Abstract
Opium poppy (Papaver somniferum) produces a diverse array of bioactive benzylisoquinoline alkaloids and has emerged as a versatile model system to study plant alkaloid metabolism. The plant is widely cultivated as the only commercial source of the narcotic analgesics morphine and codeine. Variations in plant secondary metabolism as a result of genetic diversity are often associated with perturbations in other metabolic pathways. As part of a functional genomics platform, we used 1H nuclear magnetic resonance (NMR) metabolite profiling for the analysis of primary and secondary metabolism in opium poppy. Aqueous and chloroform extracts of six different opium poppy cultivars were subjected to chemometric analysis. Principle component analysis of the 1H NMR spectra for latex extracts clearly distinguished two varieties, including a low-alkaloid variety and a high-thebaine, low-morphine cultivar. Distinction was also made between pharmaceutical-grade opium poppy cultivars and a condiment variety. Such phenotypic differences were not observed in root extracts. Loading plots confirmed that morphinan alkaloids contributed predominantly to the variance in latex extracts. Quantification of 34 root and 21 latex metabolites, performed using Chenomx NMR Suite version 4.6, showed major differences in the accumulation of specific alkaloids in the latex of the low-alkaloid and high-thebaine, low-morphine varieties. Relatively few differences were found in the levels of other metabolites, indicating that the variation was specific for alkaloid metabolism. Exceptions in the low-alkaloid cultivar included an increased accumulation of the alkaloid precursor tyramine and reduced levels of sucrose, some amino acids, and malate. Real-time polymerase chain reaction analysis of 42 genes involved in primary and secondary metabolism showed differential gene expression mainly associated with alkaloid biosynthesis. Reduced alkaloid levels in the condiment variety were associated with the reduced abundance of transcripts encoding several alkaloid biosynthetic enzymes.
The use of opium poppy (Papaver somniferum) as a source of medicine predates recorded history. Archeological evidence suggests that opium poppy was among the first domesticated plant species (Karg and Märkle, 2002; Stika, 2005), and its cultivation continues to have important medical, socioeconomic, and political implications. The plant remains the sole commercial source of the narcotic analgesics morphine, codeine, and semisynthetic derivatives of thebaine, such as oxycodone. More than 80 benzylisoquinoline alkaloids occur in opium poppy, and many have potent pharmacological properties, including morphine and codeine, the antimicrobial agent sanguinarine, the muscle relaxant papaverine, and the antitumorogenic agent noscapine. Most alkaloids accumulate in the multinucleate cytoplasm (i.e. latex) of articulated cells (i.e. laticifers) that form an internal secretory system under a positive turgor similar to sieve elements (Thureson-Klein, 1970). The traditional harvesting of opium (i.e. dried, oxidized latex) involves lancing unripe seed capsules to collect the alkaloid-rich exudate.
In opium poppy, much work has focused on the benzophenanthridine and morphinan alkaloid branch pathways, which has resulted in the identification of many biosynthetic enzymes and the isolation of several cognate cDNAs (Zulak et al., 2006; Ziegler and Facchini, 2008). However, certain key steps remain uncharacterized, including the demethylation/oxidation of thebaine and oripavine to neopinone and morphinone, respectively, and the demethylation of codeine to morphine. Additionally, the biosynthetic pathways leading to noscapine and papaverine are virtually uncharacterized. Moreover, the regulation of alkaloid metabolism remains largely unknown. The degree of regulatory complexity has been indicated by the general unpredictability of metabolic engineering involving the posttranscriptional gene silencing of key biosynthetic genes. Transformation of opium poppy with antisense berberine bridge enzyme, which catalyzes the first step in sanguinarine biosynthesis, caused an altered ratio of alkaloids in latex but not in roots (Frick et al., 2004). Similarly, the apparent shutdown of the entire morphinan pathway at the upstream precursor (S)-reticuline as a result of the RNA interference-mediated silencing of codeinone reductase, the penultimate step in morphine biosynthesis, was unexpected (Allen et al., 2004). Recently, suppression of salutaridinol 7-O-acetyltransferase in opium poppy resulted in the unexpected accumulation of salutaridine instead of the predicted substrate salutaridinol (Allen et al., 2008). Natural and induced mutant varieties of opium poppy with altered alkaloid profiles have been reported. Millgate et al. (2004) used chemical mutagenesis to generate a thebaine-rich variety that did not produce morphine or codeine. The same phenotype has also been reported to occur in natural populations (Nyman, 1978, 1980). Another naturally occurring cultivar, known as ‘Marianne’, accumulates noscapine and narcotoline in addition to morphinan alkaloids (Williams and Ellis, 1989; Frick et al., 2005). The breeding of “opium-free” varieties with extremely low alkaloid content, such as ‘Przemko’, was encouraged, in part, by the social and political ramifications of illicit opium poppy cultivation (Sharma et al., 1999; Németh et al., 2002; Bernáth et al., 2003).
Metabolite analysis in opium poppy has generally involved alkaloid profiling using traditional methods, such as thin-layer chromatography, HPLC, and liquid chromatography-mass spectrometry (LC-MS). Recently, electrospray ionization (ESI)-MS/MS and ESI-Fourier transform-ion cyclotron resonance-MS were used to examine pathway flux in opium poppy seedlings following the feeding of [ring-13C6]tyramine precursor (Schmidt et al., 2007). This approach enabled the identification of both major alkaloids and minor compounds that were previously not detected. Technological advances in the field of plant metabolomics have extended applications to include mutant classification and gene annotation, rendering metabolite profiling an integral platform for functional genomics (Schauer and Fernie, 2006). For example, a statistical comparison of metabolite profiles obtained for Arabidopsis (Arabidopsis thaliana) mutants defective in starch metabolism with uncharacterized mutants showed that plants with similar genotypes clustered together based solely on their metabolic fingerprints (Messerli et al., 2007). Nontargeted metabolite profiling of mutant phenotypes provides clues about the gene(s) involved and can reveal insights into overall pathway regulation. Available analytical technologies include MS, NMR spectroscopy, and vibrational spectroscopy. NMR spectroscopy is a specific, nonselective technology suitable to high-throughput applications and has been used for metabolite profiling for over 20 years (Dunn et al., 2005). Recently, efforts have been made to standardize the use of 1H NMR for metabolomics applications (Rubtsov et al., 2007; Sumner et al., 2007). NMR spectra contain a wealth of qualitative and quantitative information about the components of in vivo or in vitro samples (Ratcliffe and Shachar-Hill, 2005). 1H NMR has been used to characterize foodstuffs and transgenic plants (Choi et al., 2004a; Defernez et al., 2004; Manetti et al., 2004; Moing et al., 2004; Baker et al., 2006; Mattoo et al., 2006), classify species or cultivars (Choi et al., 2004b, 2005; Frédérich et al., 2004; Kim et al., 2005), and analyze stress responses (Choi et al., 2004c, 2006; Liang et al., 2006a, 2006b) and plant-herbivore interactions (Widarto et al., 2006).
As part of our functional genomics program, we have applied 1H NMR metabolite profiling to investigate opium poppy metabolism. Preliminary screening of several opium poppy varieties identified six candidates with unique alkaloid phenotypes. Common problems associated with the analysis of 1H NMR spectra are spectral overlap and low metabolite concentrations, which hinder the identification and quantification of metabolites. We have used a novel approach, known as targeted profiling, to overcome these limitations (Weljie et al., 2006). Chenomx NMR Suite version 4.6 includes a metabolite library constructed by chemically modeling compounds of interest using their peak center and J-coupling information. This library was used to analyze the spectra of plant extracts and create mathematical models for detected metabolites in a cumulative manner. Principle component analysis (PCA) was used to extract and display the systematic variation in the data sets. Chemometric strategies, such as PCA, help to circumvent the overwhelming size and complexity of metabolomic information (Eriksson et al., 2004). Multivariate techniques are routinely used to identify groupings, trends, and outliers within NMR data sets (Trygg et al., 2007). Our results show that variations in alkaloid profile were generally associated with relatively few detectable, but potentially important, differences in the steady-state levels of primary metabolites.
RESULTS
PCA of 1H NMR Spectra Shows Variance in Latex Due to Differential Alkaloid Accumulation
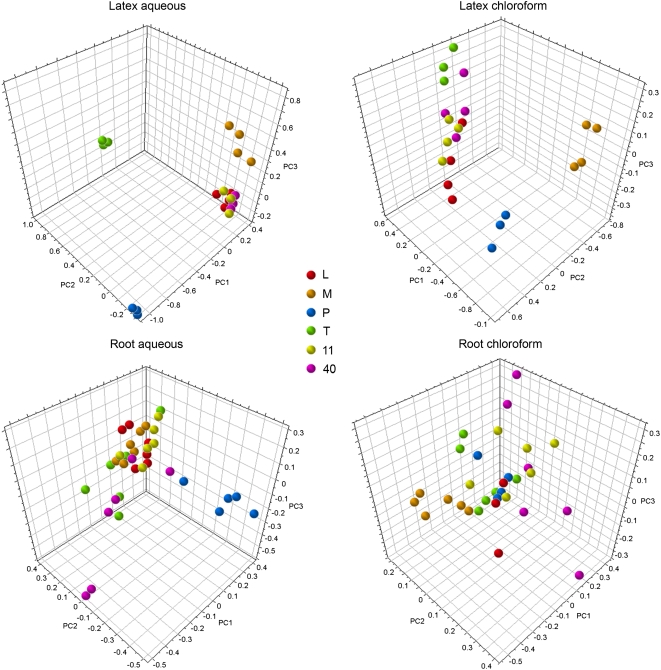
Latex and root extracts were partitioned to reduce overall spectral complexity and separate compounds based on their degree of polarity. Initial inspection of 1H NMR spectra showed greater overall abundance of metabolites in aqueous (i.e. methanol:water [1:1, v/v]) compared with chloroform extracts. Figure 1 shows the three-dimensional score plots for unsupervised PCA analyses of latex and root extracts in both D2O and CDCl3. Greater discrimination between samples of different varieties was observed in latex compared with root. Sample replicates were clustered in aqueous latex extracts, suggesting that technical error was small in comparison with biological variation. The largest degree of variance was observed between ‘Przemko’ (P) and three commercial cultivars designated L, 11, and 40, which contained high levels of morphinan alkaloids. In score plots for both aqueous and chloroform latex extracts, P separated from the commercial cultivars along the PC1 axis (Figs. 1 and 2). Similar results were obtained for aqueous root extracts, although replicates for each variety were less clustered. In aqueous latex extracts, the high-thebaine (T) variety showed the second-highest degree of variance compared with commercial cultivars by separating along PC2, whereas the third principle component distinguished ‘Marianne’ (M) from other cultivars (Fig. 1).
Figure 1.
Score plots for unsupervised, pareto-scaled three-dimensional PCA of NMR spectral data obtained for aqueous or chloroform extracts of opium poppy latex and root tissues. NMR spectra for aqueous and chloroform extracts were obtained in D2O and CDCl3, respectively. Samples representing six different varieties (L, M, P, T, 11, and 40) are color coded as indicated.
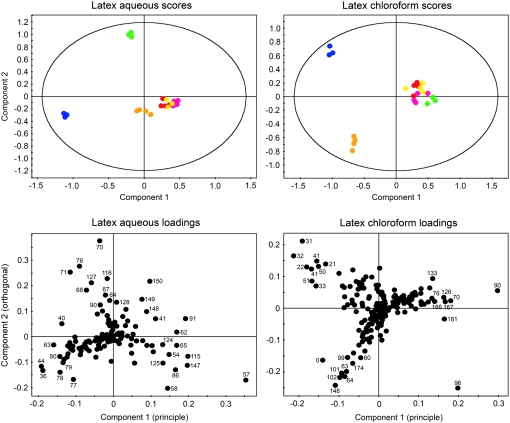
Figure 2.
Score and loading plots for unsupervised, pareto-scaled two-dimensional PCA of NMR spectral data obtained for aqueous or chloroform extracts of opium poppy latex. NMR spectra for aqueous and chloroform extracts were obtained in D2O and CDCl3, respectively. For score plots, samples representing six plant varieties are color coded as follows: L, red; M, orange; P, blue; T, green; 11, yellow; 40, pink. For loading plots, bin numbers corresponding to those listed in Table I (aqueous) and Supplemental Table S1 (chloroform) are indicated.
The loading value of a given variable, or bin, along a PC reflects the commonality between that bin and the particular component (Massart et al., 1988; Eriksson et al., 2001). For NMR data, loading plots can be employed to discern the bins containing the metabolite resonances responsible for variance. Since more variance was observed in latex compared with root tissue, loading plots were generated for NMR spectra of aqueous and chloroform latex extracts (Fig. 2). Corresponding two-dimensional PCA plots are also shown to clarify interpretation.
Latex Aqueous Loadings
Spectral bins having the highest values along component 1 were found to contain resonances of codeine, morphine, and thebaine protons (Fig. 2; Table I), indicating that an absence and/or reduced level of these alkaloids was responsible for the separation of P extracts from those of the commercial cultivars along PC1. Similarly, bins with low values along component 2 were found to contain resonances corresponding to codeine and morphine, indicating that T samples, which display high values along PC2, lack these alkaloids. Gln-containing bins 36 and 44 and tyramine-containing bin 63 had the lowest values along component 1, suggesting an increased level of these compounds in P compared with other varieties. Shifts assigned to oripavine appeared in several bins with high values along component 2, demonstrating the contribution of this alkaloid to variance in T. Other latex metabolites with shifts in bins contributing to variance included γ-aminobutyrate (GABA), Asns, Asp, and Suc.
Table I.
PCA loadings of NMR spectral data obtained for latex aqueous extracts
Bin numbers, corresponding ppm ranges, and values along axes PC1 and PC2 are given (see Fig. 2). Metabolites with proton shifts occurring within the given ppm range are indicated.
| Bin | Range | PC1 | PC2 | Metabolite(s) |
|---|---|---|---|---|
| ppm | ||||
| 36 | 2.09–2.13 | −0.18758 | −0.1322 | Gln |
| 40 | 2.25–2.29 | −0.13576 | 0.050093 | GABA |
| 41 | 2.29–2.33 | 0.112824 | 0.068253 | Codeine, morphine, thebaine |
| 44 | 2.41–2.45 | −0.1903 | −0.11936 | Gln |
| 54 | 2.81–2.85 | 0.148701 | −0.07009 | Asn, Asp, unknown |
| 57 | 2.93–2.97 | 0.354216 | −0.17082 | Codeine |
| 58 | 2.97–3.01 | 0.145427 | −0.20391 | Codeine, morphine |
| 62 | 3.13–3.17 | 0.168642 | 0.016777 | Codeine |
| 63 | 3.17–3.21 | −0.15754 | −0.03232 | Tyramine |
| 64 | 3.21–3.25 | −0.00925 | 0.143199 | Unknown |
| 65 | 3.25–3.29 | 0.168996 | −0.03562 | Codeine, morphine |
| 67 | 3.33–3.37 | −0.01974 | 0.162539 | Unknown |
| 68 | 3.37–3.41 | −0.07023 | 0.18163 | Unknown |
| 70 | 3.45–3.49 | −0.0361 | 0.375795 | Oripavine |
| 71 | 3.49–3.53 | −0.11518 | 0.252514 | Thebaine |
| 76 | 3.69–3.73 | −0.09147 | 0.274855 | Thebaine |
| 77 | 3.73–3.77 | −0.10532 | −0.16877 | Codeine |
| 78 | 3.77–3.81 | −0.14036 | −0.14064 | Codeine |
| 79 | 3.81–3.85 | −0.13255 | −0.1002 | Suc |
| 80 | 3.85–3.89 | −0.14028 | −0.07937 | Suc |
| 86 | 4.09–4.13 | 0.166037 | −0.13023 | Codeine, morphine |
| 90 | 4.25–4.29 | −0.03436 | 0.124292 | Unknown |
| 91 | 4.29–4.33 | 0.193299 | 0.06839 | Codeine, morphine |
| 115 | 5.25–5.29 | 0.199413 | −0.07834 | Codeine, morphine |
| 116 | 5.29–5.33 | −0.01603 | 0.227009 | Oripavine |
| 124 | 5.61–5.65 | 0.13222 | −0.03296 | Codeine, morphine |
| 125 | 5.65–5.69 | 0.132138 | −0.10564 | Codeine, morphine |
| 127 | 5.73–5.77 | −0.05792 | 0.208666 | Oripavine |
| 128 | 5.77–5.81 | 0.008774 | 0.136042 | Oripavine, thebaine |
| 147 | 6.53–6.57 | 0.197496 | −0.11224 | Codeine, thebaine |
| 148 | 6.57–6.61 | 0.089071 | 0.098222 | Thebaine |
| 149 | 6.61–6.65 | 0.077565 | 0.146763 | Oripavine |
| 150 | 6.65–6.69 | 0.09671 | 0.21596 | Oripavine |
Latex Chloroform Loadings
PCA of spectra acquired in CDCl3 showed a separation of P and M from other varieties along both PCs 1 and 2 (Fig. 2). The corresponding loading plot showed that this separation was largely due to the absence and/or reduced level of thebaine in P and M. Nearly all of the bins with high values along the component 1 axis contained exclusively thebaine resonances (Supplemental Table S1). Unambiguous chemical shift assignments and concentration determinations could not be made due to the unavailability of a chloroform-based Chenomx standard library. However, visual inspection of the spectra revealed the presence of thebaine, marked by two distinctive methyl proton singlets at δ 3.85 and δ 3.62, two aromatic proton doublets centered at δ 6.70 and δ 6.64, and characteristic C-ring proton resonances in the 5 to 6 ppm region. After corroborating shift assignments with inspection of 1H-1H total correlation spectroscopy and 13C-1H heteronuclear single quantum coherence spectroscopy spectra, the 1H NMR spectrum for thebaine standard was integrated into the Chenomx Profiler database. Analysis of the spectra using Chenomx software indicated that thebaine was the only detectable alkaloid and, based on PCA evidence, a major contributor to the variance observed in chloroform latex extracts.
Identification and Quantification of Metabolites Using NMR Provides a Biochemical Profile of Different Opium Poppy Varieties
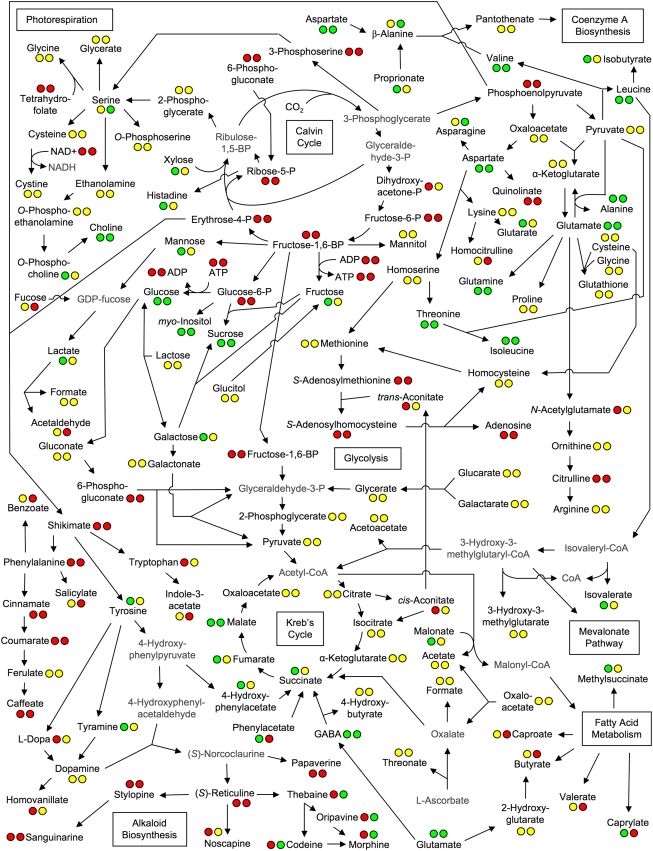
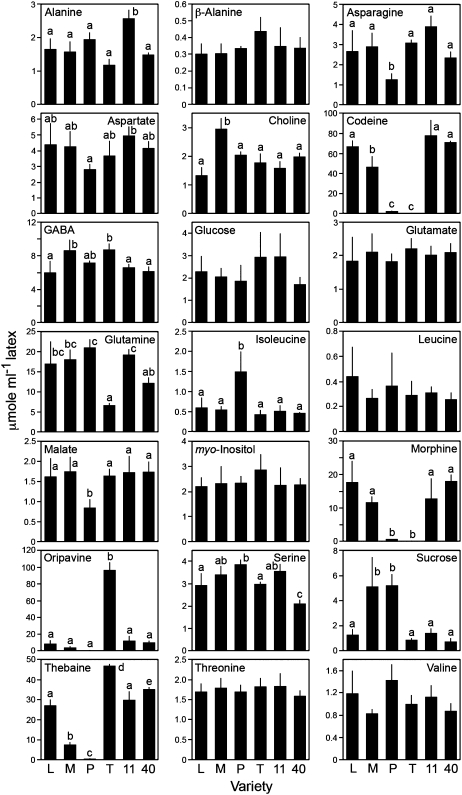
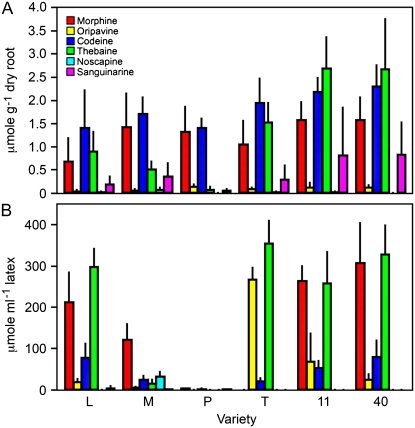
A total of 34 metabolites from root and 21 metabolites from latex were identified and quantified by 1H NMR, with 14 metabolites common to both. Among the detectable metabolites were several classes of compounds, including amino acids, sugars, polyols, fatty/organic acids, amines, and alkaloids. The respective involvement of these compounds in overall plant metabolism, along with metabolites below the limit of detection or whose signals were masked by those of other compounds, is depicted in Figure 3. Neither the presence nor the absence of masked metabolites could be determined due to the domination of spectral regions in which their proton resonances were expected by more abundant compounds, especially sugars and alkaloids. Figure 3 reveals patterns in certain biochemical pathways. For example, metabolite levels of phenylpropanoid intermediates were generally below detection limits. Calvin cycle and sugar phosphate intermediates were not observed, but their sugar end products (e.g. Glc, Fru, and Suc) were present. Fatty acids were absent in latex, and alkaloids were not detected in root. In contrast, morphinan alkaloids were abundant in latex, but sanguinarine, noscapine, and papaverine were not detected. Individual metabolite quantities in latex and roots are shown in Figure 4 and Supplemental Figure S1, respectively. The most abundant metabolites in root were sugars, which presented a challenge in the detection of compounds present in lesser quantities, particularly in the spectral region δ 2.5 to δ 4.5. This problem was not encountered in the analysis of latex spectra, as comparatively little sugar was present. Roots contained large quantities of choline and relatively low levels of its derivative O-phosphocholine. Also abundant in root were metabolites known to be involved, directly or indirectly, in nitrate assimilation, such as Gln, Glu, and malate. In both aerial and underground tissues, Gln was the most abundant amino acid, followed by Glu in roots and Asp, Asn, and Ser in latex.
Figure 3.
Metabolite linkage map representing primary and secondary plant metabolism in opium poppy. The circles associated with each metabolite indicate whether the metabolite was detected (green), not detected (red), or masked (yellow) in root and latex extracts (first and second circles, respectively). Data could not be obtained for metabolites shown in gray because information regarding their standard 1H NMR spectra was not available.
Figure 4.
Individual metabolite quantities in latex as determined by 1H NMR analysis for six varieties of opium poppy. Plant variety abbreviations are the same as in Figure 1. Data are given as means ± se, which were calculated using at least three biological replicates. Letters above the bars indicate pair-wise differences identified by Tukey-Kramer multiple comparison tests. Bars lacking letters were not significantly different. Quantification was achieved using Chenomx NMR Suite software with DSS as the internal standard.
Interestingly, whereas PCA results for aqueous latex extracts indicated that Gln levels contributed to variance in P compared with other varieties (Fig. 2; Table I), no significant differences were observed in the quantities of this compound relative to high-morphine varieties (Fig. 4). This prompted another PCA using individual latex metabolite concentrations rather than spectral bins (data not shown), which indicated that Gln was indeed a factor contributing to variance in P and confirmed that Gln levels, when examined in isolation, did not show variety-specific differences. Glutamine was consistently associated with general trends in metabolite levels, and this relationship was revealed by the metabolite concentration-specific PCA as a factor contributing to variance among the varieties. In contrast with Gln, aromatic amino acids were either present in trace amounts (e.g. 0.5 μmol g−1 Tyr in root) or undetectable. Phe-derived phenylpropanoids were also not detected or masked by chemical shifts of more abundant compounds. Tyramine, the decarboxylation product of Tyr and early precursor to benzylisoquinoline alkaloids (Schmidt et al., 2007), was detected at trace levels in roots (e.g. 0.5 μmol g−1) but was generally masked in latex. Due to a low abundance in latex, putative tyramine shifts could not be assigned with confidence. However, a tyramine signature spectrum was clearly evident in Przemko latex at a mean concentration of 4.67 ± 1.03 μmol mL−1. Other metabolites showing differential accumulation in latex were Suc (i.e. most abundant in M and P), the amino acids Asn and Ile (i.e. lower and higher, respectively, in P compared with alkaloid-accumulating varieties), and Gln (i.e. least abundant in T). Malate was also low in the latex and root of P compared with other varieties (Fig. 4; Supplemental Fig. S1). The absence of alkaloids was the major contributor to variance in P compared with other cultivars and might have masked less substantial differences in metabolites. Significant modulations in the levels of Asn, Ile, and malate were identified using targeted metabolite profiling in addition to chemometric analysis of 1H NMR spectra.
Comparison of Poppy Varieties Reveals Differences in the Alkaloid Content of Latex But Not Roots
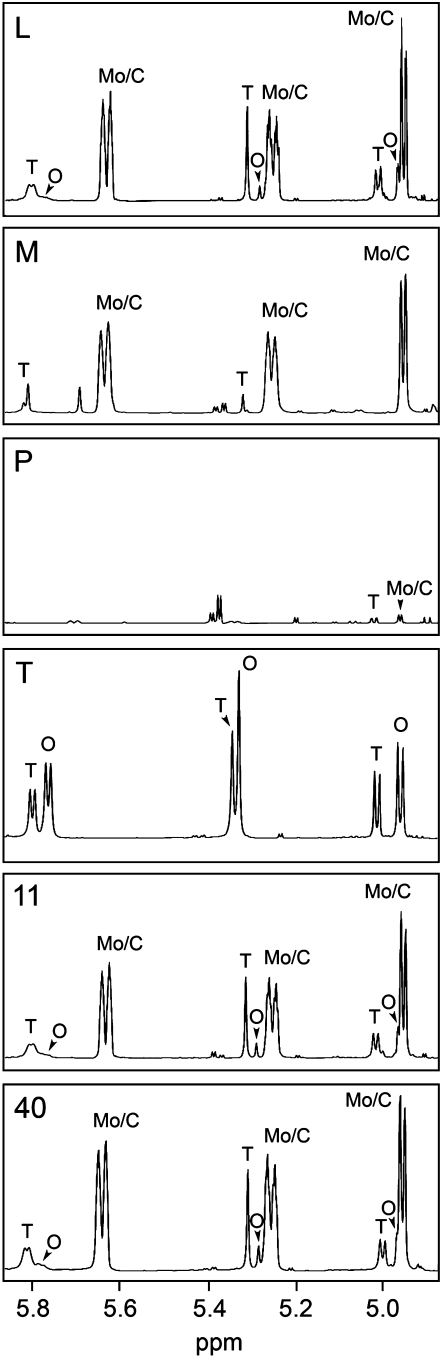
Morphinan alkaloid proton resonances clearly dominated latex 1H NMR spectra. Although a degree of signal overlap occurred in some spectral regions, those assigned to C-ring protons at δ 4.8 to δ 6.0 (Fig. 5), O- and N-methyl protons at δ 3.5 to δ 4.0 (data not shown), and aromatic protons allowed the identification and quantification of morphine, codeine, thebaine, and oripavine. As seen in Figures 4 and 5, dramatically different alkaloid profiles occurred in P and T latex compared with those of high-morphine commercial cultivars. Morphine and codeine levels were 35- to 40-fold higher in the latex of L, 11, and 40 compared with P, whereas thebaine was about 100-fold more abundant. In the latex of T, neither morphine nor codeine was detected. Thebaine and oripavine levels were significantly higher in T relative to the commercial cultivars. Differential alkaloid accumulation was less marked in M compared with the commercial cultivars, although total alkaloid levels, especially thebaine and codeine, were reduced.
Figure 5.
1H NMR spectra of latex aqueous extracts for six varieties of opium poppy. The spectral region corresponding to C-ring protons is shown, with proton resonances for morphine (Mo), codeine (C), thebaine (Th), and oripavine (O) indicated. Comparisons of relative alkaloid abundance may be made between plant varieties, whose abbreviations are the same as in Figure 1.
Proton NMR analysis revealed codeine as the most abundant alkaloid, which likely reflects its higher solubility in water compared with other alkaloids. Morphine and thebaine solubility in water are considerably lower (Yalkowsky and He, 2003). Whereas aqueous extraction was performed with 50% methanol, 1H NMR analysis was done in 100% D2O, favoring increased concentrations of codeine relative to other alkaloids. In turn, 1H NMR in CDCl3 revealed only thebaine, whose second O-methyl group confers additional hydrophobicity. The relative abundances of thebaine in chloroform and aqueous extracts of latex were identical for all six varieties (data not shown). The alkaloids noscapine and narcotoline, which have been reported in the latex of M (Frick et al., 2005), are insoluble in water and were not detected. Although sanguinarine is soluble in chloroform, this alkaloid was not detected in chloroform extracts from roots or latex.
To complement our analysis of benzylisoquinoline alkaloids by 1H NMR, we examined methanol extracts from latex and root tissues by reverse-phase HPLC. An HPLC approach circumvented the problem of low solubility, as alkaloids are generally soluble in methanol. LC-UV is approximately 1,000-fold more sensitive than NMR (Sumner et al., 2003), permitting the specific detection and accurate quantification of alkaloids. In corroboration with the 1H NMR results, large differences in alkaloid levels were observed in the latex of P and T compared with the commercial poppy varieties (Fig. 6). Only trace amounts of alkaloids were detected in P, and morphine was completely absent from T. Mean oripavine levels were 3.5-fold higher in T latex compared with variety 11, which showed the second highest level of this alkaloid. Alkaloids not seen by 1H NMR were detected using LC-UV, including sanguinarine in roots, codeine in the latex of T, and noscapine in the latex and root of M. Narcotoline was detected in the latex of M, although a lack of authentic standard prevented quantification. Representative HPLC chromatograms for latex and root extracts are shown in Supplemental Fig. S2. Compound identity was confirmed by LC-MS. The relative abundance of morphinan alkaloids measured by HPLC was somewhat different (i.e. morphine and thebaine) than that determined by 1H NMR, which likely reflects their differential solubility in D2O compared with methanol.
Figure 6.
Quantification of alkaloids determined by HPLC analysis for six varieties of opium poppy. Plant variety abbreviations are the same as in Figure 1. The different alkaloids are color coded as indicated. Data are given as means ± se, which were calculated using six biological replicates. Compounds were identified by comparing retention times and UV spectra with those of authentic standards. LC-MS analysis of each sample was also performed for further confirmation. Quantification was achieved using standard curves developed for each alkaloid using authentic standards.
An intriguing result was the lack of a significant difference between the alkaloid content in roots of the various cultivars. Although P latex was virtually alkaloid free, morphinan alkaloid levels in P roots were similar to those of the commercial cultivars. Similarly, morphine and codeine were detected in T roots but were absent from latex. Only M roots could be distinguished due to the presence of noscapine.
Different Latex Alkaloid Profiles Are Not Associated with Transcriptional Perturbations in Primary Metabolism
The expression profiles of 30 genes encoding enzymes involved in pathways linked to alkaloid metabolism were determined using quantitative real-time reverse transcription (RT)-PCR to detect possible perturbations in primary metabolism resulting from differential alkaloid production. In addition, the relative expression levels for 12 genes encoding known alkaloid biosynthetic enzymes were measured. The metabolic roles for each of these 42 enzymes are illustrated in Supplemental Figure S3. The metabolic components that were examined included the oxidative pentose phosphate pathway, ammonium fixation and photorespiration, S-adenosyl-Met metabolism, the shikimate pathway, and benzylisoquinoline alkaloid biosynthesis. The relative abundance of each gene transcript in flower buds (Fig. 7), stems (Supplemental Fig. S4), and roots (Supplemental Fig. S5) was determined. Significant differences were detected in the expression of a few primary metabolic genes in flower buds, stems, and roots. In contrast, the abundance of gene transcripts encoding alkaloid biosynthetic enzymes showed considerably more variation. In M flower buds, transcript levels were generally lower for approximately half of the alkaloid biosynthetic genes tested compared with varieties (i.e. L, T, 11, and 40) that showed higher alkaloid accumulation (Fig. 7). These genes encode key upstream enzymes (e.g. Tyr/3,4-dihydroxy-Phe [DOPA] decarboxylase [TyDC] and (S)-coclaurine N-methyltransferase [CNMT]) and downstream, morphinan pathway-specific enzymes (e.g. codeinone reductase [COR]). Variety L showed relatively high expression levels of genes encoding upstream O-methyltransferases (i.e. norcoclaurine 6-O-methyltransferase [6OMT] and 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase [4′OMT]) and morphinan branch pathway-specific enzymes (i.e. salutaridine reductase [SalR] and salutaridinol 7-O-acetyltransferase [SalAT]). Similar differences in alkaloid biosynthetic gene expression profiles were also found in stems and roots (Supplemental Figs. S4 and S5). In variety P, gene transcripts encoding all known alkaloid biosynthetic enzymes were detected in each organ despite the lack of alkaloid accumulation in the latex.
Figure 7.
Quantitative real-time RT-PCR expression analysis of 30 primary metabolic and 12 alkaloid biosynthetic genes in flower buds of six opium poppy varieties. Plant variety abbreviations are the same as in Figure 1. Each bar represents the average of six measurements (two technical replicates on each of three independent biological samples) ± sd. Letters above bars indicate pair-wise differences identified by Tukey-Kramer multiple comparison tests. Bars lacking letters were not significantly different. BBE, Berberine bridge enzyme; CYP80B3, N-methylcoclaurine 3′-hydroxylase; DAHP, deoxy-d-arabino-heptosonate-7-phosphate; EPSP, 5-enolpyruvylshikimate-3-phosphate; NCS1, norcoclaurine synthase; 7OMT, reticuline 7-O-methyltransferase; SAM, S-adenosyl-Met; TNMT, tetrahydroprotoberberine-cis-N-methyltransferase.
DISCUSSION
Benzylisoquinoline alkaloid metabolism in opium poppy is highly complex, involving at least 14 enzyme-catalyzed reactions leading to morphine and beginning with the primary metabolite Tyr. The enzymology of morphine and sanguinarine biosynthesis in opium poppy is largely known, and many biosynthetic genes have been isolated. However, key steps in the branch pathways leading to morphinan, sanguinarine, and noscapine remain uncharacterized at both the enzymatic and molecular levels. Moreover, little is known about the regulation of alkaloid biochemistry in opium poppy. Metabolite and gene transcript profiling were used to investigate metabolism in opium poppy with a focus on the effects of altered alkaloid levels on the accumulation of relevant primary metabolites. Six plant varieties were characterized in an attempt to reveal underlying biochemical perturbations and garner clues about the genes responsible for each chemotype. PCA-based chemometric analysis permitted an examination of variance between different plant varieties. Although the sensitivity of 1H NMR limits the number of compounds that can be analyzed, multivariate analysis of proton spectra revealed modulations in metabolite profile even in the absence of specific identification. The results suggested that alkaloids are the primary factors discriminating two cultivars, P and T, from several high-morphine commercial varieties. Application of Chenomx NMR Suite software enabled the identification and quantification of 34 root and 21 latex compounds within complex sample extracts, providing a novel perspective on the coordination of metabolism in opium poppy. With few exceptions, quantitative data corroborated those obtained by PCA, with significant variation in the levels of morphinan alkaloids for P and T relative to the commercial varieties L, 11, and 40. Lower levels of morphinan alkaloids, especially thebaine, were found in the latex of M compared with varieties L, T, 11, and 40, as shown in the multivariate analyses of both aqueous and chloroform extracts. Elevated Suc levels were observed in cultivars M and P, which showed correspondingly lower alkaloid accumulation in the latex relative to other varieties (Fig. 4). Suc is a distal carbon source for alkaloids (Supplemental Fig. S3), and altered levels of this carbohydrate in latex might reflect overall carbon use modulations. In contrast with M and P, Suc levels were not significantly different in T relative to high-morphine varieties L, 11, and 40. Although morphine and codeine were not detected in T latex (Figs. 4–6), levels of the upstream intermediates thebaine and oripavine were substantially higher in T compared with L, 11, and 40. Carbon requirements for alkaloid biosynthesis were unchanged in T compared with high-morphine varieties, consistent with the approximately stoichiometric replacement of morphine/codeine with thebaine/oripavine.
Despite differences in Suc abundance, genetic variations affecting alkaloid metabolism might not necessarily perturb other primary metabolic compounds, at least in terms of steady-state levels. To support alkaloid biosynthesis, depletion of key precursors such as Tyr must be avoided and metabolic homeostasis must be maintained by the enhancement of flux through certain pathways. Because changes in flux might be associated with the differential regulation of genes encoding metabolic enzymes, we examined the relative transcript abundances of 30 genes encoding enzymes involved in primary metabolic pathways with links to alkaloid biosynthesis. Substantial modulations in the abundance of available primary metabolic gene transcripts were not detected in flower buds (Fig. 7), stems (Supplemental Fig. S4), or roots (Supplemental Fig. S5). However, it cannot be ruled out that compensatory adjustment in the primary metabolism of opium poppy varieties with substantially different alkaloid accumulation profiles might occur in cell types other than laticifers, which contain latex. The metabolite profile of the latex does not necessarily reflect perturbations occurring in neighboring cells. Since the biosynthesis of alkaloids is known to occur in sieve elements (Bird et al., 2003; Samanani et al., 2006), alterations in primary metabolic networks might be more pronounced in phloem tissues.
Metabolomics has been used extensively to characterize root metabolites, especially in legumes and crop plants such as tomato (Solanum lycopersicum), barley (Hordeum vulgare), and maize (Zea mays; Hall, 2006; Ward et al., 2007). Although root metabolite levels are highly species and circumstance specific, compounds detected in abundance often include the amino acids Asn, Gln, and/or Glu. Such was the case for all opium poppy varieties tested with the exception of Asn, which could not be identified in roots due to masking. Nitrate reduction in roots provides the shoot with organic nitrogen, mostly in the form of Gln and Asn, via the transpiration stream in xylem vessels (Heldt, 1997; Satoh, 2006). Malate, which is required for the transport of nitrate assimilation substrates and products across plastid membranes (Lam et al., 1996), was also abundant in roots. Malate was lower in the latex (Fig. 4) and roots (Supplemental Fig. S1) of P compared with alkaloid-accumulating varieties, potentially reflecting the reduced demand for nitrogen in support of alkaloid metabolism. The substantially reduced biosynthesis of alkaloids in P appeared to alter the abundance of certain amino acids in the latex, with lower levels of Asn and higher levels of Ile compared with alkaloid-producing cultivars. These modulations were not observed in M, which displayed only a reduced accumulation of alkaloids compared with cultivars L, T, 11, and 40. Amino acid levels in phloem and xylem sap are tightly regulated to satisfy the nitrogen requirements of various organs, especially seeds (Lam et al., 1995). In opium poppy, alkaloid production likely increases the demand for specific amino acids, such as Glu and, more directly, Tyr. In general, aromatic amino acid levels were either low or not detectable in latex and root, suggesting a diversion of flux from the shikimate pathway toward alkaloid biosynthesis. However, few significant modulations were detected in aromatic amino acid levels in the latex of P compared with other varieties (Fig. 4).
In roots, choline was highly abundant in contrast with relatively low levels of O-phosphocholine. Although choline serves as a precursor to the osmoprotectants betaine and Gly betaine in many plants, such compounds, to our knowledge, have not been reported previously in opium poppy. Choline is also required for the production of the ubiquitous membrane component phosphatidylcholine via the CDP-choline pathway, which is known to operate in Arabidopsis, Brassica napus, and Pisum sativum (Jackowski and Fagone, 2005). The nonprotein amino acid GABA, a metabolite generally associated with the stress response in plants, was abundant in latex (Fig. 4). Multiple roles for GABA have been proposed, including defense against insects (Bouché and Fromm, 2004). The high levels of GABA, which acts as a neurotransmitter in both vertebrates and invertebrates, might play a defensive role against herbivores.
The cell-specific localization of alkaloid biosynthetic enzymes (Bird et al., 2003; Samanani et al., 2006) supports a general separation of alkaloid and primary metabolism. Nevertheless, potentially important differences in the steady-state levels of some primary metabolites accompanied major differences in alkaloid profile. However, the limited dynamic range of NMR-based methods coupled with a degree of biological variance might have precluded the identification of other affected metabolites.
Accumulation of Tyramine in Low-Alkaloid Poppy Suggests Altered Metabolism Upstream of Alkaloid Biosynthesis
Our present knowledge about the initial steps of benzylisoquinoline alkaloid biosynthesis is largely based on studies involving feeding experiments using radiolabeled precursors (Roberts et al., 1987; Rueffer and Zenk, 1987; Stadler and Zenk, 1990). The first committed step in alkaloid formation uses dopamine and 4-hydroxyphenylacetaldehyde to produce (S)-norcoclaurine. Dopamine is formed by the decarboxylation of DOPA or the hydroxylation of tyramine. Both DOPA and tyramine are incorporated into opium poppy benzylisoquinoline alkaloids, although their relative contributions in the plant are not clear. Recently, opium poppy seedlings fed with [13C]tyramine were found to accumulate dopamine and thebaine with 49% and 37% 13C content, respectively (Schmidt et al., 2007). In support of previous results involving 13C NMR spectroscopy (Roberts et al., 1987), Schmidt et al. (2007) showed that tyramine was specifically incorporated into the tetrahydroisoquinoline moiety of benzylisoquinoline alkaloids. The accumulation of tyramine in the latex of the low-alkaloid cultivar P, but not in others, suggests a perturbation early in alkaloid metabolism. The conversion of tyramine to its catecholamine derivative dopamine would require ring-hydroxylation at the meta position, but an enzyme catalyzing this reaction has not been isolated. Low alkaloid levels in P could represent a reduction, or block, in the conversion of tyramine to dopamine, resulting in increased tyramine pools.
Altered Alkaloid Accumulation in Latex, But Not Roots, Suggests Differential, Organ-Specific Regulation of Alkaloid Biosynthesis
The biosynthesis and accumulation of alkaloids in opium poppy has been localized to cell types associated with the phloem in all organs of the plant. Alkaloid biosynthetic gene transcripts were detected in companion cells, whereas the corresponding enzymes were found in sieve elements of the phloem (Bird et al., 2003; Samanani et al., 2006). The accumulation of alkaloids in laticifers suggests the intercellular translocation of pathway intermediates and/or products. The continuity of phloem tissues throughout the plant supports the occurrence of alkaloid biosynthetic gene transcripts in flower buds (Fig. 7), stems (Supplemental Fig. S4), and roots (Supplemental Fig. S5). Although alkaloid profiles are different in roots and shoots, alkaloid metabolism is generally a tissue-specific rather than an organ-dependent process.
Differences in alkaloid profile were most prevalent in the latex of P and T (Figs. 4–6). Variety T displays a chemotype similar to that of top1, a mutant obtained by induced mutagenesis (Millgate et al., 2004). Like T, top1 accumulates thebaine and oripavine but not morphine. Millgate et al. (2004) suggested that a gene encoding a single demethylase, or one regulating its activity, might be affected in the top1 mutant. This phenotype might also result from a structural or transport component that prevents thebaine and oripavine from trafficking to the subcellular compartment where O-demethylation occurs. As part of their characterization of top1, Millgate et al. (2004) used a microarray-based approach to examine global changes in gene expression relative to the high-morphine parent. Although several genes were expressed at lower levels in top1, including an ATP-dependent membrane transporter and three putative enzyme-encoding genes, the differential accumulation of transcripts encoding an oxidative, demethylating enzyme that uses thebaine or oripavine was not detected.
An important difference between the phenotypes of T and top1 is the occurrence of codeine in the latex of T. HPLC analysis showed that trace amounts of codeine accumulate in T (Fig. 6), and because codeine is purportedly produced subsequent to the demethylation of thebaine, the genetics responsible for the altered profile of alkaloids in these variants could be different. Thebaine-accumulating, morphine-free opium poppy also occurs in nature among populations of high-morphine plants (Nyman, 1978, 1980). Although the morphine-free phenotype might result from the lack of a single enzyme, a more complex mechanism cannot be ruled out. It is notable that although alkaloid phenotypes are unique in P and T compared with commercial varieties, the root alkaloid profile is unaffected, suggesting that alkaloid metabolism is regulated differently in shoot and root organs. Efforts to engineer alkaloid metabolism by overexpressing the enzyme catalyzing the penultimate step in morphine biosynthesis, COR (Supplemental Fig. S3), increased alkaloid accumulation in the carpel but not in roots, leaves, pollen, or seeds (Larkin et al., 2006). Previously, transformation of opium poppy with antisense berberine bridge enzyme cDNA altered the ratio of alkaloids in latex but not in roots (Frick et al., 2004). Despite the substantial differences in the accumulation of alkaloids in the P and T varieties, transcripts corresponding to all available biosynthetic genes were detected in flower buds, stems, and roots of all cultivars (Fig. 7; Supplemental Figs. S4 and S5). This suggests that different alkaloid accumulation profiles were the result of factors other than the differential expression of known biosynthetic genes. However, the possibility must be considered that variations in the regulation of alkaloid metabolism might result from posttranscriptional events, such as altered enzyme activities related to known biosynthetic genes, or from alterations in the transport of enzymes and/or metabolites.
Reduced Alkaloid Accumulation Is Associated with Lower Alkaloid Biosynthetic Gene Expression
Gene expression profiling revealed a lower abundance of transcripts encoding several alkaloid biosynthetic enzymes (e.g. TyDC, CNMT, and COR) in variety M compared with high-alkaloid varieties (i.e. L, T, 11, and 40). TyDC catalyzes the formation of both tyramine and dopamine, with the latter compound used directly for the norcoclaurine synthase-catalyzed formation of the basic benzylisoquinoline alkaloid skeleton (Supplemental Fig. S3). Three methyl transfer steps, including an N-methylation catalyzed by CNMT, and one hydroxylation reaction lead to (S)-reticuline formation. (S)-Reticuline undergoes conversion to (S)-scoulerine by berberine bridge enzyme or epimerization to (R)-reticuline, leading to the formation of thebaine, codeine, and morphine. Three steps along the morphinan alkaloid branch pathway have been characterized at the biochemical and molecular levels, including SalR, SalAT, and COR. Increased morphinan alkaloid content has been achieved by the overexpression of COR in opium poppy, indicating an important role for this enzyme (Larkin et al., 2006). However, 1H NMR and HPLC analyses of latex alkaloids showed that the major metabolite occurring at the lowest levels in M compared with L, T, 11, and 40 was thebaine (Figs. 4–6), which is a pathway intermediate upstream of COR. An overall reduction in the morphinan alkaloid content of M latex relative to pharmaceutical-grade varieties was observed in agreement with Frick et al. (2005), who reported lower alkaloid levels in M relative to an industrially elite opium poppy variety. The generally reduced abundance of transcripts encoding alkaloid biosynthetic enzymes likely contributes to a reduction in the alkaloid content of the latex.
Interestingly, L flower buds showed an increased relative abundance of several alkaloid biosynthetic gene transcripts, including those encoding 6OMT, 4′OMT, SalR, and SalAT (Fig. 7). However, the elevated transcript levels were not accompanied by increased accumulation of morphine, codeine, or thebaine compared with other high-alkaloid varieties (Figs. 4 and 6). Taken together with the gene expression analysis for M, this suggests that although reduced gene expression might contribute to lower alkaloid levels, elevated gene expression does not necessarily lead to increased flux through alkaloid pathways. Alkaloid biosynthesis is at least partly dependent on a minimum threshold of gene expression. Above this threshold, no further accumulation occurs, but below a minimum level of gene expression, alkaloid biosynthesis is reduced.
CONCLUSION
The nontargeted metabolite profiling analysis presented in this work represents a step toward understanding the consequences of alkaloid biosynthesis on the metabolome of opium poppy. Major differences in latex alkaloid contents of plant varieties were accompanied by significant modulations in the levels of some primary metabolites, but the metabolite profiles detected via the 1H NMR spectra of aqueous and chloroform extracts of opium poppy latex and roots were generally similar. This is in contrast with the elicitor-induced accumulation of sanguinarine in opium poppy cell cultures, in which substantial reprogramming of primary metabolism was associated with the accumulation of sanguinarine and other modulations in secondary metabolism (Zulak et al., 2007, 2008). The apparently limited effect of alkaloid biosynthesis on the primary metabolic profile of the intact plant might reflect the specific cellular localization of alkaloid biosynthetic enzymes and the separation of cell types involved in alkaloid formation and storage (Bird et al., 2003; Samanani et al., 2006). Changes in metabolic flux might also account for the observed differences in alkaloid accumulation profiles without detectable alterations in the steady-state abundance of primary metabolites. Gene expression analysis suggested that modulations in primary metabolic flux are not regulated at the transcriptional level. However, a minimum threshold of alkaloid biosynthetic gene expression appears necessary to support high levels of product accumulation.
MATERIALS AND METHODS
Plant Material
Three commercial, high-morphine varieties (L, 11, and 40) and three natural mutant varieties (M, P, and T) of opium poppy (Papaver somniferum) were cultivated in a growth chamber (Conviron) at 20°C/18°C (light/dark) under high-intensity metal halide lights with a photoperiod of 16 h. Seed capsules were lanced 1 to 2 d after anthesis, and latex was collected and flash frozen with liquid nitrogen. The same plants were used to collect root tissue, which was flash frozen. All tissue was stored at −80°C until further analysis.
Solvents and Chemicals
D2O, CDCl3, 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS), and tetramethylsilane (TMS) were purchased from Sigma-Aldrich. (R,S)-Stylopine was synthesized as described by Liscombe and Facchini (2007), and morphinan alkaloids were provided by Sanofi-Aventis. All other chemicals were purchased from Sigma-Aldrich.
Sample Preparation for NMR Spectroscopy
Immediately following harvest, all tissue was flash-frozen in liquid N2 and kept at −80°C until extraction (approximately 2–4 weeks). The method of Choi et al. (2004a) was used, with modifications, to prepare samples for NMR spectroscopy. Latex (500 μL) was mixed with 500 μL of methanol, and the extract was partitioned using 3 mL of water:methanol (1:1, v/v) and 3 mL of chloroform. The extracts were vortexed for 30 s, sonicated for 4 min at 4°C, and centrifuged at 1,000g to separate the phases and pellet insoluble material. The phases were collected separately, the pellet was reextracted once, and the fractions were pooled and reduced to dryness. For root, 10 g of tissue was ground to a fine powder using liquid nitrogen and extracted with 10 mL of methanol:water (1:1, v/v) and chloroform. Following centrifugation at 10,000g, the upper methanol-water phase was collected and insoluble material at the interface was removed. The lower chloroform phase was filtered using Whatman No. 1 filter paper to remove the remaining insoluble root material, which was then added to the material collected at the phase interface. Following reextraction of the insoluble material, the respective fractions were pooled and reduced to dryness. The insoluble material was dried and weighed. Dried latex and root extracts were suspended in either 1.0 mL of buffered D2O (100 mm KD2PO4, pH 7.0 ± 0.002, 10 mm NaN3, and 0.5 mm DSS) or CDCl3 (0.5 mm TMS).
Sample Preparation for HPLC and LC-MS
To prepare samples for HPLC and LC-MS, 10 μL of latex was suspended in 1.0 mL of methanol, vortexed and incubated at 70°C for 10 min, and centrifuged to remove insoluble material. The pellet was reextracted with methanol, and the supernatants were pooled and reduced to dryness. For root, 10 g was extracted overnight in 10 mL of methanol at 70°C. The extracts were centrifuged to pellet the tissue, which was reextracted with methanol. Supernatants were pooled and reduced to dryness, and the remaining tissue was dried and weighed. Latex and root extracts were dissolved in 500 μL of methanol for analysis.
NMR Spectroscopy
All experiments were performed on a Bruker Advance 600 spectrometer (Bruker Daltonics) operating at 600.22 MHz and equipped with a 5-mm TXI probe at 298 K for solution-state analysis. All one-dimensional 1H NMR spectra of aqueous samples (1 mL in conventional cells) were acquired using a standard Bruker noesypr1d pulse sequence in which the residual water peak was irradiated during the relaxation delay of 1.0 s and during the mixing time of 100 ms. A total of 256 scans were collected into 65,536 data points over a spectral width of 12,195 Hz, with a 5-s repetition time. A line broadening of 0.5 Hz was applied to the spectra prior to Fourier transformation, phasing, and baseline correction. Spectra of samples in chloroform were acquired using similar parameters, with no solvent suppression pulses. Additional NMR experiments were performed for the purpose of confirming chemical shift assignments, including total correlation spectroscopy and heteronuclear single quantum coherence spectroscopy, using standard Bruker pulse programs.
Chemometric Analysis
One-dimensional 1H NMR spectra were imported into Chenomx NMR Suite version 4.6 (Chenomx) for target profiling analysis to determine compound concentrations and spectral binning. All shifts related to the solvent (i.e. in the range of 4.5–5.0 ppm) and DSS were excluded, and the remaining spectral regions were divided into 0.04-ppm bins. The same approach was taken for CDCl3-solvated extracts, in which case the single chloroform proton peak was removed prior to binning. Chemometric analysis was performed using SIMCA-P version 11.5 (Umetrics) with unsupervised PCA. All variables were pareto scaled to minimize the influence of baseline deviations and noise. In addition, PCA analysis was conducted on compound concentrations from target profiling analysis, in which case all variables were autoscaled prior to PCA.
Targeted Metabolite Profiling
Metabolite identification and quantification were achieved using the Profiler feature of Chenomx NMR Suite version 4.6 for analysis of one-dimensional 1H NMR spectra. Chenomx Profiler is linked to a database of metabolites whose unique NMR spectral signatures are encoded at various spectrometer frequencies, including 600 MHz. For the purposes of this study, one-dimensional 1H NMR signatures corresponding to selected compounds not present in the standard Chenomx library, including those of several benzylisoquinoline alkaloids, were used to create a custom opium poppy database (Supplemental Table S2). All standard NMR spectra used for metabolite identifications are commercially available (www.chenomx.com) or can be obtained from the corresponding author. Comparisons of NMR spectra with this database produced a list of compounds and their respective concentrations. A combination of one- and two-dimensional proton and heteronuclear NMR techniques were employed to confirm compound identities where necessary. Metabolites were quantified by the addition of a known amount of internal standard (i.e. DSS in D2O, TMS in CDCl3), which also served as a chemical shift reference. Due to the present unavailability of a metabolite database designed for the analysis of NMR spectra acquired in CDCl3, extensive analysis could only be carried out for spectra acquired in D2O. However, the 1H NMR spectrum for thebaine in CDCl3 was integrated into the metabolite database such that access by Chenomx Profiler would permit the identification and quantification of this metabolite in chloroform extracts of latex tissue. All data used to calculate metabolite levels are provided in Supplemental Table S3.
HPLC and LC-MS
HPLC was performed using the System Gold pump and photodiode array detector (Beckman-Coulter). All separations were performed at a flow rate of 1.0 mL min−1 on a C18 reverse phase column (4.6 × 250 mm, 5 μm, Discovery; Supelco). A 50-μL injection was separated using a defined gradient of solvent A (water, 0.1% triethylamine) and solvent B (methanol, 0.1% triethylamine). Chromatography was initiated using 70% (v/v) solvent A, which was decreased to 40% over 3 min, followed by a decrease over 22 min to 30%. Subsequently, the gradient was ramped to 100% solvent B over 3 min and maintained as such for 12 min. Peaks corresponding to morphine, oripavine, codeine, thebaine, noscapine, and sanguinarine were monitored at 280 nm and identified on the basis of retention times and UV spectra compared with those of authentic standards.
The mass measurements of latex and root extracts were performed on a Bruker Daltonics Esquire 3000 ion-trap mass spectrometer equipped with an Agilent model 1100 ESI HPLC system (Agilent Technologies). Mass spectrometry in the positive ion mode with an interface flow rate set to 0.3 mL min−1 was performed at a nebulizer setting of 45.0 pounds per square inch, a dry gas flow of 10.0 L min−1, a dry gas temperature of 350°C, a capillary voltage of 4,000 V, a tap drive voltage of 38.8 V, and a target mass of 500 m/z.
Gene Expression Analysis
Total RNA was isolated with TRIzol according to the manufacturer's protocol (Invitrogen). RT was performed at 42°C for 60 min using 2.5 μm anchored oligo(dT) primer (dT20VN), 0.5 mM dNTP, 10 to 40 ng μL−1 RNA, and 5 microunits μL−1 reverse transcriptase (Fermentas) following denaturing of the RNA-primer mix at 70°C for 5 min. Real-time PCR using SYBR Green detection was performed using an Applied Biosystems 7300 real-time PCR system. Each 10-μL PCR included 1 μL of cDNA (taken directly from the RT reaction in the case of stem, or diluted 50% with water for bud and root), 300 nm forward and reverse primers, and 1× Power SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences are listed in Supplemental Table S4. The thermal cycling conditions for relative quantification included 40 cycles of template denaturation, primer annealing, and primer extension. To evaluate PCR specificity, the amplified products of all primer pairs were subjected to melt curve analysis using the dissociation method suggested by Applied Biosystems. The reported values were calculated using six independent trials per plant line (i.e. two technical replicates for each of three independent biological samples). The 2−ΔΔCt method was employed for the analysis of relative gene expression (Livak and Schmittgen, 2001), where the gene encoding elongation factor 1a (elf1a) was used as the internal control and, for each of 42 target genes, the plant line exhibiting the highest expression level served as the calibrator. All data used to calculate relative gene expression levels are provided in Supplemental Table S5.
Statistical Methods
One-way ANOVA was performed to determine significant differences in metabolite levels (i.e. μmol mL−1 latex or μmol g−1 root) and transcript levels (i.e. relative abundance) using the NCSS Statistical Analysis and Graphics 2007 software package. Tukey-Kramer multiple-comparison tests were performed to reveal pair-wise differences between the means. α was set to 0.05 in all cases.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF025430, AF108432, AF191772, AF339913, AY217333, AY217335, AY217336, AY268893, AY860500, DQ028579, DQ316261, EB740724 to EB740749, EB740751 to EB740754, and U08598.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Individual metabolite quantities in root as determined by 1H NMR analysis for six varieties of opium poppy.
Supplemental Figure S2. Typical HPLC elution profiles for latex and root extracts for six opium poppy varieties. Peaks corresponding to morphine (M, red), oripavine (O, yellow), codeine (C, dark blue), thebaine (T, green), noscapine (N, light blue), and sanguinarine (S, purple) were identified by comparison with UV absorption spectra and retention times of authentic standards.
Supplemental Figure S3. Network of metabolic pathways leading from Suc to the benzylisoquinoline alkaloids sanguinarine and morphine in opium poppy.
Supplemental Figure S4. Quantitative real-time RT-PCR expression analysis of 30 primary metabolic and 12 alkaloid biosynthetic genes in stems of six opium poppy varieties.
Supplemental Figure S5. Quantitative real-time RT-PCR expression analysis of 30 primary metabolic and 12 alkaloid biosynthetic genes in roots of six opium poppy varieties.
Supplemental Table S1. PCA loadings of NMR spectral data obtained for latex chloroform extracts.
Supplemental Table S2. List of metabolites for which one-dimensional 1H NMR signatures are available in a Chenomx NMR Suite compound database customized for opium poppy.
Supplemental Table S3. Raw data sets for the 1H NMR-based metabolite profiling of opium poppy latex and roots.
Supplemental Table S4. Primers used for real-time PCR gene expression analysis in opium poppy.
Supplemental Table S5. Raw data for the quantitative real-time PCR analysis of gene expression in opium poppy stem, root, and flower buds.
Supplementary Material
Acknowledgments
We thank Glen MacInnis, Dr. Rustem Shaykhutdinov, and Dr. Ping Zhang for technical assistance and helpful discussions. We are grateful to Sanofi-Aventis for the gift of the opium poppy varieties and the morphinan alkaloid standards used in this study.
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant (to P.J.F.). J.M.H. is the recipient of an Alberta Ingenuity Graduate Student Scholarship. A.M.W. is the recipient of an Alberta Ingenuity Industrial Fellowship. The Bio-NMR Center is supported by grants from the Canadian Institutes of Health Research and the University of Calgary. P.J.F. holds the Canada Research Chair in Plant Metabolic Processes Biotechnology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peter J. Facchini (pfacchin@ucalgary.ca).
The online version of this article contains Web-only data.
References
- Allen RS, Miller JAC, Chitty JA, Fist AJ, Gerlach WL, Larkin PJ (2008) Metabolic engineering of morphinan alkaloids by over-expression and RNAi suppression of salutaridinol 7-O-acetyltransferase in opium poppy. Plant Biotechnol J 6 22–30 [DOI] [PubMed] [Google Scholar]
- Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JAC, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22 1559–1566 [DOI] [PubMed] [Google Scholar]
- Baker JM, Hawkins ND, Ward JL, Lovegrove A, Napier JA, Shewry PR, Beale MH (2006) A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol J 4 381–392 [DOI] [PubMed] [Google Scholar]
- Bernáth J, Németh É, Petheõ F (2003) Alkaloid accumulation in capsules of the selfed and cross-pollinated poppy. Plant Breed 122 263–267 [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9 110–115 [DOI] [PubMed] [Google Scholar]
- Choi HK, Choi YH, Verberne M, Lefeber AWM, Erkelens C, Verpoorte R (2004. a) Metabolic fingerprinting of wild type and transgenic tobacco plants by 1H NMR and multivariate analysis technique. Phytochemistry 65 857–864 [DOI] [PubMed] [Google Scholar]
- Choi YH, Kim HK, Hazekamp A, Erkelens C, Lefeber AWM, Verpoorte R (2004. b) Metabolic differentiation of Cannabis sativa cultivars using 1H NMR spectroscopy and principle component analysis. J Nat Prod 67 953–957 [DOI] [PubMed] [Google Scholar]
- Choi YH, Kim HK, Linthorst HJM, Hollander JG, Lefeber AWM, Erkelens C, Nuzillard JM, Verpoorte R (2006) NMR metabolomics to revisit the tobacco mosaic virus infection in Nicotiana tabacum leaves. J Nat Prod 69 742–748 [DOI] [PubMed] [Google Scholar]
- Choi YH, Sertic S, Kim HK, Wilson EG, Michopoulos F, Lefeber AWM, Erkelens C, Kricun SDP, Verpoorte R (2005) Classification of Ilex species based on metabolomic fingerprinting using nuclear magnetic resonance and multivariate data analysis. J Agric Food Chem 53 1237–1245 [DOI] [PubMed] [Google Scholar]
- Choi YH, Tapias EC, Kim HK, Lefeber AWM, Erkelens C, Verhoeven JTJ, Brzin J, Zel J, Verpoorte R (2004. c) Metabolic discrimination of Catharanthus roseus leaves infected by phytoplasma using 1H NMR spectroscopy and multivariate data analysis. Plant Physiol 135 2398–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defernez M, Gunning YM, Parr AJ, Shepherd LVT, Davies HV, Coquhoun IJ (2004) NMR and HPLC-UV profiling of potatoes with genetic modifications to metabolic pathways. J Agric Food Chem 52 6075–6085 [DOI] [PubMed] [Google Scholar]
- Dunn WB, Bailey NJC, Johnson HE (2005) Measuring the metabolome: current analytical technologies. Analyst 130 606–625 [DOI] [PubMed] [Google Scholar]
- Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgran F, Long I, Lundstedt T, Trygg J, Wold S (2004) Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabolomics (gpm). Anal Bioanal Chem 380 419–429 [DOI] [PubMed] [Google Scholar]
- Eriksson L, Johansson E, Kettaneh-Wold N, Wold S (2001) Multi- and Megavariate Data Analysis. Umetrics Academy, Umea, Sweden, pp 43–69
- Frédérich M, Choi YH, Angenot L, Harnischfeger G, Lefeber AWM, Verpoorte R (2004) Metabolomic analysis of Strychnos nux-vomica, Strychnos icaja and Strychnos ignatii extracts by 1H nuclear magnetic resonance spectrometry and multivariate analysis techniques. Phytochemistry 65 1993–2001 [DOI] [PubMed] [Google Scholar]
- Frick S, Chitty JA, Kramell R, Schmidt J, Allen RS, Larkin PJ, Kutchan TM (2004) Transformation of opium poppy (Papaver somniferum L.) with antisense berberine bridge enzyme gene (anti-bbe) via somatic embryogenesis results in an altered ratio of alkaloids in latex but not in roots. Transgenic Res 13 607–613 [DOI] [PubMed] [Google Scholar]
- Frick S, Kramell R, Schmidt J, Fist AJ, Kutchan TM (2005) Comparative qualitative and quantitative determination of alkaloids in narcotic and condiment Papaver somniferum varieties. J Nat Prod 68 666–673 [DOI] [PubMed] [Google Scholar]
- Hall RD (2006) Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol 169 453–468 [DOI] [PubMed] [Google Scholar]
- Heldt HW (1997) Plant Biochemistry and Molecular Biology. Oxford University Press, Oxford
- Jackowski S, Fagone P (2005) CTP:phosphocholine cytidylyltransferase: paving the way from gene to membrane. J Biol Chem 280 853–856 [DOI] [PubMed] [Google Scholar]
- Karg S, Märkle T (2002) Continuity and changes in plant resources during the Neolithic period in western Switzerland. Veget Hist Archaeobot 11 169–176 [Google Scholar]
- Kim HK, Choi YH, Erkelens C, Lefeber AWM, Verpoorte R (2005) Metabolic fingerprinting of Ephedra species using 1H NMR spectroscopy and principle component analysis. Chem Pharm Bull (Tokyo) 53 105–109 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Coruzzi G (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47 569–593 [DOI] [PubMed] [Google Scholar]
- Larkin PJ, Miller JAC, Allen RS, Chitty JA, Gerlach WL, Frick S, Kutchan TM, Fist AJ (2006) Increasing morphinan alkaloid production by over-expressing codeinone reductase in transgenic Papaver somniferum. Plant Biotechnol J 5 26–37 [DOI] [PubMed] [Google Scholar]
- Liang YS, Choi YH, Kim HK, Linthorst HJM, Verpoorte R (2006. a) Metabolomic analysis of methyl jasmonate treated Brassica rapa leaves by 2-dimensional NMR spectroscopy. Phytochemistry 67 2503–2511 [DOI] [PubMed] [Google Scholar]
- Liang YS, Kim HK, Lefeber AWM, Erkelens C, Choi YH, Verpoorte R (2006. b) Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J Chromatogr A 1112 148–155 [DOI] [PubMed] [Google Scholar]
- Liscombe DK, Facchini PJ (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J Biol Chem 282 14741–14751 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Manetti C, Bianchetti C, Bizzarri M, Casciani L, Castro C, D'Ascenzo G, Delfini M, Di Cocco ME, Lagana A, Miccheli A, et al (2004) NMR-based metabolomic study of transgenic maize. Phytochemistry 65 3187–3198 [DOI] [PubMed] [Google Scholar]
- Massart DL, Vandeginste BGM, Deming SN, Michotte Y, Kauffman L (1988) Chemometrics: A Textbook. Elsevier, New York
- Mattoo AK, Sobolev AP, Neelam A, Goyal RK, Handa AK, Segre AL (2006) Nuclear magnetic resonance spectroscopy-based metabolite profiling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiol 142 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli G, Nia VP, Trevisan M, Kolbe A, Schauer N, Geigenberger P, Chen J, Davison AC, Fernie AR, Zeeman SC (2007) Rapid classification of phenotypic mutants of Arabidopsis via metabolite fingerprinting. Plant Physiol 143 1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millgate AG, Pogson BJ, Wilson IW, Kutchan TM, Zenk MH, Gerlach WL, Fist AJ, Larkin PJ (2004) Morphine-pathway block in top1 poppies. Nature 431 413–414 [DOI] [PubMed] [Google Scholar]
- Moing A, Maucourt M, Renaud C, Gaudillère M, Brouquisse R, Lebouteiller B, Gousset-Dupont A, Vidal J, Granot D, Denoyes-Rothan B, et al (2004) Quantitative metabolic profiling by 1-dimensional 1H NMR analyses: application to plant genetics and functional genomics. Funct Plant Biol 31 889–902 [DOI] [PubMed] [Google Scholar]
- Németh É, Bernáth J, Sztefanov A (2002) New results of poppy (Papaver somniferum L.) breeding for low alkaloid content in Hungary. Acta Hortic 576 151–158 [Google Scholar]
- Nyman U (1978) Selection for high thebaine/low morphine content in Papaver somniferum L. Hereditas 89 43–50 [Google Scholar]
- Nyman U (1980) Selection for high thebaine/low morphine content in Papaver somniferum L. II. Studies of the I4 and I5 generations from a spontaneous mutant. Hereditas 93 121–124 [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y (2005) Revealing metabolic phenotypes in plants: inputs from NMR analysis. Biol Rev Camb Philos Soc 80 27–83 [DOI] [PubMed] [Google Scholar]
- Roberts MR, Kutchan TM, Brown RT, Coscia CJ (1987) Implication of tyramine in the biosynthesis of morphinan alkaloids in Papaver. Planta 172 230–237 [DOI] [PubMed] [Google Scholar]
- Rubtsov DV, Jenkins H, Ludwig C, Easton J, Viant MR, Günther U, Griffin JL, Hardy N (2007) Proposed reporting requirements for the description of NMR-based metabolomics experiments. Metabolomics 3 223–229 [Google Scholar]
- Rueffer M, Zenk MH (1987) Distant precursors of benzylisoquinoline alkaloids and their enzymatic formation. Z Naturforsch [C] 42 319–332 [Google Scholar]
- Samanani N, Alcantara J, Bourgault R, Zulak KG, Facchini PJ (2006) The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J 47 547–563 [DOI] [PubMed] [Google Scholar]
- Satoh S (2006) Organic substances in xylem sap delivered to above-ground organs by the roots. J Plant Res 119 179–187 [DOI] [PubMed] [Google Scholar]
- Schauer N, Fernie AR (2006) Plant metabolomics: towards biological function and mechanism. Trends Plant Sci 11 508–516 [DOI] [PubMed] [Google Scholar]
- Schmidt J, Boettcher C, Kuhnt C, Kutchan TM, Zenk MH (2007) Poppy alkaloid profiling by electrospray tandem mass spectrometry and electrospray FT-ICR mass spectrometry after [ring-13C6]-tyramine feeding. Phytochemistry 68 189–202 [DOI] [PubMed] [Google Scholar]
- Sharma JR, Lal RK, Gupta AP, Misra HO, Pant V, Singh NK, Pandey V (1999) Development of non-narcotic (opiumless and alkaloid-free) opium poppy, Papaver somniferum. Plant Breed 118 449–452 [Google Scholar]
- Stadler R, Zenk MH (1990) A revision of the generally accepted pathway for the biosynthesis of the benzyltetrahydroisoquinoline alkaloid reticuline. Liebigs Ann Chem 6 555–562 [Google Scholar]
- Stika HP (2005) Early Neolithic agriculture in Ambrona, Provincia Soria, central Spain. Veget Hist Archaeobot 14 189–197 [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, et al (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62 817–836 [DOI] [PubMed] [Google Scholar]
- Thureson-Klein A (1970) Observations on the development and fine structure of the articulated laticifers of Papaver somniferum. Ann Bot (Lond) 34 751–759 [Google Scholar]
- Trygg J, Holmes E, Lundstedt T (2007) Chemometrics in metabolomics. J Proteome Res 6 469–479 [DOI] [PubMed] [Google Scholar]
- Ward JL, Baker JM, Beale MH (2007) Recent applications of NMR spectroscopy in plant metabolomics. FEBS J 274 1126–1131 [DOI] [PubMed] [Google Scholar]
- Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM (2006) Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem 78 4430–4442 [DOI] [PubMed] [Google Scholar]
- Widarto HR, Van Der Meijden E, Lefeber AWM, Erkelens C, Kim HK, Choi YH, Verpoorte R (2006) Metabolomic differentiation of Brassica rapa following herbivory by different insect instars using two-dimensional nuclear magnetic resonance spectroscopy. J Chem Ecol 32 2417–2428 [DOI] [PubMed] [Google Scholar]
- Williams RD, Ellis BE (1989) Age and tissue distribution of alkaloids in Papaver somniferum. Phytochemistry 28 2085–2088 [Google Scholar]
- Yalkowsky SH, He Y (2003) Handbook of Aqueous Solubility Data. CRC Press, New York
- Ziegler J, Facchini PJ (2008) Alkaloid biosynthesis: metabolism and trafficking. Annu Rev Plant Biol 59 735–769 [DOI] [PubMed] [Google Scholar]
- Zulak KG, Cornish A, Daskalchuk TE, Deyholos MK, Goodenowe DB, Gordon PM, Klassen D, Pelcher LE, Sensen CW, Facchini PJ (2007) Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta 225 1085–1106 [DOI] [PubMed] [Google Scholar]
- Zulak KG, Liscombe DK, Ashihara H, Facchini PJ (2006) Alkaloids. In A Crozier, M Clifford, H Ashihara, eds, Plant Secondary Metabolism in Diet and Human Health. Blackwell Publishing, Oxford, pp 102–136
- Zulak KG, Weljie AM, Vogel HJ, Facchini PJ (2008) Quantitative 1H NMR metabolomics reveals extensive metabolic reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures. BMC Plant Biol 8 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.