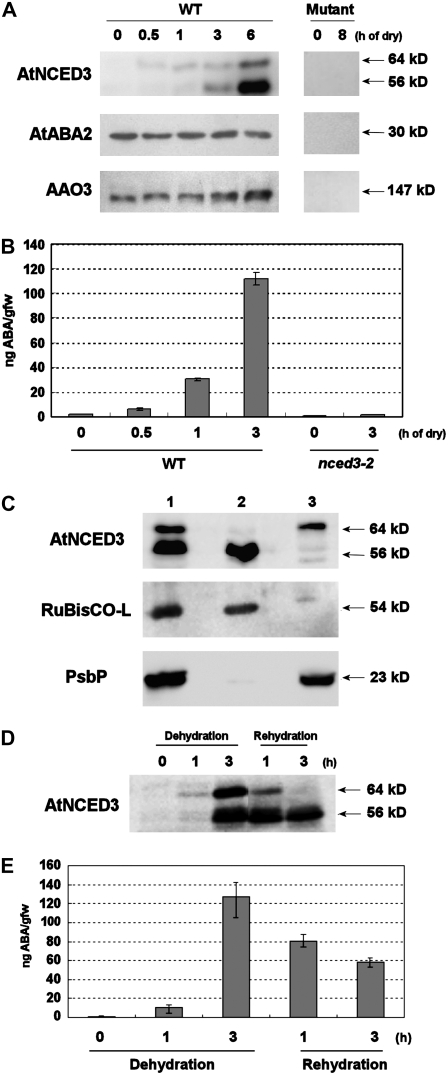
To set up the experimental system for the study of tissue-specific localization of the entire set of ABA biosynthetic enzymes, we prepared specific antibodies against AtNCED3, AtABA2, and AAO3 (see “Materials and Methods”). His-tagged full-length proteins expressed in Escherichia coli and yeast were used for the production of antibodies against AtNCED3 and AtABA2, respectively. Antibodies against AAO3 used in this study were reported previously (Koiwai et al., 2004). The specificity of these antibodies was confirmed by determining that only a single band could be detected in E. coli or yeast expressing each corresponding enzyme. These antibodies were then used for western-blot analysis on turgid and drought-stressed Arabidopsis plant extracts (Fig. 1A). The antibody against AtNCED3 did not detect any specific signals in the extracts of turgid plants. However, in dehydrated plants, a single 64-kD band was detected at 30 min after dehydration treatment, and an additional 56-kD band was detected after 1 h of dehydration (Fig. 1A, left). Both bands were not detected in the nced3 mutant (Fig. 1A, right), confirming that both are products of the AtNCED3 gene. To determine whether AtNCED3 protein accumulation was associated with increased ABA levels, we next measured ABA levels in dehydrated plants (Fig. 1B). The ABA levels were increased slightly at 30 min after the onset of dehydration, with further profound increases thereafter. This drought-induced ABA accumulation was AtNCED3 dependent, because the nced3 mutant did not show such increases (Fig. 1B). Antibodies against AtABA2 and AAO3 recognized single protein bands of 30 and 147 kD, respectively (Fig. 1A, left). These bands were not observed in the corresponding mutants, aba2 and aao3 (Fig. 1A, right). In contrast to AtNCED3, AtABA2 protein levels were similar in both turgid and drought-stressed plants. On the other hand, the AAO3 protein levels increased slightly at 6 h after dehydration, which became more pronounced after 12 h of dehydration (data not shown). In a previous study, Seo et al. (2000a) reported that the protein levels of AAO3 were not increased by dehydration stress despite the up-regulation of its gene. This difference might be due to the higher specificity of the antibody used in this study.
Figure 1.
Specific recognition of the ABA biosynthesis enzymes by polyclonal antibodies. A, Western-blot analysis of protein extracts from turgid and dehydrated plants. Two-week-old wild-type and mutant plants (AtNCED3, nced3-1; AtABA2, aba2-2; and AAO3, aao3-4) were subjected to dehydration as indicated at top. Forty micrograms of total protein was loaded in each lane and immunoreacted with antibodies against AtNCED3, AtABA2, and AAO3. The molecular masses of ABA biosynthetic enzymes are indicated along the right side. B, ABA measurement of wild-type and nced3-2 mutant plants after dry stress treatment. Two-week-old wild-type and nced3-2 mutant plants were subjected to dehydration treatment during 0 to 3 h (as indicated at bottom). ABA levels normalized by initial fresh weigh prior to dehydration are shown. Error bars indicate se (n = 3). C, Western-blot analysis of chloroplast proteins from dehydrated plants. Intact chloroplasts were isolated from wilted Arabidopsis rosette leaves using a Percoll gradient. Proteins from the intact chloroplast (lane 1), stroma plus envelope membrane (lane 2), and thylakoid fraction (lane 3) were analyzed by western blotting with anti-AtNCED3, anti-spinach Rubisco-L, and anti-PsbP antibodies, respectively. The molecular masses of AtNCED3, Rubisco-L, and PsbP are indicated along the right side. D, Western-blot analysis of AtNCED3 upon rehydration. Analysis was performed with protein samples extracted from 2-week-old plants subjected to 0 to 3 h of dehydration and 1 and 3 h of subsequent rehydration. AtNCED3 was detected using anti-AtNCED3 antibodies. Molecular masses are indicated along the right side. E, Changing endogenous ABA levels during dehydration and rehydration. Two-week-old wild-type plants were subjected to dehydration for 0, 1, and 3 h. For rehydration treatment, plants were rehydrated for 1 or 3 h after 3 h of dehydration (as indicated at bottom). ABA levels normalized by initial fresh weigh prior to dehydration are shown. Error bars indicate se (n = 3).
To understand the role of the two forms of AtNCED3, we explored the distribution of these forms in chloroplasts isolated from dehydrated rosette leaves. Thylakoid-localized NCED is thought to be the active form (Qin and Zeevaart, 1999). Antibodies against ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (Rubisco-L) and PsbP proteins were used to confirm stroma and thylakoid fractions, respectively (Nishimura and Akazawa, 1974; Ifuku et al., 2005). Immunoblot analysis of these marker proteins showed that cross-contaminations between thylakoid and stroma fractions were negligible. Western-blot analysis using the fractionated chloroplast proteins revealed that the 64-kD form was mainly located in the thylakoid membrane, while the other form (56 kD) was found in the stroma (Fig. 1C). We then examined the protein levels of two AtNCED3 forms during rehydration following dehydration. After 1 h of rehydration, levels of both the 64- and 56-kD forms started to decline (Fig. 1D). After 3 h of rehydration, the 64-kD form was not detectable, while a significant amount of the 56-kD form still remained (Fig. 1D). This result showed that the protein levels of the 64-kD form were correlated with the changing ABA levels (Fig. 1E).
Tissue-Specific Localization of AtNCED3, AtABA2, and AAO3 in Rosette Leaves
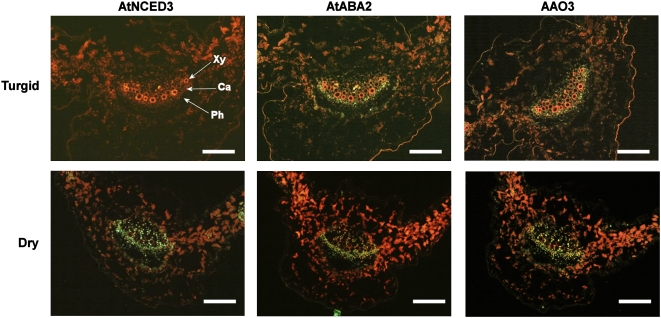
The localization of AtNCED3, AtABA2, and AAO3 in rosette leaves was examined, because several studies have suggested that leaves are the main organ of de novo ABA production in response to osmotic stresses (Cornish and Zeevaart, 1985; Thompson et al., 2007). To do this, immunohistochemical analyses were performed on serial sections of either turgid or dehydrated leaves. In turgid leaves, an intense immunofluorescent signal for AtABA2 was observed in the vascular tissues of the main and lateral veins (Fig. 2). Notably, the fluorescence pattern of AtABA2 was quite similar to that of AAO3 (Fig. 2). These signals were localized most intensively in the parenchyma cells at the boundary between the xylem and phloem bundles in the main veins. On the other hand, in the lateral veins, where the boundary between xylem and phloem was less prominent, the fluorescent signals were distributed more randomly (data not shown). As indicated in the western-blot analysis, we failed to detect a signal in turgid leaves when anti-AtNCED3 antibodies were used (Fig. 2).
Figure 2.
Immunolocalization of ABA biosynthetic enzymes in turgid and dehydrated rosette leaves. Immunostained transverse sections of the main vein of rosette leaves were prepared from turgid or 6-h dehydrated wild-type plants. Serial sections were immunostained with antibodies against AtNCED3, AtABA2, and AAO3. Ca, Cambium; Ph, phloem; Xy, xylem. Green fluorescence indicates specific signals of each ABA biosynthetic enzyme. Red fluorescence indicates autofluorescence from chloroplasts or xylem vessels. Bars = 100 μm. The reproducibility of this experimental result was confirmed by three independent experiments using independent samples prepared at different times.
We then analyzed the localization of ABA biosynthetic enzymes in the dehydrated rosette leaves (Fig. 2, bottom row). The spatial patterns of fluorescent signals of AtABA2 and AAO3 in 6-h dehydrated leaves did not change compared with those of AtABA2 and AAO3 in turgid leaves. In contrast, an intense AtNCED3-dependent signal was observed in stressed leaves. Images of serial sections revealed that AtNCED3 was predominantly localized in the vascular parenchyma cells, where AtABA2 and AAO3 were also localized. In each section, an intense immunofluorescent signal was observed in cambial region and parenchyma cells adjacent to xylem and phloem vessels (Fig. 2).
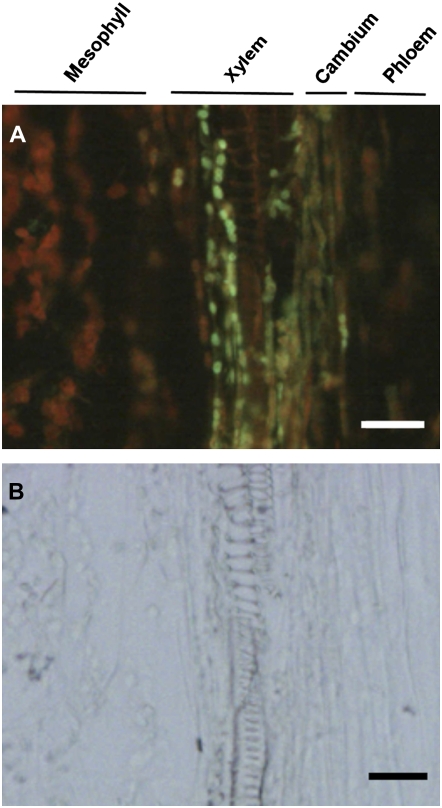
Longitudinal sections of the main vein of wilted rosette leaves were also immunostained with antibodies against AtNCED3. AtNCED3 was detected in the chloroplast of xylem parenchyma next to xylem vessels and cambial tissue between the xylem vessel and phloem sieve tube (Fig. 3). Interestingly, there were no detectable signals in mesophyll cells, where a large amount of carotenoids could be produced (Figs. 2 and 3). We also failed to detect AtNCED3 protein in guard cells of the drought-stressed plants. Immunofluorescent signal for AtNCED3 was not detected in the nced3 mutant (Supplemental Fig. S1), which is consistent with the results of western blots (Fig. 1A). These results clearly show that the ABA biosynthetic enzymes are predominantly expressed in vascular parenchyma of dehydrated leaves.
Figure 3.
Immunolocalization of AtNCED3 in longitudinal sections of dehydrated rosette leaves. Longitudinal sections of the main vein of rosette leaves were prepared from wild-type plants subjected to 6 h of dehydration treatment. Sections were immunostained with anti-AtNCED3 antibodies. Immunofluorescence of AtNCED3 (A) and the corresponding bright-field images (B) are shown. Bars = 20 μm. The reproducibility of these experiments was confirmed by repeating each experiment at least three times.
Tissue-Specific Localization of the AtNCED3 mRNA in the Water-Stressed Leaf Veins
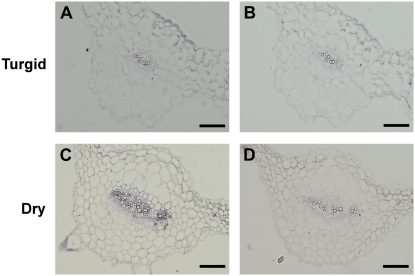
Drought induction of the AtNCED3 protein was tightly restricted to the vascular tissue (Fig. 2). Because drought induction of the AtNCED3 gene is thought to be the committed step in the activation of ABA-dependent stress responses, it is important to investigate whether the activation of the AtNCED3 gene by drought stress is also spatially restricted. To examine this possibility, in situ hybridizations were conducted on sections of drought- or water-treated rosette leaves. When AtNCED3 antisense probe was used, no hybridization signal was observed in the nonstressed rosette leaves (Fig. 4A). On the other hand, intense AtNCED3-dependent signal was observed in the veins of water-stressed rosette leaves (Fig. 4C). We did not find any detectable signal in other tissues of dehydrated leaves, including stomata (data not shown). Only negligible signal was detected when the sense RNA probe was used for hybridization (Fig. 4, B and D). These results show that the spatial pattern of the AtNCED3 transcript reflected that of the AtNCED3 protein and was restricted to the leaf veins after water stress.
Figure 4.
Localization of AtNCED3 mRNA in rosette leaves. In situ hybridization was performed on transverse sections of rosette leaves from nonstressed (A and B) or 3-h drought-stressed (C and D) plants using AtNCED3 antisense (A and C) and sense (B and D) digoxigenin-labeled RNA probes. Bars = 100 μm. The reproducibility was confirmed by repeating identical experiments using three independent sample preparations.
Tissue Specificity of Other Drought-Responsive Gene Expression
Induction of AtNCED3 was observed in vascular parenchyma in response to drought stress. Are the expression patterns of other stress-responsive genes also restricted to vascular cells? To answer this question, we analyzed the tissue specificity of drought-responsive gene expression in mesophyll and vascular cells. To do this, a laser microdissection (LM) technique was used to collect cells from these tissues.
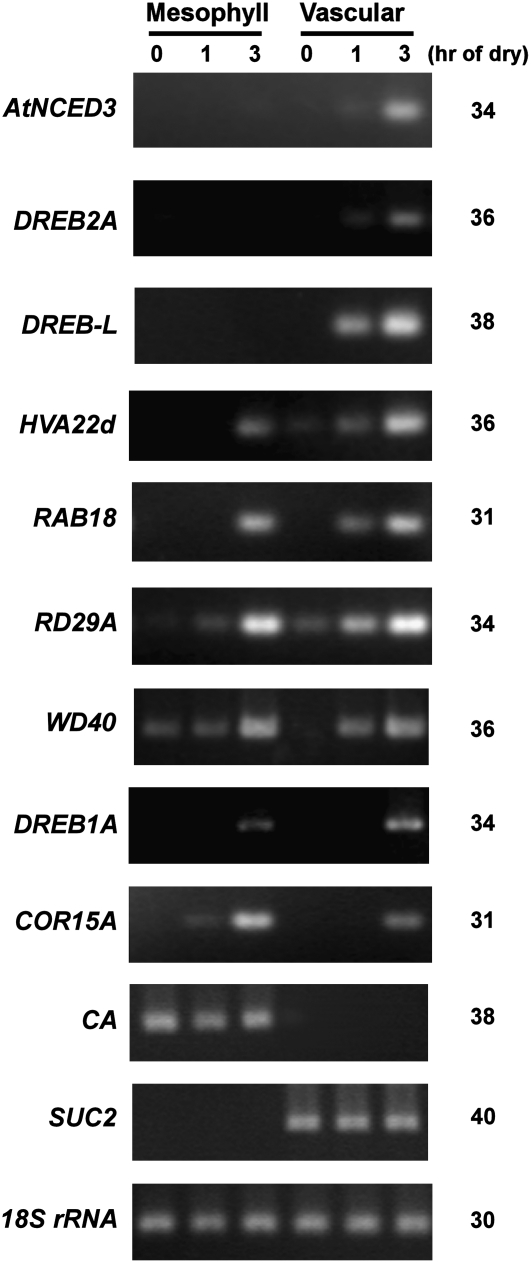
Correct sampling during LM operation was evaluated by monitoring the expression of vascular and mesophyll marker genes by semiquantitative reverse transcription (RT)-PCR. SUC2 (for Suc transporter 2) and CA (for carbonic anhydrase) genes were used as vascular and mesophyll markers, respectively (Ivashikina et al., 2003; Inada and Wildermuth, 2005). The expression of SUC2 was detected only in vascular samples, whereas CA was detected only in mesophyll samples, indicating negligible levels of cross-contamination (Fig. 5). The expression pattern of AtNCED3 was consistent with the in situ hybridization experiment, in which rapid and predominant induction was observed in vascular cells (Fig. 5). Other drought-responsive genes were selected based on their different degrees of ABA inducibility (Seki et al., 2002; Sakuma et al., 2006). Among nine drought-inducible genes tested, six genes (AtNCED3, DREB2A, DREB-L, HVA22d, RAB18, and WD40) showed significant drought induction in vascular cells at 1 h. The drought induction of RD29A was observed in both vascular and mesophyll tissues. Interestingly, COR15A showed its drought induction in the mesophyll at 1 h (Fig. 5). It is also notable that the late induction at 3 h of dehydration was observed more frequently in both vascular and mesophyll cells (six of nine genes examined). The constraint pattern of AtNCED3 expression might be key to the systemic change in drought-induced gene expression.
Figure 5.
Expression patterns of drought-responsive genes on LM mesophyll and vascular cells. RNA samples were extracted from laser microdissected cells and normalized against 18S rRNA (see “Materials and Methods”). Normalized templates were used for semiquantitative RT-PCR. The number of PCR cycles is indicated at right. PCR products were loaded on 4% agarose gels and visualized with ethidium bromide staining. This experiment was repeated twice using independent plant materials, and similar results were obtained.
DISCUSSION
Drought-induced ABA acts as a mobile second messenger for systemic drought stress adaptation responses. Osmotic stress in the root increases the ABA levels in the shoot (Christmann et al., 2005). This systemic response can be mediated by the activation of de novo ABA biosynthesis or by the hydrolysis of ABA Glc ester (Lee et al., 2006). Although the involvement of ABA in the systemic drought response has been reported (Christmann et al., 2005), tissues and cell types that express ABA biosynthesis enzymes in water-stressed plants were not investigated. This study focused on the de novo ABA biosynthesis under dehydration conditions to determine the localization of three important ABA biosynthetic enzymes, AtNCED3, AtABA2, and AAO3. Using immunohistochemical methods, we demonstrated that these three enzymes were localized in vascular parenchyma cells. Furthermore, we showed that the drought stress induction of AtNCED3 occurs in vascular parenchyma, suggesting that these are the cells that perceive the osmotic stress signal to induce the de novo ABA biosynthesis.
Western-blot analysis using the anti-AtNCED3 antibody identified two forms of AtNCED3, a 64-kD thylakoid form and a 56-kD stromal form (Fig. 1C). This result is different from the previous report for the import assay of AtNCED3 using isolated pea (Pisum sativum) chloroplasts (Tan et al., 2003). This is presumably due to the difference in the experimental systems. We analyzed the AtNCED3 levels in vivo, whereas Tan et al. (2003) used an in vitro heterologous system.
The levels of the 64-kD form were correlated with the changing ABA levels during dehydration and subsequent rehydration (Fig. 1D). The thylakoid form was first observed at 30 min and diminished rapidly upon rehydration. On the other hand, the stromal 56-kD form was detected after 1 h of dehydration and remained even at 3 h after rehydration. The protein levels of the stromal form were not correlated with ABA levels (Fig. 1E). It is worth noting that the AtNCED3 detected by immunohistochemical analysis in 6-h dehydrated leaves was mostly the 56-kD form, which is possibly the truncated version of the 64-kD form. Nonetheless, considering the result from in situ hybridization experiments depicted in Figure 4, we assume that the site of the 64-kD form of the AtNCED3 protein is also primarily in vascular parenchyma cells.
Dehydration stress-induced de novo ABA biosynthesis is a common stress avoidance/adaptation mechanism in plants. In Arabidopsis, the dehydration-induced AtNCED3 mRNA accumulation is thought to be the committed step to trigger the ABA-dependent stress signaling (Shinozaki and Yamaguchi-Shinozaki, 2007). In this study, we showed that AtNCED3 mRNA accumulation was restricted spatially to vascular tissues (Figs. 4 and 5). The immunohistochemical analysis also indicated that drought-induced AtNCED3 protein was localized in the vascular parenchyma cells (Fig. 3). It is noteworthy that the early induction of other drought-responsive genes at 1 h of dehydration was observed predominantly in vascular cells (Fig. 5), suggesting that vascular tissues play a key role in the perception of drought stress as well as the systemic responses. Vascular parenchyma cells have been implicated in multiple plant systemic responses, such as auxin polar transport (Galweiler et al., 1998), blue light perception (Sakamoto and Briggs, 2002), and the transport of macromolecules, minerals, and nutrients (Okumoto et al., 2002; Kataoka et al., 2004; Sunarpi et al., 2005; Lough and Lucas, 2006). Elucidation of the mechanisms that regulate stress-induced ABA biosynthesis in this cell type will be the next challenge for a full understanding of the systemic stress adaptation machinery of plants.
We also showed that the AtABA2 and AAO3 proteins are localized in the vascular parenchyma cells in water-stressed plants. The AAO3 protein localization in vascular parenchyma cells of turgid plants was reported previously (Koiwai et al., 2004). Here, we show that this localization of AAO3 was also observed in the drought-stressed plants (Fig. 2). Localization of the AtABA2 and AAO3 proteins in vascular parenchyma cells reinforces the importance of this cell type in drought stress-induced ABA synthesis (Fig. 2). Both in turgid and in water-stressed plants, AtABA2 and AAO3 proteins are most abundant in parenchyma cells at the boundary of xylem and phloem bundles, presumably the cambium cells. These proteins are also present in the parenchyma cells around xylem and phloem bundles, although the parenchyma cells that expressed ABA biosynthetic enzymes tended to be densely located at the cambium side. Interestingly, the localization of drought-induced AtNCED3 coincided with this pattern (Fig. 2). This suggests that these ABA biosynthetic enzyme genes are spatially regulated by a similar or common mechanism. It is noteworthy that these genes are differentially induced by different stimuli, such as sugars, osmotic stress, and ABA itself (Cheng et al., 2002). The spatial constraint of a set of enzymes responsible for the ABA biosynthesis pathway might rely on the efficient production of this hormone rapidly responding to the water stress. Furthermore, we showed that gene expression patterns of many drought-inducible genes changed drastically under temporal and spatial regimes. The spatial constraint of ABA biosynthesis in vascular tissues provides new insight into the direction and kinetics of ABA movement, which is key in the systemic stress response.
MATERIALS AND METHODS
Plant Materials
Wild-type and mutant plants used in this study were Arabidopsis (Arabidopsis thaliana) of the Columbia accession. The nced3-1 (T5004), nced3-2, aba2-2, and aao3-4 mutants have been described previously (Nambara et al., 1998; Iuchi et al., 2001; Seo et al., 2004; Umezawa et al., 2006). Seeds were surface sterilized and sown on GM medium (0.5% [w/v] gelangum, 1× Murashige and Skoog salt mixture, 2.5 mm MES, and 100 mg L−1 myoinositol, pH 5.8). Seeds sown on the medium were incubated at 4°C for 4 d and then grown in a growth chamber at 22°C for 2 weeks under continuous light.
Antibodies
For the preparation of antibodies against AtNCED3 and AtABA2, recombinant proteins of AtNCED3 and AtABA2 containing His6 tags were constructed. The full-length AtNCED3 open reading frame was cloned into the NdeI and BamHI sites of the expression vector pET16b (Novagen). The His6-tagged AtNCED3 protein was expressed in Escherichia coli (BL21 DE3) and was purified according to the manufacturer's instructions (Novagen). The expression and purification of His6-tagged AtABA2 were performed as described by Cheng et al. (2002). These antigens were injected into rabbits (Qiagen) to generate antisera that were affinity purified by membrane purification (Sauer and Stadler, 1993). The generation of AAO3-specific antibody has already been described (Koiwai et al., 2004).
To reduce nonspecific signals in immunoblot analysis, affinity-purified AtNCED3 and AtABA2 antibodies were preabsorbed in an equal volume of phosphate-buffered saline (PBS) containing 0.1% (v/v) Tween 20, complete protease inhibitor cocktail (Roche Diagnostics), and 5% (w/v) acetone powder from 2-week-old plants of nced3-1 and aba2-2 mutants, respectively. The mixtures were gently rotated at 4°C overnight and centrifuged to remove insoluble debris. The clear serum was collected and stored at −30°C until use.
Protein Extraction and Immunoblot Analysis
For drought stress treatment, 2-week-old plants were transferred to filter paper to absorb excess water, and then eight to 10 plantlets were placed on a clear plastic tray (14 × 10 × 1 cm [width × length × depth]). The trays were transferred in a clear plastic container with a cover (22 × 16 × 4.2 cm) to avoid rapid dehydration. The container was incubated in the growth chamber under illumination. The average water loss ratios at 0.5, 1, 3, and 6 h were 8%, 12%, 17%, and 39%, respectively. For rehydration, dehydrated plants were transferred to wet filter paper placed on a plastic tray (14 × 10 × 1 cm). The dehydrated or rehydrated plants were frozen in liquid nitrogen and stored at −80°C. Frozen plant tissue was pulverized in liquid nitrogen, and total protein was extracted in extraction buffer (50 mm Tris-HCl, pH 6.8, 2% [w/v] SDS, and 10% [v/v] glycerol) containing complete protease inhibitor (Roche Diagnostics) and 0.1 mm phenylmethanesulfonyl fluoride. The extracts were cleared by centrifugation (8,000g for 5 min at 4°C), and the amounts of extracted proteins were measured using the Bio-Rad DC protein assay kit. The extracted proteins were mixed with SDS sample buffer and heated at 95°C for 5 min. Forty micrograms of proteins of each sample was separated by SDS-PAGE with 7.5%, 10%, or 12.5% (w/v) polyacrylamide gels for AAO3, AtNCED3, or AtABA2, respectively, and the separated samples were electroblotted to a polyvinylidene fluoride membrane (ATTO). The antigen detection procedure was performed using the ECL Plus kit (Amersham Biosciences) according to the manufacturer's instructions, with minor modifications. Anti-AtNCED3, anti-AtABA2, and anti-AAO3 antibodies were diluted in Can Get Signal solution I (Toyobo) at dilutions of 1:100, 1:200, and 1:5,000 (v/v), respectively. For the detection of fraction marker proteins, anti-Rubisco-L and anti-PsbP antibodies were diluted in PBS containing 0.1% (v/v) Tween 20 at dilutions of 1:5,000 and 1:10,000 (v/v), respectively. In the secondary immunoreaction, horseradish peroxidase-linked IgG (Amersham Biosciences) was used at a dilution of 1:5,000 (v/v) in PBS containing 0.1% (v/v) Tween 20. ECL Plus western-blotting detection reagents (Amersham Biosciences) were used for chemiluminescent signal detection, and the signal was recorded on BioMax XAR film (Kodak). Can Get Signal (Toyobo) was used to enhance antigenic signal.
ABA Measurement
Two-week-old plants (100–200 mg) were used for ABA measurements. Deuterium-labeled d6-ABA (Icon Services) was added to each sample prior to extraction. Procedures for ABA extraction and purification have been described previously (Saika et al., 2007). Purified samples were subjected to liquid chromatography-tandem mass spectrometry (MS/MS) analysis. A binary solvent system was used for separation by liquid chromatography comprising acetonitrile containing water (solvent A) and 0.05% (v/v) acetic acid (solvent B) at a flow rate of 0.2 mL min−1 with a linear gradient of solvent B from 3% to 98% in 10 min. The retention time of ABA and d6-ABA was 4.18 min. MS/MS conditions were as follows: capillary, 2.8 kV; source temperature, 80°C; desolvation temperature, 400°C; cone gas flow, 0 L h−1; desolvation gas flow, 500 L h−1; collision energy, 8.0; MS/MS transition, 263/153 m/z for unlabeled ABA and 269/159 m/z for d6-ABA. A calibration curve was calculated using MassLynx version 4.1 (Micromass). ABA levels were normalized by initial fresh weight measured prior to dehydration treatment. The ratio of dry weight to initial fresh weight did not change remarkably after drought stress treatment (n = 3): average ± sd values were 0.094 ± 0.02, 0.098 ± 0.03, and 0.100 ± 0.004 for turgid, 1-h dehydrated, and 3-h dehydrated wild-type plants, respectively.
Chloroplast Isolation from Arabidopsis Leaves
Chloroplast isolation and fractionation were performed according to the method of Robinson and Mant (2002). Briefly, chloroplasts were isolated from 2-week-old Arabidopsis plants grown on GM medium. Two grams of rosette leaves from 3-h drought-stressed plants was homogenized (Physcotron) in 30 mL of ice-cold grinding buffer, and intact chloroplasts were fractionated on a Percoll gradient as described.
Immunohistochemistry
Sample preparation, cryosectioning, and indirect immunofluorescence staining were performed according to Koiwai et al. (2004). Minor modifications included using PBS containing 3% (w/v) bovine serum albumin to block primary antibodies and 2% (w/v) skim milk to block secondary antibodies. Immunofluorescence images were captured on an epifluorescence microscope (model BX51; Olympus) equipped with a digital camera (model DP50; Olympus).
In Situ Hybridization
Fully expanded leaves of 30-d-old plants were cut from the shoot after 3 h of dehydration treatment. For the turgid controls, shoots were kept on wet filter paper in sealed petri dishes for 3 h. After each treatment, leaves were separated from the shoot, cut into small pieces, and fixed overnight at 4°C in 50 mm sodium phosphate buffer (pH 7.2) containing 4% (w/v) paraformaldehyde and 0.25% (v/v) glutaraldehyde. The tissues were dehydrated with a graded series of ethanol. Ethanol was replaced by t-butyl alcohol followed by liquid paraffin (Paraplast+; Sigma). Embedded tissue was cut into 8-μm sections and mounted on APS-coated glass slides (Matsunami). The paraffin was removed using 100% xylene. The sections were rehydrated with a series of decreasing concentrations of ethanol and then treated with proteinase K (5 μg mL−1) at 37°C for 15 min. Tissues were hybridized with digoxigenin-labeled antisense or sense riboprobes prepared from a 0.95-kb fragment of AtNCED3 (1–950 of AtNCED3 full-length cDNA; AY056255) cloned in pBluescript SK+ (Stratagene). Hybridization signals were visualized using chromogenic substrates, nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate, for alkaline phosphatase-conjugated anti-digoxigenin antibodies (Roche Diagnostics).
Sample Preparation and LM
Arabidopsis plants were grown on soil for 1 month. Shoots were detached and subjected to dehydration treatments for 0, 1, or 3 h. Fully expanded leaves were cut into small pieces (5 × 5 mm2) and fixed in Farmer's fixative (ethanol:acetate, 3:1 [v/v]) overnight at 4°C. Dehydration and paraffin embedding were performed as described by Inada and Wildermuth (2005). Paraffin-embedded samples were sectioned at 16 μm and mounted on PEN membrane glass slides (Molecular Devices) for LM using Veritas laser capture microdissection and the LCC1704 laser cutting system (Molecular Devices). To remove paraffin, slides were placed in 100% xylene, 50% xylene/50% ethanol, and 100% ethanol (v/v) for 5 min at each step and then air dried completely. Vascular (main vein) and mesophyll tissues were dissected from approximately 60 to 80 transverse sections of each sample.
RNA Extraction and Amplification
Total RNA was extracted from LM cells using the PicoPure RNA isolation kit (Molecular Devices). The Quant-iT RiboGreen RNA reagent and kit (Invitrogen) was used for RNA quantification. One nanogram of total RNA was amplified using the WT-Ovation RNA amplification system (NuGEN).
RT-PCR Expression Analysis
Amplified samples were normalized against 18S rRNA levels. Quantitative RT-PCR was performed with the QuantiTect SYBER Green PCR kit (Qiagen) to quantify the AtNCED3 and 18S rRNA mRNA levels. AtNCED3 and 18S rRNA primer sequences were as described by Seo et al. (2004) and Kushiro et al. (2004).
For semiquantitative RT-PCR, normalized samples were diluted and subjected to RT-PCR. PCR products were loaded on 4% agarose gels and visualized by ethidium bromide. Primer sequences were as follows: for DREB2A (At5g05410), forward (5′-AGACTATGGTTGGCCCAATGAT-3′) and reverse (5′-ACACATCGTCGCCATTTAGGT-3′); for DREB-like (At2g20880), forward (5′-GAGGCCACAGCGCAACAC-3′) and reverse (5′-TGGCGTTTCAGGTTCTTTCTG-3′); for HVA22d (At4g24960), forward (5′-TCCATTGTACGCATCGGTGAT-3′) and reverse (5′-CCGTGAGTGAGAGGAACGAATAT-3′); for RAB18, forward (5′-TTGTAACGCAGTCGCATTCG-3′) and reverse (5′-GCCAGATGCTCATTACACACTCA-3′); for RD29A (At5g52310), forward (5′-ACTGTTGTTCCGGTGCAGAAG-3′) and reverse (5′-ACATCAAAGACGTCAAACAAAACAC-3′); for WD-40 repeat protein (At1g78070), forward (5′-ATGGGAGCTATCAGAGCCTTGAG-3′) and reverse (5′-TAACCAGCTTCCGTGTCAAACA-3′); for DREB1A (At4g25480), forward (5′-CCGGAATCAACTTGCGCTAA-3′) and reverse (5′-CAACAAACTCGGCATCTCAAAC-3′); for COR15A (At2g42540), forward (5′-AAAAAACAGTGAAACCGCAGATACA-3′) and reverse (5′-ACTCTGCCGCCTTGTTTGC-3′); for CA (At3g01500), forward (5′-CCTCTCTCCGGCTTCTTTCT-3′) and reverse (5′-GGCAAAAAACTGGCTCGTTAC-3′); and for SUC2 (At1g22710), forward (5′-TTGTGCCTTTCACGATGACTGA-3′) and reverse (5′-GCAAATGGCGAGGATGAAGTTA-3′).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Negative controls of immunohistochemical analysis on cross sections from dehydrated rosette leaves of the corresponding mutants.
Supplementary Material
Acknowledgments
We thank Dr. Dario Bonetta (University of Ontario Institute of Technology) for critical reading of the manuscript; Dr. Makoto Hayashi (National Institute for Basic Biology), Drs. Fumihiko Sato and Kentaro Ifuku (Kyoto University), and Dr. Teruhiro Takabe (Meijo University) for providing anti-spinach Rubisco-L, anti-PsbP, and anti-cytochrome f antibodies, respectively; Dr. Yusuke Jikumaru and Mr. Atsushi Hanada (RIKEN Plant Science Center) for ABA determination; Drs. Katsuhiro Shiono and Nobuhiro Tsutsumi (University of Tokyo) for supporting LM experiments; and Takashi Okamoto (Tokyo Metropolitan University) and Kiyoshi Tatematsu and Shinjiro Yamaguchi (RIKEN Plant Science Center) for valuable discussion. We also thank Ms. Yoshiko Kashiwagi (Yamagata University) and Kunimi Matsumi (Tokyo Metropolitan University) for experimental support and Ms. Yuko Doi for general assistance.
This work was supported by a Grant-in-Aid for Scientific Research B (grant no. 16370026) to T.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eiji Nambara (eiji.nambara@utoronto.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion: tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol 125 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Hoffmann T, Teplova I, Grill E, Müler A (2005) Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol 137 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Weiler EW, Steudle E, Grill E (2007) A hydraulic signal in root-to-shoot signalling of water shortage. Plant J 52 167–174 [DOI] [PubMed] [Google Scholar]
- Cornish K, Zeevaart JAD (1985) Abscisic acid accumulation by roots of Xanthium strumarium L. and Lycopersicon esculentum Mill. in relation to water stress. Plant Physiol 79 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrini M, Vernieri P, Toncelli ML, Rossi VD, Pugliesi C (1995) Characterization of a wilty sunflower (Helianthus annuus L.) mutant. III. Phenotypic interaction in reciprocal grafts from wilty mutant and wild-type plants. J Exp Bot 46 525–530 [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook NM, Shashidhar VR, James RA, Munns R (2002) Stomatal control in tomato with ABA-deficient roots: response of grafted plants to soil drying. J Exp Bot 53 1503–1514 [PubMed] [Google Scholar]
- Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N, Wildermuth MC (2005) Novel tissue preparation method and cell-specific marker for laser microdissection of Arabidopsis mature leaf. Planta 221 9–16 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 325–333 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashikina N, Deeken R, Ache P, Kranz E, Pommerrenig B, Sauer N, Hedrich R (2003) Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J 36 931–945 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004) Root-to-shoot transport of sulfate in Arabidopsis: evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T (2004) Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol 134 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Kim HY, Piao HL, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57 203–232 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39 853–858 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56 165–185 [DOI] [PubMed] [Google Scholar]
- Nishimura M, Akazawa T (1974) Studies on spinach leaf ribulose biophosphate carboxylase. Carboxylase and oxygenase reaction examined by immunochemical methods. Biochemistry 13 2277–2281 [DOI] [PubMed] [Google Scholar]
- North HM, Almeida AD, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50 810–824 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277 45338–45346 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XQ, Zeevaart JAD (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C, Mant A, editors (2002) Import of Protein into Isolated Chloroplast and Thylakoid Membranes, Vol 2. Oxford University Press, Oxford
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48 287–298 [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer A, Stadler A (1993) A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: cloning and heterologous expression in baker's yeast. Plant J 4 601–610 [DOI] [PubMed] [Google Scholar]
- Sauter A, Dietz KJ, Hartung W (2002) A possible stress physiological role of abscisic acid conjugates in root-to-shoot signalling. Plant Cell Environ 25 223–228 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52 627–658 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin XQ, Zeevaart JAD (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, McCarty DR, Zeevaart JAD (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276 1872–1874 [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2 282–291 [DOI] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45 1694–1703 [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T (2000. a) Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23 481–488 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7 41–48 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JAD, Koornneef M, Kamiya Y, Koshiba T (2000. b) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58 221–227 [DOI] [PubMed] [Google Scholar]
- Sindhu RK, Walton DC (1987) Conversion of xanthoxin to abscisic acid by cell-free preparations from bean leaves. Plant Physiol 85 916–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44 928–938 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35 44–56 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Mulholland BJ, Dadswell AR, Blake PS, Symonds RC, Burbidge A, Taylor IB (2000) Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J 23 363–374 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Mulholland BJ, Jackson AC, McKee JM, Hilton HW, Symonds RC, Sonneveld T, Burbidge A, Stevenson P, Taylor IB (2007) Regulation and manipulation of ABA biosynthesis in roots. Plant Cell Environ 30 67–78 [DOI] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, et al (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146 1368–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K (2006) CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J 46 171–182 [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ (1997) Xylem sap pH increase: a drought signal received at the apoplastic face of the guard cell that involves the suppression of saturable abscisic acid uptake by the epidermal symplast. Plant Physiol 113 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem 277 8588–8596 [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JA, Boyer GL (1984) Accumulation and transport of abscisic acid and its metabolites in Ricinus and Xanthium. Plant Physiol 74 934–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39 439–473 [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.