Abstract
A predominantly nuclear RNA-binding protein, HuR translocates to the cytoplasm in response to stress and proliferative signals, where it stabilizes or modulates the translation of target mRNAs. Here, we present evidence that HuR phosphorylation at S202 by the G2-phase kinase Cdk1 influences its subcellular distribution. HuR was specifically phosphorylated in synchronous G2-phase cultures; its cytoplasmic levels increased by Cdk1-inhibitory interventions and declined in response to Cdk1-activating interventions. In keeping with the prominently cytoplasmic location of the nonphosphorylatable point mutant HuR(S202A), phospho-HuR(S202) was shown to be predominantly nuclear using a novel anti-phospho-HuR(S202) antibody. The enhanced cytoplasmic presence of unphosphorylated HuR was linked to its decreased association with 14–3–3 and to its heightened binding to target mRNAs. Our findings suggest that Cdk1 phosphorylates HuR during G2, thereby helping to retain it in the nucleus in association with 14–3–3 and hindering its post-transcriptional function and anti-apoptotic influence.
Keywords: RNA-binding protein, nucleocytoplasmic shuttling, 14–3–3 proteins, post-transcriptional gene regulation, cell division cycle, elav
Proliferative and damaging stimuli activate signaling pathways that enable mammalian cells to respond appropriately. Many such signaling cascades activate or inhibit transcription factors that collectively change subsets of expressed genes. In addition, the stimulus-triggered changes in gene expression patterns are decisively influenced by post-transcriptional regulatory processes, notably alterations in mRNA turnover and translation rates (Mitchell and Tollervey 2000; Orphanides and Reinberg 2002; Moore 2005). Among the post-transcriptional factors governing the expression of subsets of genes are RNA-binding proteins (RBPs) that associate with distinct groups of target mRNAs bearing specific motifs and influence their stability and translation (Keene 2007). RBPs recognizing U- or AU-rich elements (collectively known as AREs) comprise a distinct family of proteins capable of promoting mRNA decay (e.g., AUF1/hnRNP D, TTP, BRF1, KSRP) (Zhang et al. 1993; Carballo et al. 1998; Chen et al. 2001; Stoecklin et al. 2002) or inhibiting translation (e.g., TIAR, TIA-1) (Anderson and Kedersha 2002). The best known RBPs that stabilize and in some instances influence the translation of ARE-bearing mRNAs are the members of the Hu/ELAV RBP family. Hu/ELAV proteins comprise the primarily neuronal proteins HuB/Hel-N1, HuC, HuD, and the ubiquitous protein HuR/HuA (Ma et al. 1996; Antic et al. 1999; Brennan and Steitz 2001).
Like other Hu members, HuR contains three RNA-recognition motifs (RRMs) through which it binds target mRNAs. HuR target mRNAs encode proteins such as cyclins (A, B1, D1), tumor suppressors (p53), proto-oncogenes (c-fos, c-myc), growth factors (VEGF), cytokines (TGF-β, TNF-α), cyclin-dependent kinase (cdk) inhibitors (p21, p27), and anti-apoptotic factors (prothymosin α [ProTα], Bcl-2, Mcl-1) (Lafon et al. 1998; Levy et al. 1998; Wang et al. 2000a, b; Nabors et al. 2001; Kullmann et al. 2002; Mazan-Mamczarz et al. 2003; Lal et al. 2004, 2005; Abdelmohsen et al. 2007b); many such HuR targets contain one or several copies of an RNA signature motif (López de Silanes et al. 2004). Ribonucleoprotein (RNP) associations comprising HuR and its target transcripts are controlled by extracellular cues, notably proliferative and damaging signals. Given the functions of its target mRNAs, HuR has been implicated in processes such as cell proliferation, carcinogenesis, differentiation, and responses to stress and immune stimuli (Brennan and Steitz 2001; López de Silanes et al. 2003; van der Giessen et al. 2003; Katsanou et al. 2005; Lal et al. 2005; Abdelmohsen et al. 2007b).
Located between RRM2 and RRM3 of HuR is a hinge region that encompasses a nucleocytoplasmic shuttling sequence (HNS, spanning residues 205–237). Since the decay of ARE-bearing mRNAs is thought to occur in the cytoplasm, HuR has been proposed to elicit its mRNA-stabilizing function by binding target mRNAs in the nucleus, exporting and protecting them during cytoplasmic transit, and facilitating their recruitment to the translational machinery (Fan and Steitz 1998; Keene 1999). The nuclear export of HuR is directly mediated via its association with transportin 1 (Trn1) and Trn2, and also by the association of HuR with nuclear ligands pp32 and APRIL, which contain nuclear export signals that are recognized by the export receptor CRM1 (Gallouzi and Steitz 2001; Rebane et al. 2004). The import of HuR is also mediated by Trn1 and Trn2, interacting with the HNS (Guttinger et al. 2004); in addition, under conditions of metabolic stress causing reduced energy levels, phosphorylation of importin α by the AMP-activated protein kinase (AMPK) also contributes to the nuclear import of HuR (Wang et al. 2002, 2004). Other signaling kinases that regulate HuR subcellular abundance and/or association with specific target mRNAs have also been reported. Activation of the MAPK (mitogen-activated protein kinase)-activated protein kinase 2 (MK2), a downstream target of the MAPK p38, elevated cytoplasmic HuR levels and increased its ability to bind target mRNAs encoding urokinase-type plasminogen activator and its receptor (Tran et al. 2002). In addition, heat shock promoted the nuclear export of HuR bound to hsp70 mRNA, an event that was facilitated by HuR’s association with pp32 and APRIL (Gallouzi and Steitz 2001; Gallouzi et al. 2001).
Recently, the checkpoint kinase Chk2 was shown to phosphorylate HuR at S88, S100, and T118 (located within and between RRM1 and RRM2), modifications that influenced HuR’s ability to bind target transcripts in response to oxidative stress (Abdelmohsen et al. 2007a), and by PKCα, which phosphorylated HuR at S158 and S221 and elevated its cytoplasmic abundance (Doller et al. 2007). A chemical-genetic screen to identify targets of Cdk1 (also known as cell division cycle 2, Cdc2) identified serine 202 of HuR as a phosphorylation substrate for Cdk1 (Blethrow et al. 2008), but did not assign a function to this modification. Further, HuR(S202) has been shown to be phosphorylated in vivo in a phosphoproteomic survey (Olsen et al. 2006). Here, we provide evidence that Cdk1 phosphorylation of HuR at S202, near the HNS, prevents the cytoplasmic accumulation of HuR. Using chimeric HuR proteins and a novel antibody that recognizes phospho (p)-HuR(S202), we investigate the biological significance of this modification. Our findings indicate that p-HuR(S202) preferentially associates with a previously unknown nuclear ligand, 14–3–3, and suggest that this association results in the nuclear accumulation of HuR. Accordingly, the nonphosphorylatable mutant HuR(S202A) has a more pronounced cytoplasmic presence leading to the increased expression of HuR target mRNAs implicated in cell proliferation and survival; in turn, HuR(S202A) elicits an enhanced anti-apoptotic phenotype. Taken together, our results reveal that Cdk1 phosphorylation of HuR regulates its subcellular localization and function, and point to HuR as a downstream effector of gene expression programs by Cdk1.
Results
Cdk1 interacts with HuR
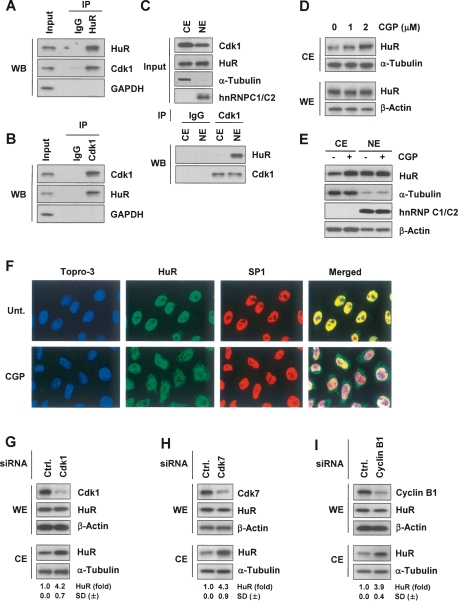
The identification of HuR(S202) as a Cdk1 phosphorylation site prompted us to study if HuR and Cdk1 formed protein–protein associations by coimmunoprecipitation (co-IP) analysis. Using whole-cell extracts (WE) prepared from asynchronous proliferating HeLa (human cervical carcinoma) cells, Cdk1 was detected by Western blot analysis in IP samples obtained using an anti-HuR antibody (Fig. 1A); conversely, HuR was detected in Cdk1 IP samples (Fig. 1B). This association did not require the presence of nucleic acids, as it was not disrupted by RNase or DNase treatments (not shown). Cdk1 was present in both nuclear and cytoplasmic extracts (NE, CE, respectively), but it associated with HuR primarily in NE (Fig. 1C).
Figure 1.
Inhibition or silencing of Cdk1, an HuR-interacting protein, elevates cytoplasmic HuR levels. (A) HeLa whole-cell lysates were used in immunoprecipitation (IP) assays with either control mouse IgG or anti-HuR antibodies, followed by Western blot (WB) detection of Cdk1, HuR, and the negative control GAPDH. (B) Assays were performed as in A except that control rabbit IgG or anti-Cdk1 antibodies were used for IP. (C) Cytosolic extracts (CE) and nuclear extracts (NE) were immunoprecipitated with either control rabbit IgG or anti-Cdk1 antibodies, followed by Western blot detection of HuR and Cdk1. The levels of Cdk1, HuR, α-Tubulin (cytosolic marker), and hnRNP C1/C2 (nuclear marker) were detected by Western blot analysis. (D) Following treatment for 2 h with the indicated CGP concentrations, HeLa cytoplasmic extracts (CE) were prepared and the levels of HuR and loading controls α-Tubulin and β-Actin were monitored by Western blot analysis. (E) After treatment with CGP (2 h, 10 μM), CE (10 μg) and NE (5 μg) were prepared and the levels of HuR, the cytoplasmic marker α-Tubulin, and the nuclear marker hnRNP C1/C2 were assessed by Western blot analysis. (F) Immunofluorescence microscopy to assess HuR distribution (green) in cells that were either left untreated (Unt.) or were treated with CGP (2 μM for 2 h). Nuclei were visualized using Topro-3 (blue) and the transcription factor SP1 (red). (Merged) Overlay of Topro-3, HuR, and SP1 signals. (G–I) Cells were transfected with control siRNAs (Ctrl.) or with siRNAs targeting Cdk1, Cdk7 or cyclin B1; 48 h after transfection, the levels of HuR, Cdk1, Cdk7, Cyclin B1, and loading controls β-Actin and α-Tubulin in whole-cell extracts (WE, 5 μg) and CE were monitored by Western blot analysis. Shown are fold changes in HuR levels as measured by densitometry (±SD, standard deviation from at least three experiments).
Cdk1 inhibition increases cytoplasmic HuR levels
Since HuR function is linked to its translocation to the cytoplasm, it was important to investigate if Cdk1 activity influenced the cytoplasmic HuR abundance. After treatment of cells with the Cdk1-specific inhibitor CGP74514A (CGP, Yu et al. 2003) for 2 h, cytoplasmic HuR levels increased in a dose-dependent manner (Fig. 1D), while HuR signals in WE and NE remained unchanged (Fig. 1D,E); it was virtually impossible to see a reciprocal decrease in nuclear HuR, since HuR is much more abundant in the nucleus. A similar CGP-triggered increase in cytoplasmic HuR was seen in other carcinoma lines (data not shown). The effect of CGP on HuR subcellular localization was also examined by immunofluorescence microscopy (Fig. 1F; Supplemental Fig. S1); as shown, cytoplasmic HuR signals increased markedly in CGP-treated cells. The influence of Cdk1 activity upon the cytoplasmic HuR levels was further tested following other interventions to inhibit Cdk1 function. The levels of Cdk1, Cdk7 (a kinase that phosphorylates Cdk1 at T161 and thereby activates it, Fig. 2A) or Cyclin B1 (which binds to and activates Cdk1) were reduced by using specific siRNAs. Silencing of each protein increased cytoplasmic HuR levels by about fourfold (Fig. 1G–I). Since cytoplasmic HuR levels were found previously to change in a cell-cycle-dependent manner in other cell types (Atasoy et al. 1998; Wang et al. 2000a), it was possible that the cytoplasmic HuR increases were due to gene silencing interventions that might have arrested cells in S and G2/M phases (as shown by Yuan et al. 2002); however, this did not appear to be the case, as silencing was incomplete and did not elicit distinct arrests in S or G2/M under the conditions tested (Supplemental Fig. S2). In addition, Cyclin B1 silencing did not disrupt the interaction between Cdk1 and HuR (Supplemental Fig. S3).
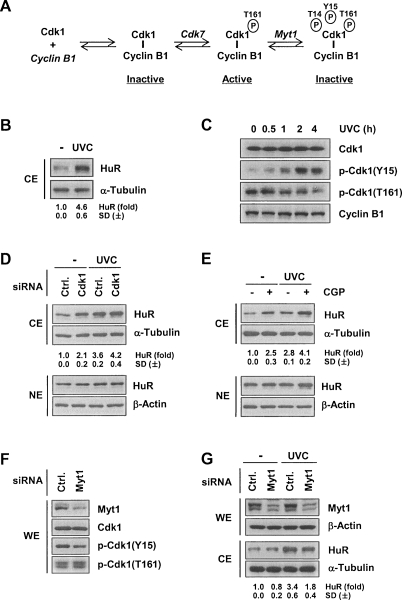
Figure 2.
UVC-mediated elevation of cytoplasmic HuR levels concomitant with Cdk1 inhibition. (A) Schematic of the modifications activating and inhibiting Cdk1. (B) HeLa cells were irradiated with UVC (25 J/m2) and collected 2 h later to assess the levels of cytosolic (CE) HuR by Western blot analysis. (C) At the times shown after UVC irradiation, the levels of total Cdk1, Cdk1 phosphorylated at Tyr 15 [p-Cdk1(Y15)] or Thr 161 [p-Cdk1(T161)], and Cyclin B1 were examined by Western blot analysis of WE. (D,E) After silencing Cdk1 for 48 h (D) or treating with CGP (2 μM) for 1 h (E), cells were either left without further treatment (−) or were irradiated with UVC (25 J/m2); 2 h later, HuR and α-Tubulin levels in CE were assessed by Western blot analysis. (F) Cells were transfected with Myt1 siRNA; 48 h after transfection, WE were prepared and the levels of Cdk1, p-Cdk1(Y15), and p-Cdk1(T161) were studied by Western blot analysis. (G) Following Myt1 silencing with a specific siRNA, cells were either left untreated (−) or were irradiated with UVC (25 J/m2) and collected 2 h later for the detection of HuR (and loading control α-Tubulin) in CE. HuR signals in A, C, D, and F were quantified by densitometry; ±SD from at least three experiments are indicated.
Cells that were irradiated with short-wavelength ultraviolet light (UVC, 25 J/m2) were also studied. UVC stress elevates cytoplasmic HuR levels (Fig. 2B; Wang et al. 2000b) in part by acting on karyopherins, but it also inhibits Cdk1 (Poon et al. 1996; Zhan et al. 1999; data not shown). Cdk1 activity is regulated by two sets of phosphorylation events: In addition to the aforementioned activation by Cdk7-mediated phosphorylation on T161, it is inactivated by phosphorylation on Tyr 15 (Y15) or Thr 14 (T14) by the kinase Wee1/Myt1. Accordingly, UVC irradiation increased Cdk1 phosphorylation at Y15 and decreased its phosphorylation at T161 (Fig. 2C). Further evidence that Cdk1 function influenced the UVC-triggered increase in cytoplasmic HuR was obtained from cells that were either transfected with Cdk1 targeting siRNA or pretreated with CGP; as shown, each intervention augmented the cytoplasmic levels of HuR, and the effects were further accentuated by irradiation of cells with UVC (Fig. 2D,E). Moreover, silencing of Wee1/Myt1, which inactivates Cdk1, reduced the cytoplasmic HuR levels in both untreated and UVC-treated cells (Fig. 2F,G). Together, these findings strongly support the view that inhibition of Cdk1 activity increased the cytoplasmic HuR levels.
HuR phosphorylation by Cdk1 linked to its reduction in the cytoplasm
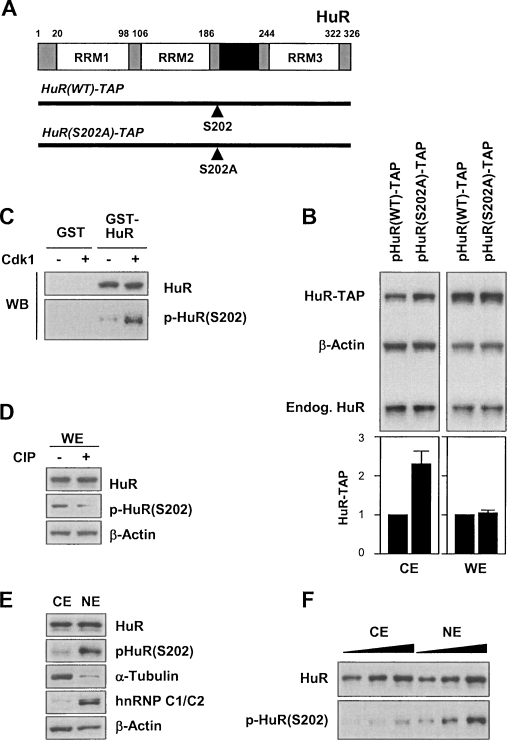
Given the physical association between HuR and Cdk1, the inverse correlation between Cdk1 activity and cytoplasmic HuR levels, and the identification of HuR(S202), adjacent to the nuclear shuttling sequence HNS, as a substrate of Cdk1 (Blethrow et al. 2008), we hypothesized that Cdk1-mediated phosphorylation of HuR might promote its nuclear localization. To test this possibility, a nonphosphorylatable Ser-to-Ala (S202A) point mutant was made in a mammalian vector that expresses HuR fused to a TAP (tandem affinity purification) tag (Lal et al. 2005). The resulting chimeric proteins (Fig. 3A), HuR(WT)-TAP and HuR(S202A)-TAP, were ∼55 kDa and could thus be distinguished from the ∼36-kDa endogenous HuR. As shown (Fig. 3B), HuR(S202A)-TAP was greater than twofold more abundant than HuR(WT)-TAP (Fig. 3B, left graph) in CE, but not in WE (Fig. 3B, right graph) or in NE (data not shown). The changes in HuR-TAP abundance were not due to changes in protein half-life, as both the wild-type and S202A variants were stable (Supplemental Fig. S4).
Figure 3.
HuR phosphorylation at Ser-202 enhances its nuclear localization. (A) HuR schematic showing the position of S202, the residue phosphorylated by Cdk1, and the vectors that were constructed to express a chimeric wild-type HuR protein linked to TAP [HuR(WT)-TAP] and a chimeric protein with nonphosphorylatable S202 [HuR(S202A)-TAP]. (RRM) RNA-recognition motif; (HNS) HuR nucleocytoplasmic shuttling sequence. (B) HeLa cells were transfected with plasmids to express the chimeric proteins depicted in A and the levels of ectopic [HuR(WT)-TAP and HuR(S202A)-TAP] and endogenous HuR in CE and WE were monitored by Western blot analysis. Shown are fold changes in ectopic HuR-TAP levels from three independent experiments, as assessed by densitometry. (C) HuR phosphorylation was assessed after an in vitro kinase reaction using GST-HuR as substrate (Materials and Methods); the levels of total HuR-GST and phosphorylated HuR-GST were monitored by Western blotting using antibodies that recognized either total HuR or phospho-HuR [p-HuR(S202)], respectively. (D) WE were prepared from HeLa cells and treated with CIP for 1 h; the levels of HuR and p-HuR(S202) were detected by Western blotting using antibodies that recognized either total HuR or p-HuR(S202). (E) CE and NE were prepared and the levels of total HuR and p-HuR(S202), as well as those of cytoplasmic marker α-Tubulin and nuclear marker hnRNP C1/C2, were detected by Western blot analysis. (F) Increasing amounts of CE and NE lysates were tested in order to achieve comparable total HuR signals and then compare p-HuR(S202) signals.
To examine the subcellular localization and protein interactions of the HuR(S202) phosphoisoform [p-HuR(S202)], we used a phosphopeptide spanning amino acids 196–208 [Ac-CLSQLYH(pS)PARRFG-amide (Supplemental Fig. S5)] to generate an anti-p-HuR(S202) polyclonal antibody. This antibody specifically recognized GST-HuR that had been phosphorylated in vitro in the presence of Cdk1 (Fig. 3C). Endogenous phosphorylated HuR was also detected in WE preparations, since treatment with alkaline phosphatase (CIP) potently reduced p-HuR(S202) signals (Fig. 3D). When the amounts of lysate tested by Western blot analysis were adjusted to achieve comparable HuR signals in CE and NE, p-HuR(S202) signals were markedly higher in NE compared with CE (Fig. 3E,F). These results strongly suggested that HuR phosphorylated at S202 resides predominantly in the nucleus.
Preferential interaction of phospho-HuR(S202) with various 14–3–3 isoforms in the nucleus
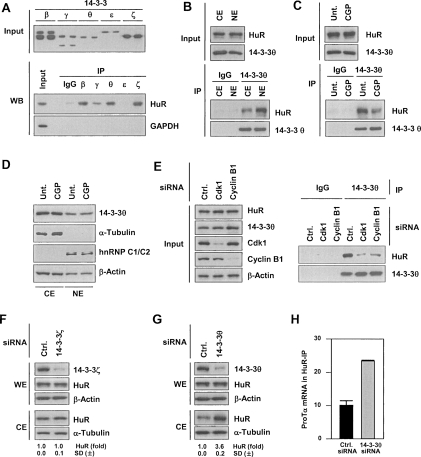
The hypothesis that phosphorylation of HuR on S202 influences its subcellular localization was further tested by examining possible nuclear ligands that could mediate the nuclear retention of p-HuR(S202). We studied the interaction of HuR with nuclear proteins, including several transport machinery proteins reported to interact with HuR (karyopherin β2 [Kap β2], also called transportin [Trn1, Trn2]; importin α1 [Imp α1]; and the nuclear export receptor CRM1 [data not shown]), as well as with 14–3–3 proteins. Screening of the latter family of proteins by co-IP analysis revealed that HuR interacted with various 14–3–3 isoforms (Fig. 4A; Supplemental Material). 14–3–3θ showed a distinct association with HuR and was studied in greater detail. 14–3–3θ was present in both CE and NE (Fig. 4B, Input), but it bound HuR predominantly in the nucleus (Fig. 4B, IP). This association decreased by approximately threefold when cells were treated with CGP before IP analysis (Fig. 4C), indicating that the interaction of 14–3–3θ with HuR was enhanced in the presence of active Cdk1. The reduced [HuR-14–3–3θ] complexes did not arise from changes in the levels of 14–3–3θ in CE or NE (Fig. 4D). Active Cdk1 was required for the formation of the [HuR-14–3–3θ] complex, as silencing of Cdk1 or cyclin B1 (Fig. 4E, left) largely abrogated the association of HuR with 14–3–3θ (Fig. 4E, right).
Figure 4.
Phosphorylated HuR binds 14–3–3θ and localizes in the nucleus. (A, top) Western blot analysis of the indicated 14–3–3 isoforms (paired samples). (Bottom) Various 14–3–3 isoforms were immunoprecipitated using specific antibodies, and the presence of HuR and negative control GAPDH in the IP materials was tested by Western blot analysis. (B) CE and NE were prepared from HeLa cells, used for IP with control rabbit IgG or anti-14–3–3θ antibodies, and HuR and 14–3–3θ in the IP samples was detected by Western blot analysis. (C) Following treatment with CGP (2 μM, 2 h), WE were prepared and assayed by IP using control rabbit IgG or anti-14–3–3θ antibodies; bound HuR and 14–3–3θ were assessed by Western blotting. (D) After CGP treatment (2 μM, 2 h), CE and NE were prepared and the levels of 14–3–3θ, the cytosolic marker α-Tubulin, the nuclear marker hnRNP C1/C2, and the loading control β-Actin were analyzed by Western blotting. (E) Forty-eight hours after transfection of cells with the siRNAs shown, WE were prepared and used for IP in the presence of rabbit IgG or anti-14–3–3θ antibodies. (Left) The levels of HuR, Cdk1, cyclin B1, and loading control β-Actin in the Input material were tested by Western blotting. (Right) The presence of HuR and 14–3–3θ in the IP samples was assessed by Western blotting. (F,G) Cells were transfected with either control (Ctrl.) siRNA or with an siRNA targeting 14–3–3ζ or 14–3–3θ. Forty-eight hours after transfection, CE and WE were prepared and the levels of HuR and 14–3–3ζ (F) and 14–3–3θ (G) were detected by Western blotting; β-Actin and α-Tubulin were used as loading controls; ±SD of HuR signals from at least three experiments are indicated. (H) In cells transfected with 14–3–3θ siRNA, ProTα mRNA levels in HuR IP were determined by RT–qPCR (Materials and Methods); graph depicts the means and SD from three independent experiments.
The interaction of HuR with another member of this family, 14–3–3ζ, had no measurable consequences on HuR localization (Fig. 4F). By contrast, silencing 14–3–3θ greatly elevated HuR in the cytoplasm (Fig. 4G), supporting the views that 14–3–3θ specifically contributed to retaining HuR in the nucleus (as did Kap β2) (Supplemental Fig. S6) and that the different 14–3–3 isoforms could have a distinct influence on HuR levels in CE. Along with an increase in cytoplasmic HuR levels, HuR binding to ProTα mRNA was also greater than twofold higher after 14–3–3θ silencing, as assessed by RNP IP analysis (Fig. 4H), indicating that the increase in HuR cytoplasmic presence was associated with enhanced HuR function.
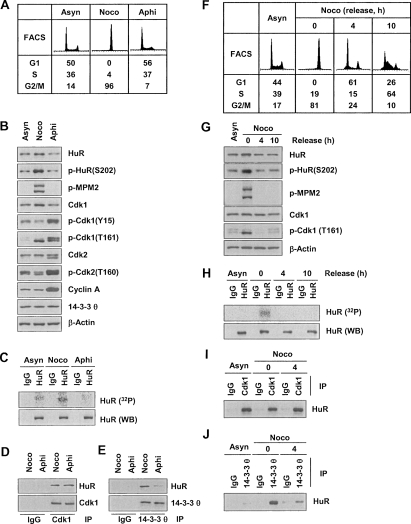
Specific HuR phosphorylation and binding to 14–3–3 during G2/M
The identification of Cdk1 as an HuR kinase prompted a series of experiments to determine if HuR phosphorylation occurred during the G2/M phase, when Cdk1 is active. First, the levels of p-HuR(S202), HuR, the mitotic epitope p-MPM2, other G1, S, and G2/M markers, as well as the percentages of cells in each cell cycle compartment were studied (Fig. 5A,B; Supplemental Fig. S7). In keeping with the findings that G2/M cells were most abundant after nocodazole (Noco) treatment and least abundant after aphidicolin (Aphi) treatment, HuR phosphorylation in vivo, as measured by monitoring the incorporation of inorganic phosphate (32Pi) into HuR, was highest in the Noco population and lowest in the Aphi group (Fig. 5C). The reduced HuR phosphorylation in the Aphi cultures occurred despite unchanged association of HuR with Cdk1 (Fig. 5D), and was linked to a decrease in the binding of HuR to 14–3–3θ (Fig. 5E).
Figure 5.
Specific HuR phosphorylation and binding to 14–3–3 during G2/M. (A) FACS analysis of HeLa cells that were either growing asynchronously (Asyn) or synchronized in G2/M using nocodazole (Noco, 100 ng/mL, 16 h) or in G1 and S using aphidicolin (Aphi, 2 μg/mL, 16 h). (B) WE lysates were prepared from the populations in A, and the levels of proteins and phosphoproteins (including G1 and S markers p-cdk1 [Y15], p-cdk2 [T160], and cyclin A; G2/M marker p-cdk1 [T161], and mitotic marker p-MPM2) were assessed by Western blot analysis. (C) In vivo HuR phosphorylation was studied by incubating the populations described in A for 2 h with 32Pi and assessing the incorporation of the radiolabel into HuR by IP using anti-HuR antibody (or IgG in control IP reactions). Total HuR in IP materials was detected by Western blot analysis (WB). Co-IP analysis was used to study the association of HuR with Cdk1 (D) and with 14–3–3θ (E). (F) FACS analysis of HeLa cells synchronized with nocodazole (100 ng/mL for 16 h), released by removing nocodazole, and examined 4 and 10 h later. (G) Using cells that were processed as explained in F, the levels of the proteins shown were studied by Western blot analysis of WE lysates. (H) In vivo HuR phosphorylation was studied by incubating the populations described in F for 2 h with 32Pi and assessing the incorporation of the radiolabel into HuR by IP using anti-HuR antibody (or IgG in control IP reactions). Total HuR in IP materials was detected by Western blotting (WB). WE lysates from the populations in F were used in co-IP assays to study the association of HuR with Cdk1 (I) and with 14–3–3θ (J).
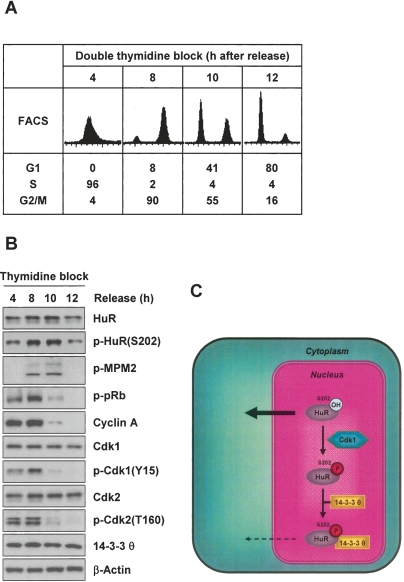
When nocodazole was washed from the culture medium and cells were released from the G2/M arrest, again p-HuR(S202) was highest when G2/M cells were most abundant, G2/M markers were most prominent, and in vivo HuR phosphorylation was highest (Fig. 5F–H). These differences in phosphorylation arose despite continued interaction between HuR and Cdk1, but 14–3–3θ binding was observed primarily in populations with phosphorylated HuR (Fig. 5I,J). Similarly, cells released from synchronization in S phase by double thymidine block (Fig. 6A) showed the highest p-HuR(S202) signals when transversing through G2/M (at 8 and 10 h after release, Fig. 6B). Collectively, the cell cycle data indicate that HuR(S202) is phosphorylated in G2/M, leading to an increased interaction with 14–3–3θ in the nucleus. Together with the prominent cytoplasmic localization of the nonphosphorylatable mutant and the predominantly nuclear p-HuR(S202), we propose a model whereby HuR phosphorylation at S202 promotes its association with 14–3–3 and favors a nuclear localization of the complex, while unphosphorylated HuR(S202) is readily exported to the cytoplasm and modulates the expression of target mRNAs (Fig. 6C).
Figure 6.
Synchronous phosphorylation of HuR and model. (A) FACS analysis of HeLa cells at 4, 8, 10, and 12 h after release from double thymidine block (Supplemental Material). (B) WE lysates from cells that were treated as in A were collected for Western blot analysis of the proteins shown. (C) Schematic of the proposed influence of Cdk1 on HuR localization; see text for details.
Ectopic HuR(S202A) and inhibition of Cdk1 promote the expression of HuR target mRNAs encoding anti-apoptotic proteins and enhance cell survival
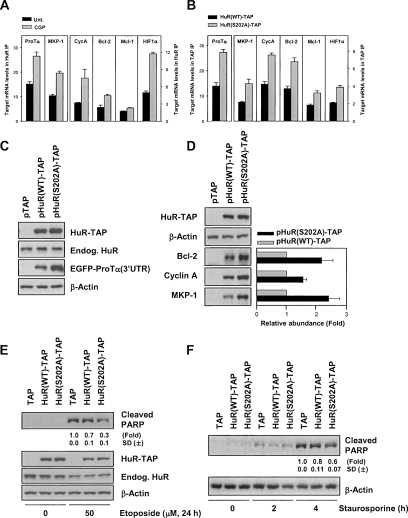
The consequences of HuR phosphorylation at S202 leading to higher cytoplasmic HuR levels were further assessed by monitoring the expression of HuR target mRNAs. First, the association of HuR with target transcripts encoding ProTα, MKP-1, cyclin A, Bcl-2, Mcl-1, and HIF-1α was tested by RNP IP analysis. Compared with untreated cells, CGP-treated populations showed an enhanced association of these transcripts with HuR (Fig. 7A); likewise, these mRNAs were significantly enriched in HuR(S202A)-TAP IP samples compared with HuR(WT)-TAP IP samples (Fig. 7B). Second, the enhanced levels of HuR RNP complexes were linked to increased expression of the encoded proteins. For ProTα, the levels of translated product were tested by measuring the abundance of reporter pEGFP-ProTα(3′UTR), given the lack of suitable anti-ProTα antibodies (Lal et al. 2005). As shown in Figure 7C, EGFP expressed from the EGFP-ProTα(3′UTR) chimeric mRNA was greater than twofold more highly expressed in the pHuR(S202)-TAP transfection group than in the pHuR(WT)-TAP transfection group (when testing a reporter that lacked the ProTα 3′UTR, EGFP levels were the same in both transfection groups) (data not shown). Similarly, the levels of MKP-1, Cyclin A, and Bcl-2 proteins implicated in the cell’s response to stress-causing and proliferative agents were greater than twofold elevated in cells expressing HuR(S202A)-TAP (Fig. 7D); examples of HuR-elicited stabilization and translational up-regulation of target mRNAs is shown (Supplemental Figure S8).
Figure 7.
S202 phosphorylation influences the levels of cytoplasmic HuR RNP complexes and the levels of proteins encoded by HuR target mRNAs. HeLa cells were treated with CGP (2 μM, 2 h) (A) or transfected with either pHuR-TAP or pHuR(S202A)-TAP and collected 48 h later (B); CE were prepared (Materials and Methods) and immunoprecipitated with mouse IgG or anti-HuR antibodies (A) or with IgG agarose beads (B). The levels of prothymosin α (ProTα), MKP-1, cyclin A (CycA), Mcl-1, Bcl-2, and HIF1α were determined by RT–qPCR; the data (means and SEM from three independent experiments) were normalized to the abundance of GAPDH mRNA (a housekeeping mRNA present at background levels) in each set of IP samples. (C) Cells were cotransfected with pTAP, pHuR(WT)-TAP, or pHuR(S202A)-TAP along with plasmid pEGFP-ProTα(3′UTR); 48 h after transfection, the levels of endogenous HuR, ectopic HuR-TAP, EGFP, and loading control β-Actin were detected in WE by Western blotting. (D) Cells were transfected as in C, WE were prepared, and the levels of MKP-1, Cyclin A, and Bcl-2 were detected by Western blotting, quantified by densitometry (graph), and normalized to the levels of loading control β-Actin. (E) After transfection with pTAP, pHuR(WT)-TAP, or pHuR(S202A)-TAP, cells were treated with etoposide for 24 h (50 μM) (left) or with staurosporine (1 μM) for the times shown (right) and WE were prepared for Western blot analysis and densitometric quantification of cleaved PARP. Data are representative of three independent experiments.
Given the anti-apoptotic influence of ProTα, Mcl-1, and Bcl-2, the effect of expressing HuR(WT)-TAP and HuR(S202A)-TAP was tested in cells responding to apoptotic agents. As shown in Figure 7E, exposure of cells to 50 μM etoposide for 24 h triggered the cleavage of PARP most prominently in the control group (TAP); by contrast, PARP cleavage was reduced to ∼70% in cells expressing HuR(WT)-TAP, and was further reduced to ∼30% of control TAP populations when the nonphosphorylatable mutant HuR(S202A) was expressed. HuR(S202A) engendered a similar protection against treatment with staurosporine (Fig. 7F). In sum, Cdk1 can elicit changes in gene expression and cell integrity by phosphorylating HuR and thereby altering HuR’s subcellular localization, its stabilizing influence upon target mRNAs encoding stress-response proteins, and its impact on cell survival.
Discussion
Interaction of HuR with 14–3–3
The studies presented here suggest a regulatory paradigm whereby HuR phosphorylation by Cdk1 promotes its nuclear localization mediated by 14–3–3 proteins (e.g., 14–3–3θ), as depicted schematically in Figure 6C. In unstressed, actively dividing cell populations, Cdk1 phosphorylates HuR at S202, thereby maintaining reduced levels of HuR in the cytoplasm; this reduction is accentuated by interventions to elevate Cdk1 activity, such as through silencing of an upstream Cdk1 inhibitor like Wee1/Myt1 (Fig. 2G). Interventions to diminish Cdk1 activity, including Cdk1 silencing, chemical inhibition, UVC irradiation, silencing of the Cdk1 cofactor cyclin B1, or silencing of the upstream activating kinase Cdk7, all increased the levels of cytoplasmic HuR (Figs. 1, 2), where HuR(S202) is almost completely unphosphorylated (Fig. 3F). The subcellular distribution and stoichiometry of unphosphorylated HuR(S202) and p-HuR(S202) could not be precisely quantified, but the use of an antibody recognizing specifically p-HuR(S202) suggested that p-HuR(S202) resides in the nucleus and is largely absent from the cytoplasm, and that p-HuR(S202) is only a fraction of the total nuclear HuR pool.
Interestingly, 14–3–3θ was found to interact only in the nucleus and with p-HuR(S202) (Figs. 4, 5); limitations to further co-IP analysis of HuR and 14–3–3θ included the lack of sufficiently sensitive and specific anti-14–3–3θ antibodies and the fact that HuR-TAP migrates close to the heavy IgG chain. Instead, the dependence of HuR phosphorylation for binding to 14–3–3θ was tested following interventions to reduce Cdk1 activity. This analysis revealed that silencing of either Cdk1 or Cyclin B1 potently inhibited the association of HuR with 14–3–3θ (Fig. 4). The regulatory paradigm proposed for [HuR-14–3–3] partly resembles those of other phosphorylated RBPs interacting with 14–3–3 proteins. For example, 14–3–3σ binds to p37AUF1 and retains it in the cytoplasm, enhancing the degradation of target mRNAs (He and Schneider 2006); various 14–3–3 proteins interact with phosphorylated BRF1 and thereby block its ability to promote target mRNA decay (Schmidlin et al. 2004); phosphorylated TTP associates with 14–3–3 and alters its cytoplasmic distribution, at least in part by excluding [TTP-14–3–3] complexes from stress granules (Johnson et al. 2002; Stoecklin et al. 2004); and Akt-phosphorylated KSRP associates with 14–3–3 and is unable to recruit target mRNAs to the exosome (Gherzi et al. 2006). The biological consequences of these [RBP-14–3–3] interactions are diverse, in keeping with the functional heterogeneity of 14–3–3 proteins (Yaffe 2002), but they suggest that the RBP localization, RNA-binding activity, and post-transcriptional influence upon target mRNAs are broadly linked to the RBP phosphorylation status and interaction with 14–3–3 proteins.
The interaction between HuR and 14–3–3θ raises several questions for immediate consideration: Does 14–3–3θ binding prevent the binding of HuR to components of the nuclear export machinery? Does the [HuR-14–3–3θ] complex form initially in the nucleus? Or, does it form in the cytoplasm as a prerequisite for rapid nuclear import? Is the [HuR-14–3–3θ] complex recognized by components of the nucleocytoplasmic shuttling apparatus? In this regard, we detected an interaction between HuR and karyopherin β2 (Kap β2 [Trn1 and Trn2]) and between HuR and importin α1, but did not find that these associations were influenced by HuR phosphorylation or by 14–3–3θ in vivo or in vitro (data not shown). Similarly, use of leptomycin B, which inhibits the export factor and reported HuR ligand CRM1 (Gallouzi et al. 2001) failed to influence binding of HuR to 14–3–3θ, regardless of HuR(S202) phosphorylation status (data not shown).
Cdk1 regulating HuR function during the cellular response to stress and apoptosis
In cells responding to damaging stimuli, the regulation of HuR localization and function through phosphorylation by Cdk1 is predicted to have important physiologic implications. Many stress stimuli transiently inhibit cell division, and in particular progression through G2/M (Stark and Taylor 2006); a temporary inhibition of Cdk1 would, in turn, facilitate the export of unphosphorylated HuR to the cytoplasm at a time when the injured cell needs to implement a new gene expression program and synthesize stress-response proteins. Supporting this view is the fact that numerous HuR target mRNAs that encode stress-response proteins (e.g., p21, p53, ProTα, Bcl-2, Mcl-1) are stabilized and/or translated more actively by HuR (Wang et al. 2000b; Mazan-Mamczarz et al. 2003; Lal et al. 2005; Abdelmohsen et al. 2007a, b). The functional effects of Cdk1 upon the cytoplasmic localization of HuR were recapitulated by point mutation of S202. Compared with HuR(WT), the nonphosphorylatable mutant HuR(S202A) had a more prominent cytoplasmic presence. Accordingly, S202 phosphorylation-dependent changes in HuR subcellular levels directly impacted on HuR’s binding to mRNAs encoding proliferative and anti-apoptotic proteins (Fig. 7A–D). While the relative contribution of changes in mRNA stability, protein translation, and protein stability remain to be studied in detail for these proteins, their enhanced expression contributed to reducing apoptosis in response to etoposide and staurosporine (Fig. 7E). Given HuR’s involvement in differentiation, carcinogenesis, and the immune response, the functional implications of HuR phosphorylation at S202 await direct analysis in these important biological processes.
HuR kinases Chk2 and Cdk1
Recent studies from our laboratory (Abdelmohsen et al. 2007a) describe HuR phosphorylation at S88, S100, and T118 by the checkpoint kinase Chk2. Chk2 was activated by treatment with H2O2, an agent that elevated cytoplasmic HuR levels (Wang et al. 2000b). However, HuR phosphorylation by Chk2 in response to the oxidant did not directly influence HuR subcellular levels, but instead affected HuR’s ability to bind mRNAs: It triggered the dissociation of HuR from certain targets (e.g., SIRT1 mRNA), and enhanced the association of other targets (e.g., ProTα mRNA), suggesting that HuR phosphorylation by Chk2 broadly affects HuR’s RNA-binding activity. While HuR(S202) phosphorylation in response to H2O2 was not tested directly, we anticipate that it will be reduced, given the general decline in Cdk1 activity that follows exposure to genotoxins, likely contributing to the observed increase in cytoplasmic HuR.
Interestingly, these two paradigms of HuR phosphorylation are closely linked, as Chk2 is an upstream regulator of Cdk1 activity in cells responding to damaging stimuli. Genotoxic stresses such as UVC, oxidants, and ionizing radiation activate Atm (the ataxia telangiectasia mutated protein) and Atr (the Atm- and Rad3-related protein), which in turn activate Chk2 and the functionally related kinase Chk1. Chk1 and Chk2 phosphorylate and thereby inactivate Cdc25, the phosphatase that activates Cdk1 (for review, see Stark and Taylor 2006). A model is thus envisioned wherein Chk2 and Cdk1 function in tandem to regulate HuR function: Stress agents inactivate Cdk1, thereby permitting the increased presence of HuR in the cytoplasm, and activate Chk2, thus allowing HuR to appropriately elevate or reduce its association with target mRNAs. Efforts are underway to test this model experimentally.
HuR at the convergence of cell proliferation and survival
In mammalian cells, the proliferative and stress responses are intricately connected, both functionally and molecularly. On a functional level, stress stimuli often suppress cell cycle progression, an adaptive response that is viewed as affording the cell with additional time to repair the damage and survive the harmful hit. On a molecular level, many factors mediating both sets of responses are shared, as they operate through common networks of cellular effectors. These signaling paths often begin at the same initial sensor proteins (e.g., growth factor receptors), are frequently transduced through the same signaling pathways (e.g., MAPK, PKC), and culminate in the utilization of many common transcription factors (e.g., p53, pRB, c-Myc, E2F-DP, C/EBP, AP-1) that implement adaptive gene transcription programs. In addition to the shared transcriptional regulators, our results suggest that HuR functions as a joint downstream effector of post-transcriptional gene expression changes in response to stress and proliferative agents. Thus, Cdk1 function lies at the convergence of these two response pathways, increasing during G2 progression and decreasing by stress, and in turn Cdk1 coordinates gene expression programs via downstream effectors like HuR. That Cdk1-mediated phosphorylation of HuR influences its localization and activity constitutes important new insight into the mechanisms controlling the function of HuR, a pivotal regulator of homeostasis in mammalian cells responding to environmental cues.
Materials and methods
Cell culture, treatment, and transfection
Human cervical carcinoma HeLa cells were cultured in DMEM containing 5% fetal bovine serum. The dose of UVC irradiation was 25 J/m2. CGP74514A (CGP, Calbiochem) and cycloheximide (CHX, Sigma) were added directly into the culture medium. siRNAs targeting Cdk1, Cdk7, cyclin B1 (Santa Cruz Biotechnologies), or the 3′UTR of HuR (Supplemental Material) were used at 100 nM. Plasmid pTAP-HuR(S202A) was prepared by site-directed mutagenesis of pTAP-HuR (Lal et al. 2005). Oligofectamine and Lipofectamine 2000 (Invitrogen) were used for transfection of siRNAs and plasmids, respectively. Unless otherwise specified, all transfected cells were analyzed 48 h later. HeLa cells (50% confluence) were synchronized by treatment with nocodazole (100 ng/mL) or aphidicolin (2 μg/mL) for 16 h to induce G2/M or G1/S arrest, respectively, and released from the arrest by incubation in fresh medium.
Cell fractionation and protein analysis
Cytosolic and nuclear fractionation, as well as immunofluorescence to study the subcellular distribution of HuR and SP1, were performed as described (Supplemental Material; Lal et al. 2004). For co-IP analysis, CE, NE, and WE (∼1 mg) were incubated with antibodies recognizing HuR, Cdk1, 14–3–3θ, or 14–3–3ζ prebound to Protein A-Sepharose beads (Sigma). Mouse IgG (BD Pharmingen) or rabbit IgG (Santa Cruz Biotechnologies) were used in control IP reactions. Protein–protein interactions were studied by Western blot analysis of IP samples. For Western blot analysis, CE (10 μg), NE (5 μg), and WE (5 μg) were size-fractionated (12% SDS-PAGE) and transferred onto PVDF membranes. Following incubation (16 h, 4°C) with primary monoclonal antibodies (Supplemental Material), blots were washed and incubated with appropriate HRP-conjugated secondary antibodies for 1 h. Signals were detected by enhanced luminescence (Amersham Biosciences).
Evaluation of HuR phosphorylation
An antibody recognizing the phosphopeptide CLSQLYH(pS) PARRFG (Supplemental Fig. S5) was custom-made (Quality Controlled Biochemicals). To test its suitability in detecting HuR, a Cdk1 kinase reaction mixture (2.5 μL of Cdk1/Cyclin B, 10 μL of ATP, 2.5 μL of either HuR-GST or GST) was incubated for 10 min at 30°C and stopped by adding SDS sample buffer. Total HuR-GST and phosphorylated HuR-GST were detected by Western blot analysis using HuR and phospho-HuR antibodies, respectively. Control incubations with calf intestine phosphatase (CIP) were performed by pre-incubating WE lysates with CIP (10 units) for 1 h at 30°C. To check the level of p-HuR(S202) in the cytoplasm and nucleus, CE and NE were prepared, a range of lysate amounts loaded on the gel, and total HuR and p-HuR(S202) detected by Western blot analysis using anti-HuR and anti-p-HuR(S202) antibodies, respectively.
In vivo HuR phosphorylation was assessed by incubating with 32Pi (0.5 mCi) for 2 h, whereupon cells were lysed using RIPA buffer. IP assays were carried out for 1 h at 4°C using either mouse IgG or anti-HuR antibodies. After extensive washes in RIPA buffer, IP materials were resolved by 12% SDS-PAGE, transferred onto PVDF filters, and visualized with a PhosphorImager (Molecular Dynamics).
IP of RNP complexes
For IP of RNP complexes, cytoplasmic fractions were prepared as described (Lal et al. 2006). Briefly, cells were either left untransfected or were transfected with pTAP, pHuR(WT)-TAP, or pHuR(S202A)-TAP (the efficiency of TAP IP is shown [Supplemental Fig. S9]), and CE prepared in cytoplasmic lysis buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.3% NP-40) for 5 min, and centrifugation at 10,000g for 10 min at 4°C. The supernatants were used for IP by incubation for 1 h with protein A-Sepharose beads coated with mouse IgG (BD Pharmingen) or anti-HuR antibody (Santa Cruz Biotechnologies); rabbit IgG-agarose was used for TAP IP (Sigma). After the beads were washed with NT2 buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 0.05% NP-40), the complexes were treated with 20 units of RNase-free DNase I (15 min at 37°C) and 0.1% SDS/0.5 mg/mL Proteinase K (15 min, 55°C) to remove DNA and protein, respectively. RNA was isolated by phenol-chloroform extraction and subjected to RT using oligo-dT and SSII RT (Invitrogen) followed by qPCR analysis as previously described (Lal et al. 2006), using specific primer pairs to detect ProTα, cyclin A, MKP-1, Bcl-2, Mcl-1, and GAPDH mRNAs (Supplemental Material).
Acknowledgments
We are grateful to F.E. Indig, B.P. Frank, W. Kim, and C. Sasaki for assistance with these studies. This research was supported in part by the Intramural Research Program of the NIA, NIH. J.B. and K.M.S. were supported by NIH EB001987.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1645808.
References
- Abdelmohsen K., Pullmann R., Lal A., Kim H.H., Galban S., Yang X., Blethrow J.D., Walker M., Shubert J., Gillespie D.A., et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007a;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K., Lal A., Kim H.H., Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007b;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stressful initiations. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- Antic D., Lu N., Keene J.D. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes & Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U., Watson J., Patel D., Keene J.D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Blethrow J.D., Glavy J.S., Morgan D.O., Shokat K.M. Covalent capture of kinase-specific phosphopeptides reveals novel Cdk1–Cyclin B substrates. Proc. Natl. Acad. Sci. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M., Steitz J.A. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU-binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Doller A., Huwiler A., Muller R., Radeke H.H., Pfeilschifter J., Eberhardt W. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: Implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C., Steitz J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi I.E., Steitz J.A. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- Gallouzi I.E., Brennan C.M., Steitz J.A. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherzi R., Trabucchi M., Ponassi M., Ruggiero T., Corte G., Moroni C., Chen C.Y., Khabar K.S., Andersen J.S., Briata P. The RNA-binding protein KSRP promotes decay of β-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 2006;5:e5. doi: 10.1371/journal.pbio. 0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guttinger S., Muhlhausser P., Koller-Eichhorn R., Brennecke J., Kutay U. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. 2004;101:2918–2923. doi: 10.1073/pnas.0400342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Schneider R. 14–3–3σ is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Stehn J.R., Yaffe M.B., Blackwell T.K. Cytoplasmic localization of tristetraprolin involves 14–3–3-dependent and -independent mechanisms. J. Biol. Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- Katsanou V., Papadaki O., Milatos S., Blackshear P.J., Anderson P., Kollias G., Kontoyiannis D.L. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Keene J.D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kullmann M., Gopfert U., Siewe B., Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes & Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon I., Carballes F., Brewer G., Poiret M., Morello D. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J.L., Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Kawai T., Yang X., Mazan-Mamczarz K., Gorospe M. Anti-apoptotic function of RNA-binding protein HuR effected through prothymosin α. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Abdelmohsen K., Pullmann R., Kawai T., Galban S., Yang X., Brewer G., Gorospe M. Posttranscriptional derepression of GADD45α by genotoxic stress. Mol. Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Levy N.S., Chung S., Furneaux H., Levy A.P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- López de Silanes I., Fan J., Yang X., Zonderman A.B., Potapova O., Pizer E.S., Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.J., Cheng S., Campbell C., Wright A., Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Galban S., López de Silanes I., Martindale J.L., Atasoy U., Keene J.D., Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Tollervey D. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- Moore M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nabors L.B., Gillespie G.Y., Harkins L., King P.H. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to AU-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Poon R.Y., Jiang W., Toyoshima H., Hunter T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage. J. Biol. Chem. 1996;271:13283–13291. doi: 10.1074/jbc.271.22.13283. [DOI] [PubMed] [Google Scholar]
- Rebane A., Aab A., Steitz J.A. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 2004;10:590–599. doi: 10.1261/rna.5224304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin M., Lu M., Leuenberger S.A., Stoecklin G., Mallaun M., Gross B., Gherzi R., Hess D., Hemmings B.A., Moroni C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Taylor W.R. Control of the G2/M transition. Mol. Biotechnol. 2006;32:227–248. doi: 10.1385/MB:32:3:227. [DOI] [PubMed] [Google Scholar]
- Stoecklin G., Colombi M., Raineri I., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W.F., Blackwell T.K., Anderson P. MK2-induced tristetraprolin:14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H., Brunet A., Grenier J.M., Datta S.R., Fornace A.J., DiStefano P.S., Chiang L.W., Greenberg M.E. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- van der Giessen K., Di-Marco S., Clair E., Gallouzi I.E. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J. Biol. Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- Wang W., Caldwell M.C., Lin S., Furneaux H., Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000a;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Furneaux H., Cheng H., Caldwell M.C., Hutter D., Liu Y., Holbrook N., Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000b;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Fan J., Yang X., Fürer-Galban S., López de Silanes I., von Kobbe C., Guo J., Georas S.N., Foufelle F., Hardie D.G., et al. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang X., Kawai T., López de Silanes I., Mazan-Mamczarz K., Chen P., Chook Y.M., Quensel C., Köhler M., Gorospe M. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1: Involvement in the nuclear import of RNA-binding protein HuR. J. Biol. Chem. 2004;279:48376–48388. doi: 10.1074/jbc.M409014200. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. How do 14–3–3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- Yu C., Rahmani M., Dai Y., Conrad D., Krystal G., Dent P., Grant S. The lethal effects of pharmacological cyclin-dependent kinase inhibitors in human leukemia cells proceed through a phosphatidylinositol 3-kinase/Akt-dependent process. Cancer Res. 2003;63:1822–1833. [PubMed] [Google Scholar]
- Yuan J., Eckerdt F., Bereiter-Hahn J., Kurunci-Csacsko E., Kaufmann M., Strebhardt K. Cooperative phosphorylation including the activity of polo-like kinase 1 regulates the subcellular localization of cyclin B1. Oncogene. 2002;21:8282–8292. doi: 10.1038/sj.onc.1206011. [DOI] [PubMed] [Google Scholar]
- Zhan Q., Antinore M.J., Wang X.W., Carrier F., Smith M.L., Harris C.C., Fornace A.J. Association with Cdk1 and inhibition of Cdk1/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- Zhang W., Wagner B.J., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]