Abstract
Phospholipid hydroperoxide glutathione peroxidase (GPx4) is a moonlighting selenoprotein, which has been implicated in basic cell functions such as anti-oxidative defense, apoptosis, and gene expression regulation. GPx4-null mice die in utero at midgestation, and developmental retardation of the brain appears to play a major role. We investigated post-transcriptional mechanisms of GPx4 expression regulation and found that the guanine-rich sequence-binding factor 1 (Grsf1) up-regulates GPx4 expression. Grsf1 binds to a defined target sequence in the 5′-untranslated region (UTR) of the mitochondrial GPx4 (m-GPx4) mRNA, up-regulates UTR-dependent reporter gene expression, recruits m-GPx4 mRNA to translationally active polysome fractions, and coimmunoprecipitates with GPx4 mRNA. During embryonic brain development, Grsf1 and m-GPx4 are coexpressed, and functional knockdown (siRNA) of Grsf1 prevents embryonic GPx4 expression. When compared with mock controls, Grsf1 knockdown embryos showed significant signs of developmental retardations that are paralleled by apoptotic alterations (TUNEL staining) and massive lipid peroxidation (isoprostane formation). Overexpression of m-GPx4 prevented the apoptotic alterations in Grsf1-deficient embryos and rescued them from developmental retardation. These data indicate that Grsf1 up-regulates translation of GPx4 mRNA and implicate the two proteins in embryonic brain development.
Keywords: Glutathione peroxidase 4, guanine-rich sequence-binding factor 1, apoptosis, brain development, embryogenesis
Embryonic brain development is characterized by programmed cell death (PCD), which is essential for tissue homeostasis and morphogenesis (for review, see Yeo and Gautier 2004). PCD is precisely controlled in a temporal and spatial manner by a complex network of factors. Phospholipid hydroperoxide glutathione peroxidase (phGPx/GPx4) has been recognized as a regulatory component of the apoptotic machinery, and the anti-apoptotic character has been related to its peroxidase activity (Imai and Nakagawa 2003). GPx4 is a multifunctional selenoprotein, which has been classified as glutathione peroxidase (GPx) because of its enzymatic activity and its structural relatedness to other GPx isoforms (Brigelius-Flohe 1999). It is capable of reducing hydroperoxides at the expense of glutathione or other reducing equivalents (Ursini and Bindoli 1987). In contrast to other GPx isoforms, GPx4 reduces complex lipid hydroperoxides even if they are incorporated in biomembranes or lipoproteins (Thomas et al. 1990). In addition to its anti-oxidative activity, GPx4 has been implicated as a structural protein in sperm maturation (Ursini et al. 1999; Roveri et al. 2002) and as a regulator of eicosanoid biosynthesis (Schnurr et al. 1996) and of cell signaling pathways (Brigelius-Flohe et al. 2000). GPx4-null mice die in utero at midgestation (Imai et al. 2003; Yant et al. 2003), and intrauterine lethality may be related to extensive PCD leading to abnormal embryonic hindbrain development (Borchert et al. 2006). The role of GPx4 in the central nervous system is not restricted to cerebral embryogenesis. In adult brain, the enzyme is expressed in neurons of cerebral cortex, hippocampus, and cerebellum (Savaskan et al. 2007), protecting them from oxidative injury (Ran et al. 2006). Interestingly, following brain injury, GPx4 is strongly induced in reactive astrocytes rescuing the cells from apoptosis, preventing further cell damage (Savaskan et al. 2007).
Transcriptional regulation of GPx4 is as complex as its functional multiplicity. Three functionally distinct isozymes [mitochondrial (m-GPx4), cytosolic (c-GPx4), and nuclear (n-GPx4) isoforms] originate from a single GPx4 gene. The start codons for m-GPx4 and c-GPx4 as well as the targeting sequence that directs m-GPx4 into the mitochondria are localized in the first exon. Expression of the 34-kDa nuclear isoform (n-GPx4) involves transcription of an alternative first exon (exon 1B) that encodes for a nuclear targeting sequence (Pfeifer et al. 2001). The cytosolic isoform is ubiquitously expressed at moderate levels in most mammalian cells. Mitochondrial and nuclear isoforms are found in large quantities in spermatoid cells (Tramer et al. 2002; Puglisi et al. 2003). Expression of c-GPx4 and m-GPx4 involves activation of general transcription factors, such as Sp1 and NF-Y (Ufer et al. 2003), and binding of the activated cAMP-response element modulator-tau to the 5′-flanking region of exon 1B has also been implicated (Borchert et al. 2003; Tramer et al. 2004).
Post-transcriptional mechanisms are a central process of gene expression and range from co-transcriptional processes such as 5′-capping to splicing, mRNA editing, polyadenylation, nuclear export, translation, and subsequent mRNA degradation (for review, see Hieronymus and Silver 2004). Most of these processes are conferred by mRNA/protein (mRNP) complexes. Research on post-transcriptional elements of GPx4 expression regulation has been focused on co-translational incorporation of selenocysteine. This catalytically essential amino acid is encoded for by an opal codon, and premature translational termination is prevented by binding of regulatory proteins to the selenocysteine insertion sequence (SECIS) localized in the 3′-untranslated region (UTR) of the GPx4 mRNA (Copeland et al. 2000). To shed light on translational mechanisms in expression regulation of GPx4, we investigated the interaction of regulatory proteins with the 5′UTR of the GPx4 mRNA. After identifying Grsf1 as the GPx4 mRNA-binding protein using the yeast three-hybrid system, we explored its role in embryonic brain development and found that knockdown of Grsf1 induced similar developmental retardations as GPx4 silencing. Since these alterations were prevented by GPx4 overexpression, one can conclude that Grsf1-dependent translational regulation of GPx4 expression is essential for cerebral embryogenesis.
Results
Identification of proteins binding to the 5′UTR of m-GPx4 mRNA
To explore post-transcriptional elements of GPx4 expression regulation, we searched for proteins capable of binding to the 5′UTR of the m-GPx4 mRNA using the yeast three-hybrid system (see the Supplemental Material). After several rounds of highly stringent positive and negative selections, we identified a single positive yeast clone. The high binding specificity was confirmed by co-transformation experiments (Supplemental Fig. S1), which indicated that activation of the lacZ reporter gene (blue staining) was only observed when all hybrids were correctly expressed, and a BLAST search of the sequence data obtained revealed 100% identity with the C-terminal 361 amino acids (75% of the full-length clone) of the guanine-rich sequence-binding factor 1 (Grsf1, NM_178700). The N-terminal 27 amino acids of the coding sequence were derived from an alternative splicing variant of Grsf1 designated K6-Grsf1. Grsf1 has recently been described as cytoplasmic RNA-binding protein involved in translation regulation of influenza proteins in virus-infected cells (Park et al. 1999).
Grsf1 binds to a 27-nucleotide (nt) motif in the 5′UTR of m-GPx4 mRNA
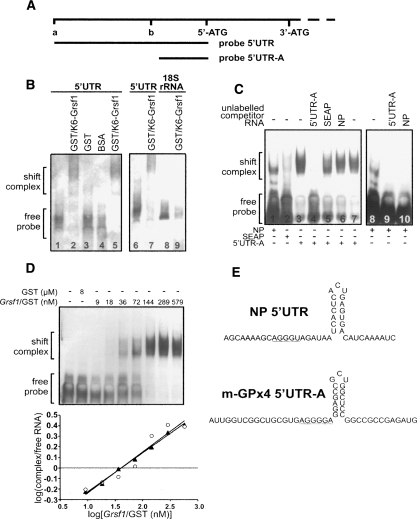
To characterize the binding region, we performed RNA mobility gel shift assays (Fig. 1). K6-Grsf1 was first expressed in Escherichia coli as a GST fusion protein and purified to homogeneity. Then two labeled RNA probes (entire 5′UTR of m-GPx4 mRNA named 5′UTR [144 nt] and the 3′-region of the 5′UTR named 5′UTR-A [50 nt]) (Fig. 1 A) were incubated with recombinant K6-Grsf1, and the protein/RNA complexes were analyzed (Fig. 1B,C). When the labeled 5′UTR probe was incubated with GST/K6-Grsf1 fusion protein, high-molecular-weight shift signals appeared (Fig. 1B,C, lanes 2,5,7), and similar signals were observed for recombinant Grsf1 (no GST fusion) (see Supplemental Fig. S2). No shift signals were detected with pure GST or BSA (Fig. 1B, lanes 3,4) and with labeled control RNA probes of 18S rRNA (Fig. 1B, lanes 8,9) and SEAP (secreted embryonic alkaline phosphatase) mRNA (Fig. 1C, lane 2).
Figure 1.
Grsf1 specifically binds to the 5′UTR of the m-GPX4 mRNA. Protein binding to the 5′UTR of m-GPx4 mRNA was studied by RNA mobility gel shift assays. For this purpose, two labeled RNA probes representing different parts of the 5′UTR of m-GPx4 mRNA were incubated in vitro with different amounts of purified recombinant K6-Grsf1/GST fusion proteins. Aliquots of this incubation mixture were loaded on a 5% polyacrylamide gel (native conditions) and the separated protein–RNA complexes were then transferred to a nylon membrane. The blots were visualized as described in the Materials and Methods. (A) 5′UTR of the m-GPx4 mRNA and RNA probes (5′UTR and 5′UTR-A) used for initial shift assays. The 5′-ATG represents the start codon for m-GPx4 and the 3′-ATG that for the cytosolic enzyme. The sequence between these two ATG codons represents the mitochondrial insertion sequence. Letters a and b indicate the position of the two major transcription initiation sites for m-GPx4 (Nam et al. 1997; Knopp et al. 1999). (B) Biotin-labeled probes of the m-GPx4 5′UTR or 18S rRNA (negative control) were incubated with 4.5 μg of the indicated proteins. (C) Digoxigenin-labeled probes of m-GPx4 5′UTR (5′UTR-A), influenza NP 5′UTR (NP), or SEAP 5′UTR (SEAP) were incubated with K6-Grsf1/GST fusion protein in the absence or presence of 50 pmol of unlabeled competitor RNA. (D, top part) A digoxigenin-labeled probe, 5′UTR-A, was incubated with varying amounts of free GST or Grsf1/GST fusion protein. (Bottom part) The ratio of signal intensities of the RNA shift band/free RNA were plotted against Grsf1/GST concentration. The intercept with the X-axis represents the Kd. (E) Sequence and predicted secondary structure of influenza NP 5′UTR (Park et al. 1999) and m-GPx4 5′UTR.
Similar shift patterns were observed when the shorter probe 5′UTR-A was incubated with the recombinant GST/K6-Grsf1 fusion protein (Fig. 1C, lanes 3,7). When K6-Grsf1 was omitted or free GST was used, no shift complexes were detected (Fig. 1D). Performing competitive gel shift assays, we found that the specific probe (Fig. 1C, lane 4) but not control SEAP mRNA (Fig. 1C, lane 5) or 18S rRNA (data not shown) competed for binding. Taken together, the results shown in Figure 1 indicate that the two mRNA probes (5′UTR and 5′UTR-A) exhibited a similar binding behavior, suggesting that the K6-Grsf1-binding sequence may be located in the 3′-region of the 5′UTR.
To further narrow down the essential binding sequence, we constructed truncated RNA probes representing various parts of the 5′UTR of the m-GPx4 mRNA (Supplemental Fig. S3). From these results, we concluded that the Grsf1-binding sequence is represented by a 27-nt motif located close to the translational initiation site of m-GPx4. This sequence contains the A(G)4A motif and exhibits a high degree of structural similarity with the Grsf1-binding sequence identified for the influenza NP 5′UTR (Park et al. 1999). In addition, the calculated secondary structures of the two mRNA sequences were also very similar, revealing a 15-nt hairpin closely preceding the A(G)4A motif (Fig. 1E). Cross-binding studies indicated that a labeled probe containing the influenza NP 5′UTR also forms a shift complex with Grsf1 (Fig. 1C, lane 1), but a molar excess of unlabeled NP 5′UTR RNA did not alter the intensity of the Grsf1/5′UTR-A (m-GPx4) shift signal (Fig. 1C, lane 6). In contrast, the Grsf1/5′UTR (NP) shift complex was competed off by a molar excess of both, the unlabeled influenza NP 5′UTR probe and the unlabeled m-GPx4 5′UTR-A probe (Fig. 1C, lanes 8–10). These data suggested a higher binding affinity of Grsf1 to the m-GPx4 5′UTR than to the influenza NP 5′UTR. To test this conclusion experimentally, we quantified the binding affinity of Grsf1 to the m-GPx4 5′UTR by RNA shift assays using variable nonsaturating amounts of Grsf1. Then the intensities of the shift signals (ratio of free vs. bound RNA) were quantified by densitometry (Fig. 1D) and plotted against Grsf1 concentration. This algorithm revealed a linear correlation and the intercept with the X-axis indicated a Kd-value of 40 nM. Similar experiments performed with the influenza NP 5′UTR revealed only low-affinity binding with an apparent Kd of 1.4 μM (data not shown).
Grsf1 up-regulated expression of luciferase reporter genes driven by the m-GPx4 mRNA 5′UTR
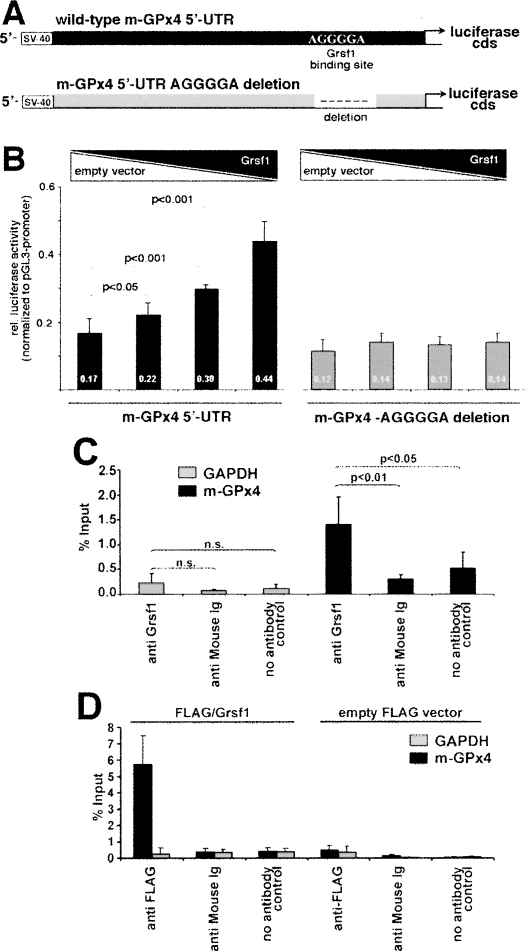
To explore the functional impact of Grsf1 binding to m-GPx4 mRNA, we performed luciferase-based reporter gene studies. For this purpose, transfection plasmids (Fig. 2A) were created consisting of the luciferase coding sequence driven by the SV40 promoter (pGL-3 promoter vector). In addition, we cloned in either the wild-type 5′UTR of m-GPx4 mRNA or a mutated version of this sequence lacking the G-rich consensus Grsf1-binding motif [A(G)4A] (Kash et al. 2002; Schaub et al. 2007). These constructs were then co-transfected into mouse embryonic fibroblasts (MEF cells) with increasing concentrations of a Grsf1 expression plasmid. To exclude transcriptional effects due to different promoter doses, the total plasmid concentration was kept constant by adding appropriate amounts of empty expression vector. Figure 2B shows that Grsf1 up-regulated (∼2.6-fold) expression of m-GPx4–5′UTR-driven luciferase constructs depending on the amount of co-transfected Grsf1 expression vector. In contrast, no increase in luciferase activity was observed when the Grsf1-binding motif was deleted. To exclude methodological artifacts on RNA turnover, we also quantified the steady-state levels of the reporter gene mRNA (qRT–PCR). In these experiments, the absence of contaminating plasmid DNA was ensured by extensive DNase treatment. Here we found that luciferase mRNA levels were not significantly altered (1.2-fold increase) in the presence of Grsf1. Taken together, these results suggested that Grsf1 binding to the m-GPx4 mRNA is capable of activating m-GPx4 expression.
Figure 2.
Grsf1 expression up-regulates expression of UTR-dependent reporter gene constructs and interacts with m-GPx4 mRNA in vivo. UTR-dependent reporter gene assays were carried out as described in the Supplemental Material. (A) For co-transfection of embryonic fibroblasts with Grsf1, two different luciferase-based reporter gene constructs containing the 5′UTR of m-GPx4 were designed. One construct contained the consensus Grsf1-binding sequence (wild-type m-GPx4 5′UTR), and the other lacked this motif (m-GPx4 5′UTR AGGGGA deletion). Increasing concentrations of Grsf1 expression plasmid were co-transfected with the reporter gene constructs. To adjust a constant quantity of pGL3-promoter, all samples were supplemented with empty vector (the proportion of Grsf1 construct was 0%, 25%, 50%, and 100%, respectively, as indicated by the bar graphics above the diagram). After co-transfection, cells were kept in culture for 6 h and luciferase activity was measured in the lysates. (B) The relative luciferase activities (numbers at the bottom of each column) were calculated. The activities measured were corrected for transfection efficiency (Renilla luciferase activity) and normalized to equal amounts of pGL3-promoter. (C) RNA immunoprecipitation in murine N2a cells was carried out as described in the Supplemental Material. RNA recovered from “Input” samples was set at 100%. Data are given as means of three independent experiments ±SD (Student’s t-test). (D) RNA immunoprecipitations in N2a cells transfected with a Grsf1/Flag expression plasmid or an empty Flag vector were carried out as described in the Supplemental Material. RNA recovered from “Input” samples was set at 100%. Data are given as means of three independent experiments ±SD.
In vivo binding of Grsf1 to m-GPx4 mRNA
To test whether Grsf1 also binds the m-GPx4 messenger under in vivo conditions, we performed RNA immunoprecipitation. For this purpose, cytosolic extracts of murine neuroblastoma N2a cells, which express both Grsf1 and m-GPx4, were prepared and protein/RNA complexes were cross-linked. Using a specific anti-Grsf1 antibody for immunoprecipitation, m-GPx4 mRNA was pulled down (Fig. 2C). Much lower amounts of m-GPx4 mRNA were precipitated when an unrelated antibody (anti-mouse Ig) was used or when the antibody was omitted. In contrast, GAPDH mRNA, which was used as negative control, was not significantly immunoprecipitated. To further support our finding of in vivo Grsf1/m-GPx4 mRNA interaction, we transfected N2a cells with an expression plasmid encoding a Flag/Grsf1 fusion protein. Using an anti-Flag antibody, we also immunoprecipitated significant amounts of m-GPx4 mRNA (Fig. 2D). In contrast, when cells were transfected with the empty Flag expression vector, neither m-GPx4 mRNA nor GAPDH mRNA was significantly pulled down. These results indicated in vivo interaction of Grsf1 with m-GPx4 mRNA.
Grsf1 recruits m-GPx4 mRNA to translationally active polysomal fractions
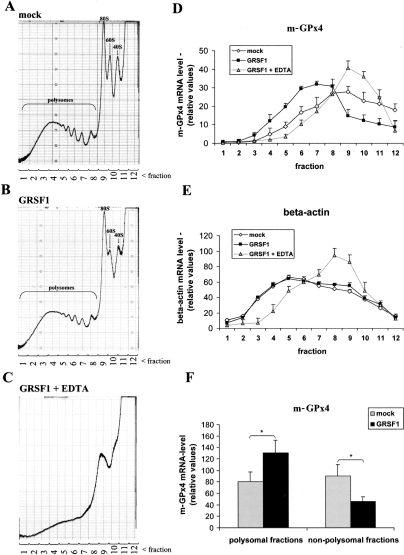
Transient Grsf1 overexpression in MEFs resulted in a moderate (threefold) increase in Grsf1 expression (data not shown). To test whether m-GPx4 ex-pression is controlled on translational level, we performed polysome fractionation studies on sucrose gradients (Fig. 3A–C). Overexpression of Grsf1 did not significantly alter ribosomal profiles of the cells, nor did it change total m-GPx4 mRNA levels (16 ± 3 molecules m-GPx4/1000 GAPDH in mock-transfected cells vs. 15 ± 3 molecules m-GPx4/1000 GAPDH in Grsf1-transfected cells). When we quantified the m-GPx4 mRNA content in different polysome fractions, we detected the highest amounts in fractions 6 and 7, which correspond to small polysomes (Fig. 3D). In contrast, in mock-transfected cells, the highest amounts were found in monosomes and translationally inactive fractions 8, 9, and 10. This difference is even more visible when the m-GPx4 mRNA content in the pooled polysomal fractions was compared with that of nonpolysomes (Fig. 3F). In Grsf1-transfected cells, significantly more m-GPx4 copies were recovered from polysomal fractions than from nonpolysomes. In mock-transfected controls, this ratio was the other way around. Translocation of m-GPx4 mRNA into active polysomes following Grsf1 transfection strongly suggests translational activation of m-GPx4 mRNA by Grsf1. To exclude unspecific effects and methodological artifacts, similar analyses were carried out with β-actin mRNA (Fig. 3E) as well as with 28S and 18S rRNA (data not shown). Here we did not observe significant differences between Grsf1-transfected and mock-transfected cells. To exclude the possibility that m-GPx4 mRNA is recruited to nonribosomal mRNP complexes, polysome fractionations were carried out in the presence of 25 mM EDTA. EDTA induces dissociation of polysomes (Fig. 3C) and thus releases m-GPx4 and β-actin mRNA into nonpolysomal fractions (Fig. 3D,E), indicating specificity of the elution profile.
Figure 3.
Polysomal gradient analysis. MEF cells were transiently transfected for 24 h using either a Grsf1 expression or a control vector (no Grsf1 insert). (A–C) Typical polysome profiles after sucrose gradient ultracentrifugation monitored at 254 nm from the bottom (51% sucrose, left) to top (17% sucrose, right) are shown. Each gradient was separated into 12 fractions. The ribosomal profile in C was determined in the presence of 25 mM EDTA. (D) Quantification of m-GPx4 mRNA concentration per fraction by RT–PCR. (E) β-Actin mRNA levels per fraction according to D. (F) Statistical evaluation of m-GPx4 mRNA levels summarized for polysomal and nonpolysomal fractions. Grsf1 overexpression resulted in a shift of m-GPx4 mRNA into polysomal fractions. Mean values are given and error bars represent the standard deviation. n = 3; (*) P < 0.05.
Grsf1 and m-GPx4 exhibit a similar tissue specific distribution pattern and are coexpressed during murine embryogenesis
To investigate whether Grsf1 binding to the 5′UTR of m-GPx4 mRNA is of biological relevance, we first explored the tissue-specific expression patterns of the two mRNA species. It has been reported before (Puglisi et al. 2003) that m-GPx4 is expressed at a high level in murine testis. Using our qPCR approach, we quantified 2949 ± 521 copies of m-GPx4 mRNA per 103 copies GAPDH mRNA in testis. In addition, we observed relatively high transcript concentrations in lung (49 ± 31 copies per 103 copies GAPDH). In other tissues (kidney, brain, muscle, colon, liver, heart, and stomach), an average m-GPx4 expression of about 16 ± 6 copies of m-GPx4 mRNA per 103 GAPDH was found. For Grsf1 mRNA a similar tissue distribution was observed. High-level expression was found in testis (126 ± 10 copies per 103 copies GAPDH) followed by lung (52 ± 28 copies per 103 copies GAPDH). In the other tissues, only low steady-state concentrations of Grsf1 mRNA (average of 16 ± 15 copies per 103 copies GAPDH) were measured. This similar distribution pattern suggested a functional relation between the two proteins.
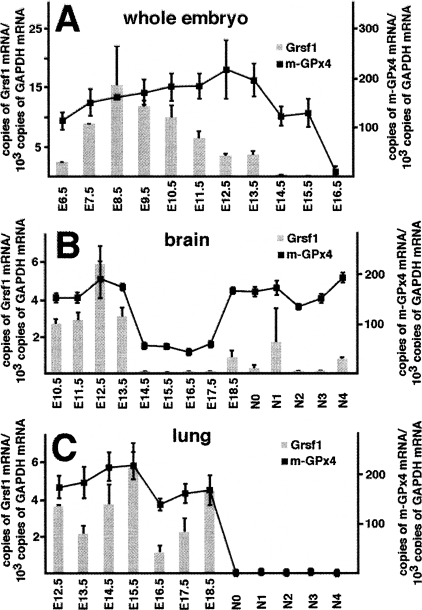
Recent studies on the role of GPx4 isoforms in murine embryogenesis indicated unique expression kinetics during embryo development (Borchert et al. 2006). If Grsf1 is involved in GPx4 expression regulation, similar expression kinetics for the two proteins during the time course of embryogenesis were expected. When we quantified the two mRNA species during embryo development, we observed parallel expression profiles but m-GPx4 expression peaks at later developmental stages (Fig. 4A). Because of their possible role in embryonic brain development, we next profiled expression of Grsf1 and m-GPx4 mRNA during the time course of perinatal brain maturation (Fig. 4B). Here again we observed parallel expression profiles of the two mRNA species, although the level of m-GPx4 mRNA exceeds that of Grsf1 mRNA by about 10-fold. For embryonic lung development, we also observed similar expression kinetics (Fig. 4C). Immediately after birth, the mRNA levels for the two proteins suddenly dropped down to undetectable levels and remained low during early postnatal development.
Figure 4.
Grsf1 and m-GPx4 are coexpressed during murine embryogenesis. Murine embryos were prepared from pregnant mice at different developmental stages E6.5–E18.5 and postnatal stages N0–N4, and total RNA was extracted. Steady-state concentrations of Grsf1 and m-GPx4 mRNA were quantified by qRT–PCR using GAPDH as an internal standard. To explore expression of the two genes during brain and lung development, these organs were prepared at different developmental stages and the two mRNA species were quantified in total RNA extracts. The numbers indicate the days of gestation. E6.5 means 6.5 d post-conception. N0 indicates the day of birth, and N1 means 24 h after birth.
Grsf1 knockdown inhibits m-GPx4 expression in embryonic brain
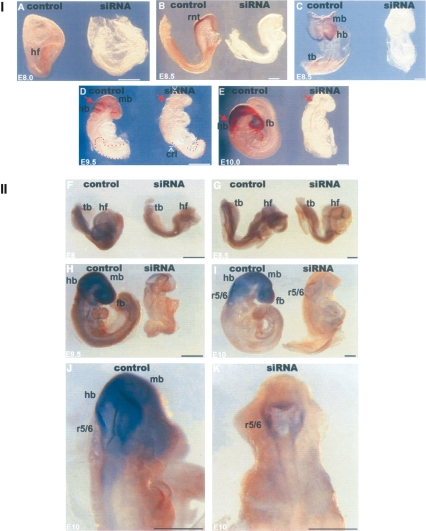
If Grsf1 is involved as a regulator in m-GPx4 expression, two consequences of Grsf1 silencing may be predicted: (1) Grsf1 knockdown should impair expression of m-GPx4. (2) Silencing of Grsf1 expression should induce similar developmental alterations as m-GPx4 knockdown. To test these hypotheses, we first explored the impact of Grsf1 siRNA constructs on m-GPx4 expression during in vitro embryogenesis. To test the suitability of our knockdown probes, we first made sure that siRNA treatment of living embryos in culture strongly reduced Grsf1 expression as indicated by in situ hybridization (Fig. 5, panel I, A–E) and RT–PCR (data not shown). When early (embryonic day 8.0, E8.0) siRNA-treated embryos were stained for m-GPx4 mRNA (Fig. 5, panel II, F,G), its expression was not significantly altered in the headfold region, in the rostal to caudal neural tube, and in the tailbud. However, at later developmental stages starting with the early somite stage, impairment of m-GPx4 mRNA expression was observed. At E9.5 and E10, m-GPx4 signals in the neuroepithelium of developing forebrain, midbrain, and hindbrain were reduced (Fig. 5, panel II, H,I), and similar alterations were detected in posterior neuroepithelium of the developing hindbrain, particularly in rhombomers 5 and 6 (Fig. 5, panel II, J,K).
Figure 5.
Targeted knockdown of Grsf1 impairs expression of m-GPx4 during murine embryogenesis and induces developmental retardation. Mouse embryos were prepared at gestational day E8, treated with control siRNA duplex (labeled as Control) or Grsf1-specific siRNA constructs (labeled as siRNA), and then cultured in vitro for up to 72 h. After different time points of the culturing period, the embryos were used for in situ hybridization using Grsf1- and m-GPx4-specific antisense probes. Dark areas indicate regions with intense hybridization signals. The different panels represent different stages of embryonic development at the end of the in vitro culturing period. Each panel consists of a left image (control embryo) and a right image (siRNA-treated embryo). Bar, 300 μm (E8–E8.5); 800 μm (E9.5–E10). (hf) Headfold; (tb) tailbud; (fb) forebrain; (mb) midbrain; (hb) hindbrain; (r) rhombomere. (Panel I) Grsf1 in situ hybridization. siRNA treatment induced abnormal mid/hindbrain development (red arrows indicate retarded mid/hindbrain boundary), posterior truncation (red dotted areas indicate shorted and twisted tail bud), and general growth retardation (white arrows indicate shorted crl). (Panel II) m-GPx4 in situ hybridization. siRNA treatment impaired m-GPx4 mRNA expression from E9.5 and induced retarded hindbrain segments at r5/6 levels.
Knockdown of Grsf1 expression induced cerebral developmental retardations
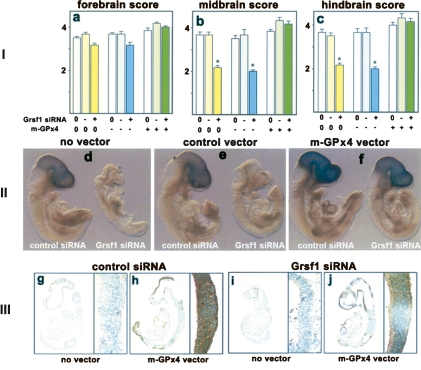
Grsf1 has been implicated in the Wnt/β-catenin signaling pathway, which is important for embryogenesis (Lickert et al. 2005). To explore whether knockdown of Grsf1 has an impact on embryonic brain development, we evaluated in vitro development of siRNA-treated embryos and corresponding controls in whole-embryo cultures. The morphological changes induced by siRNA treatment were judged by morphometric assessment of key parameters characterizing normal embryogenesis (see the Supplemental Material). Brain development was assessed according to a standard scoring procedure (Maele-Fabry et al. 1990). Applying these algorithms, the following conclusions could be drawn: (1) Silencing of Grsf1 expression induced significant growth retardations (see Supplemental Table S2) as well as impairment of midbrain and hindbrain development. In contrast, forebrain maturation was hardly impacted (Fig. 6, panel I, a–c). (2) At early stages of in vitro embryogenesis, these changes were rather subtle (Fig. 5A–C,F,G), and this may be related to the low abundance of Grsf1 expression at these developmental stages. However, at E9.5 and E10 (Fig. 5D,E,H,I), we observed severe truncation of the posterior axis and abnormal mid/hindbrain structures. (3) Shortening of the tailbud region started at E8.5, when the mid/hindbrain region still looked morphologically normal (Fig. 5B,C). Truncation of the posterior axis was well visible at E9.5 (Fig. 5D,H), although more subtle alterations were already observed at earlier stages. These morphological differences were mainly restricted to those areas of the embryos in which Grsf1 and m-GPx4 expression was detected (posterior epiblast and mid/hindbrain).
Figure 6.
m-GPx4 overexpression rescues developmental abnormalities induced by Grsf1 knockdown. Grsf1 siRNA-treated embryos (E7.5) were transfected with a mammalian overexpression vector containing or lacking (control) the m-GPx4 coding sequence and then cultured in vitro for 48 h. Then the embryos were recovered and analyzed. (Panel I) Quantification of developmental characteristics according to the scoring procedure described in the Supplemental Material. (Panel II) In situ hybridization of cotransfected embryos using a m-GPx4 specific probe. Each panel consists of a left image (control embryo) and a right image (siRNA treated embryo). (Panel III) The embryos were sectioned for immunohistochemistry analysis using a specific m-GPx4 antibody. Positive staining is indicated as dark brown areas. Each panel consists of left images (no vector) and right images (m-GPx4 overexpression vector). (Right panels) Magnified neuroepithelium regions from the brain are shown. Shown are mean values and error bars represent the standard deviation. n = 6; (*) P < 0.001.
Developmental changes induced by Grsf1 knockdown are prevented by m-GPx4 overexpression
If Grsf1 mediates its developmental effects via up-regulation of m-GPx4, recombinant overexpression of the enzyme should rescue Grsf1-siRNA-treated embryos from developmental retardations. To test this hypothesis, we overexpressed m-GPx4 during embryonic development by injecting an m-GPx4 overexpression plasmid into the yolk sac of the developing embryos and monitored m-GPx4 expression on messenger and protein level by in situ hybridization and immunohistochemistry, respectively. Overexpression of m-GPx4 in transfected embryos was confirmed by in situ hybridization and RT–PCR (data not shown), and we observed this regardless of whether the embryos were treated with the Grsf1 siRNA or not (Fig. 6, panel II). To show overexpression of the m-GPx4 protein, we performed immunohistochemical staining of the embryos using a GPx4-specific antibody (Fig. 6, panel III). Here again we also observed strong staining of the embryos transfected with the m-GPx4 overexpression plasmid regardless of whether or not they haven been cotransfected with the Grsf1 knockdown construct (Fig. 6, panel III). This finding is not surprising since the overexpression plasmid contains only the m-GPx4 coding sequence and 3′UTR but lacks the m-GPx4 5′UTR. The m-GPx4-transfected embryos did not show signs of abnormal growth characteristics or structural alterations (data not shown). Most interestingly, however, was our finding that embryos that were co-transfected with the m-GPx4 overexpression plasmids and the Grsf1 knockdown construct did not show signs of developmental retardations in the midbrain and hindbrain (Fig. 6, panel I). These data indicate that m-GPx4 overexpression rescued the embryos from developmental retardations induced by Grsf1 knockdown.
Grsf1 knockdown induced lipid peroxidation, and apoptosis is prevented by m-GPx4 overexpression
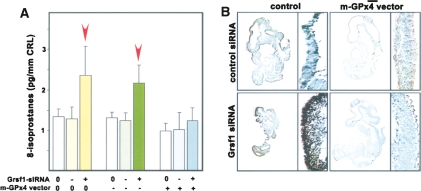
Next we attempted to explore the mechanistic basis of developmental retardation induced by Grsf1 knockdown. Since m-GPx4 exhibits anti-oxidative and anti-apoptotic properties, we hypothesized that Grsf1-deficient embryos might show signs of increase lipid peroxidation and apoptosis. To test this assumption, we quantified the isoprostane (general marker of lipid peroxidation [Wang et al. 2007]) content of embryos and found that treatment with Grsf1 knockdown constructs induced lipid peroxidation (Fig. 7A, left columns). Similar effects were obtained when the embryos were co-transfected with an overexpression plasmid lacking the m-GPx4 sequence (Fig. 7A, middle columns). However, when the embryos were co-transfected with Grsf1 knockdown and m-GPx4 overexpression constructs, no significant increase in isoprostane levels was observed. Similar results were obtained when we quantified the degree of apoptosis using the TUNEL assay (Fig. 7B). Treatment of the embryos with the Grsf1 knockdown construct induced strong apoptosis as indicated by black staining. In contrast, after co-transfection with the m-GPx4 overexpression plasmid, apoptosis was strongly reduced.
Figure 7.
m-GPx4 overexpression reduces cerebral lipid peroxidation and apoptosis induced by Grsf1 knockdown. Grsf1 siRNA-treated embryos (E7.5) were transfected with a mammalian overexpression vector containing or lacking (control) the m-GPx4 coding sequence. After a culturing period of 48 h, the embryos were recovered and analyzed for lipid peroxidation (isoprostane content, A) and apoptosis (TUNEL assay, B). Shown are mean values, and error bars represent the standard deviation. n = 5; (*) P < 0.01.
Discussion
GPx4 is a moonlighting selenoprotein, which has been implicated in anti-oxidative defense (Ursini and Bindoli 1987), sperm development (Ursini et al. 1999), apoptosis (Imai and Nakagawa 2003), and murine embryogenesis (Imai et al. 2003; Yant et al. 2003). Several mechanisms of transcriptional regulation (Borchert et al. 2003; Maiorino et al. 2003) have been suggested for various GPx4-isoforms, but cell activation studies (Kuhn and Borchert 2002; Tramer et al. 2002; Sneddon et al. 2003) and nonsense-mediated decay experiments (Sun et al. 2001; Muller et al. 2003) suggested additional post-transcriptional elements. Besides GPx4, other enzymes involved in eicosanoid metabolism such as 15-lipoxygenase and cyclooxygenase-2 are known to be regulated by post-transcriptional mechanisms, and these regulatory processes depend on characteristic structural motifs within the untranslated mRNA regions (Reimann et al. 2002; Hall-Pogar et al. 2007). Here we report that the 5′UTR of m-GPx4 mRNA contains cis-regulatory sequences and identified Grsf1 as a trans-acting factor modulating translation of m-GPx4 mRNA. Grsf1 was originally identified as an mRNA-interacting protein capable of binding to G-rich sequences (Qian and Wilusz 1994). Using the yeast three-hybrid system, we identified a Grsf1 isoform (K6-Grsf1) as an m-GPx4 mRNA-binding protein. K6-Grsf1 contains three RNA-recognition motives (RRM-domain) that are essential for RNA binding (Kash et al. 2002). Sequence alignments indicated that the RRM-domain is one of the most abundant protein domains in eukaryotes (Maris et al. 2005), and RRM-containing proteins have been implicated in numerous post-transcriptional events, such as pre-mRNA processing, splicing, mRNA stability, mRNA export, and translation regulation (Birney et al. 1993). The human ortholog of Grsf1 has been implicated in translational activation of influenza virus nucleocapsid protein (NP) in infected cells (Park et al. 1999; Kash et al. 2002). Here we found that 5′UTRs of influenza NP and m-GPx4 compete for binding at murine Grsf1, but the m-GPx4 5′UTR binds with much higher affinity. Our results indicate that Grsf1 binds to m-GPx4 mRNA reconstituted in in vitro systems as well as in in vivo conditions. Moreover, our data suggest that Grsf1 up-regulates m-GPx4 expression at the translational level by binding to its A(G)4A recognition sequence in the 5′UTR and recruiting m-GPx4 mRNA to the polysomal fractions and thereby translational activation. This A(G)4A motif and the successive stem/loop structure are conserved in the GPx4 gene of human and rat, whereas in pig and cow, the A(G)4A motif is conserved but is lacking the successive stem/loop structure, suggesting conserved regulatory mechanisms for m-GPx4 expression in mammals. The related D(G)3D consensus motif has recently been described as a recognition sequence for members of the hnRNP H (heterogeneous nuclear ribonucleoprotein H) family including Grsf1, and this has been implicated in the formation of spliceosomes (Schaub et al. 2007). Our yeast three-hybrid screen did not reveal binding of other hnRNP H family members, likely to be due to the highly stringent screening parameters used throughout that procedure. Putative Grsf1-binding motifs have been identified in a broad range of target sequences involved in development (Lickert et al. 2005), cell proliferation, apoptosis, and inflammation (Kash et al. 2002). Performing an ungapped BLAST search for the A(G)4A recognition motif in a nonredundant database of UTRs (Altschul et al. 1997), we found 12 murine cDNAs containing this Grsf1-binding motif in their UTRs (see Supplemental Table S3) including Krox20 and targets involved in spermatogenesis. In contrast, no Grsf1-binding motif is pres-ent in the 5′UTR of any other member of the GPx family.
To test the biological role of the Grsf1/GPx4-mRNA interaction, we used a multiple approach: (1) We assayed the tissue distribution of Grsf1 and compared the expression pattern with that of m-GPx4. Here we found that both proteins are preferentially expressed in the male reproductive system and in the lung. Since GPx4 has been implicated in spermatogenesis, a similar involvement might be predicted for Grsf1. Interestingly, the Grsf1 messenger was recently identified as substrate for mDAZL, an RNA-binding protein implicated in germ cell generation (Jiao et al. 2002). (2) We profiled the expression of Grsf1 and m-GPx4 during murine embryogenesis and observed coexpression. Moreover, we found parallel expression of m-GPx4 and Grsf1 during perinatal organogenesis of brain and lung, suggesting a functional relation between them. (3) Finally, we silenced Grsf1 expression during in vitro embryogenesis and observed subsequent knockdown of m-GPx4 expression and a strong induction of apoptosis. The phenotype of Grsf1 knockdown embryos resembled that of m-GPx4 silencing, suggesting that m-GPx4 constitutes a downstream target of Grsf1. The knockdown phenotype can be overcome by m-GPx4 overexpression, indicating that Grsf1 knockdown effects are dominantly due to impaired m-GPx4 expression rather than other potential Grsf1 target mRNAs.
Grsf1 is an important mediator in Wnt/β-catenin signaling, which is essential for embryogenesis (Lickert et al. 2005). Here, we confirmed this finding by showing that midbrain and hindbrain development is particularly sensitive against silencing of Grsf1 expression. RNA-binding proteins are potent modulators of post-transcriptional processes throughout embryogenesis (Webster et al. 1997; Niessing et al. 2002; Nakamura et al. 2004), and their expression during brain development follows distinct kinetic and spatial patterns (McKee et al. 2005). RNA-binding proteins, such as ELAV/Hu or Nova1, have been implicated in brain development and function (Perrone-Bizzozero and Bolognani 2002), but little is known on the molecular mechanisms of their cellular activities. Our results indicate that specific silencing of an mRNA-binding protein induces retardation of brain development. Previous studies suggested that m-GPx4 exhibits anti-apoptotic properties (Imai and Nakagawa 2003; Schnabel et al. 2006), and silencing of m-GPx4 expression during embryogenesis induces cerebral apoptosis (Borchert et al. 2006). Here, Grsf1 knockdown also induced apoptosis, and this mechanism may contribute to the observed developmental retardations.
An unsolved problem, which may be addressed in follow-up studies, is the question about the role of Grsf1 in the male reproductive system. The high Grsf1 expression in testis and the unusual tissue distribution suggest a role in sperm development. Preliminary immunohistochemical staining and expression profiles of the two proteins during sperm development (data not shown) suggested coexpression of Grsf1 and m-GPx4. These data are consistent with the finding that Grsf1 is involved in regulation of m-GPx4 translation. However, for a regulatory protein, the expression level in testis is surprisingly high (126 ± 10 copies of Grsf1 mRNA per 103 copies GAPDH mRNA), which opens the possibility that the Grsf1 protein may exhibit an additional undefined function in the male reproductive system. A similar moonlighting activity has been suggested for GPx4 (Ursini et al. 1999).
Taken together, using a yeast three-hybrid approach under highly stringent experimental conditions, we isolated a single positive yeast clone that expressed Grsf1. In vitro recombinant Grsf1 binds to an A(G)4A motif in the 5′UTR of the m-GPx4 mRNA, and in vivo interaction was indicated by RNA immunoprecipitation. Induction of Grsf1 up-regulates 5′UTR-dependent expression of reporter gene constructs and recruits m-GPx4 mRNA to the translationally active polysomal fraction as shown by polysomal gradient analysis. Moreover, Grsf1 knockdown during embryogenesis impaired m-GPx4 expression and induced similar retardations in cerebral development accompanied by strong induction of apoptosis as shown by TUNEL assay as direct silencing of m-GPx4 (Borchert et al. 2006). On the other hand, m-GPx4 overexpression relieves apoptosis in the developing neuroepithelium and rescues the Grsf1 knockdown phenotype, revealing the significance of this specific protein/RNA interaction for normal embryonic brain development. Since the Grsf1-binding sequence also occurs in other mRNA species including markers of brain development such as Krox20 (Supplemental Table S3), the mechanisms described here may be of more general importance for developmental processes.
Materials and methods
Chemicals
The chemicals used were from the following sources: SuperScript III reverse transcriptase and RNaseOUT from Invitrogen; BD Advantage 2 Polymerase Mix from BD Biosciences (Pharmingen); dNTPs from Carl Roth GmbH; QuantiTect SYBR Green PCR Kit from Qiagen; and PCR primers from BIOTEZ. The yeast strains YBZ-1 and R40coat as well as the plasmid pIIIA/MS2-1 were a generous gift from D. Bernstein (University Wisconsin–Madison, Madison).
Yeast three-hybrid system
The yeast three-hybrid system constitutes a complex screening method for detection of RNA-binding proteins (Bernstein et al. 2002). We used this method to screen a murine testis expression library for proteins capable of binding to the 5′UTR of the m-GPx4 mRNA. A detailed description of the assay system and the experimental protocol is given in the Supplemental Material.
RNA extraction, reverse transcription, and in vitro transcription
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN), and contaminating DNA was digested using Turbo DNase (Ambion). RNA was then reversely transcribed according to standard protocols with oligo d(T)18 primers. RNA probes were generated using the T7 Megashortscript Kit (Ambion) in the presence or absence of small amounts of Biotin-16-UTP (Kon3) or Digoxigenin-11-UTP (Roche). The template cDNA encoding for the 5′UTR of the m-GPx4 mRNA was generated by PCR using the primer combination specified in Supplemental Table S1 fusing the T7 promoter to the forward primer. In vitro transcription products were purified using the MEGAclear kit from Ambion.
RNA mobility shift assays
For RNA/protein-binding studies, different amounts of protein (400 ng if not stated otherwise) were incubated with 0.5–1 pmol of biotin- or digoxigenin-labeled RNA probe for 20 min at 30°C in binding buffer (10 mM HEPES-NaOH at pH 7.9, 2.5 mM KCl, 1.5 mM EDTA, 4% glycerol, 0.25 mM DTT, 7.5 μg/μL heparin, 25 ng/μL yeast tRNA) in a reaction volume of 15 μL. Afterward the samples were analyzed by native 5% polyacrylamide gel electrophoresis. Protein/RNA complexes were transferred to a nylon membrane, and blots were visualized using Biotin or DIG Luminescent Detection Kits (Roche).
UTR-dependent reporter gene assays and qRT–PCR
UTR-dependent reporter gene assays were carried out as described before (Fahling et al. 2006) in MEFs (see the Supplemental Material) using a modified pGL3-promoter vector (Promega). For our purpose, the vector-specific 5′UTR of luciferase mRNA was replaced by the m-GPx-4 5′UTR. For the deletion experiments, the AGGGGA motif was removed by PCR. qRT–PCR was carried out as described before (Borchert et al. 2006) with a Rotor Gene 3000 system (Corbett Research) using the QuantiTect SYBR Green PCR Kit from Qiagen. A detailed description of the translation assay and of the qPCR protocol including the primers is given in Supplemental Table S1.
RNA immunoprecipitation
In order to detect in vivo RNA/protein interactions, RNA immunoprecipitation was performed as described before (Gilbert et al. 2004) with some modifications, and a detailed outline of the experimental procedures is given in the Supplemental Material. N2a cells were transfected with pCMV-3Tag-1 (Invitrogen) containing the Grsf1 coding sequence or without insert (mock-treatment) using Lipofectamine 2000 (Invitrogen) according to the vendor’s instructions.
Polysomal gradient assay
MEFs were transfected with Grsf1 expression vector or empty vector using the RotiFect (Carl Roth GmbH) transfection reagent. After 24 h, cells were incubated with cycloheximide (100 μg/mL) for 10 min prior to lysis with buffer containing 20 mM Tris (pH 7.4), 150 mM KCl, 30 mM MgCl2, 100 μg/mL cycloheximide, 1 mM DTT, 1× proteinase inhibitor cocktail (Roche Diagnostics GmbH), 100 U/mL RNase inhibitor (Fermentas GmbH), and 0.5% Nonidet P-40. After 10 min on ice, cytosolic extracts were obtained after centrifugation at 10,000g for 10 min. The cytoplasmic supernatant was then layered onto 11 mL of a linear 17%–51% sucrose gradient (0.5 to 1.5 M sucrose, 20 mM Tris at pH 7.4, 150 mM KCl, 5 mM MgCl2, 1 mM DTT) and centrifuged for 2 h at 36,000 rpm using a Beckman SW-41 rotor. Following centrifugation, the gradient solution was pumped out from the bottom with a peristaltic pump, and the ribosomal profile was continuously determined at an absorbance of 254 nm using a 2138 UVICORD-S UV monitor (LKB Bromma). Each sucrose gradient was fractionated into 12 fractions. RNA was isolated using the E.Z.N.A. RNA total kit according to the manufacturer’s protocol.
Recombinant expression and purification of Grsf1
Recombinant Grsf1 was expressed in E. coli as GST fusion protein. It was purified from the bacterial lysate supernatant by affinity chromatography on a glutathione agarose column (see the Supplemental Material for details). GST-tagged recombinant proteins were eluted from the column using a step gradient of reduced glutathione and used for electrophoretic mobility shift assays without further purification.
Preparation and in vitro culture of murine embryos
All animal experiments were performed in strict adherence to the guidelines for experimentation with laboratory animals set in institutions. Inbred ICR pregnant mice were obtained from the animal house, and embryos in different developmental stages (E6.5 to postnatal day 5, N5) were prepared under a stereo-microscope (Olympus). For qRT–PCR, preparations were kept in PBS (0.1% DECP) for separation of extraembryonic tissues. At later developmental stages, embryonic brain and lung (from E10.5 to N4) were separately dissected. Different embryonic tissues from the same litter were pooled, and at least three dams were collected independently. Tissue samples were kept in RNAlater solution (Qiagen) at 4°C overnight and stored at –80°C prior to RNA extraction. For in vitro culture, the embryos were dissected in PB1 medium (5% FBS) and then placed in a whole embryo culture roller incubator (BTC Engineering). The embryos at early gastrulation stage (E7.5) were allowed to develop for up to 72 h in 100% heat-inactivated rat serum with a continuous flow of gas mixtures (Wang et al. 2007).
siRNA experiments and in situ hybridization
Grsf1-specific siRNA probes were designed using the Stealth RNAi program (BLOCK-iT RNAi Designer; Invitrogen), and the following sequences were selected: Grsf antisense probe, 5′-UA UUUCAUACACUUCCACAUACCGC-3′ and Grsf1 sense probe, 5′-GCGGUAUGUGGAAGUGUAUGAAAUA-3′. Control siRNA duplex with no homology with any vertebrate transcriptome was used as the control siRNA group (Stealth RNAi Negative Controls; Invitrogen). Murine embryos were removed from pregnant mice at E7.5 of pregnancy and transfected with siRNA constructs. For transfection, the annealed double-stranded siRNA (50–100 nM) was mixed with 0.01% Lipofectamine 2000 (Invitrogen) and then microinjected with an ASTP micromanipulator (Leica) into the amniotic cavity of the gastrulating embryos at E7.5. After microinjection, the embryos were placed in a whole embryo culture roller incubator as indicated above. For whole-mount in situ hybridization (Borchert et al. 2006), siRNA-treated and control siRNA-treated embryos were fixed in 4% p-formaldehyde (in PBS), dehydrated in graded methanol, and stored at −20°C. Suitable riboprobes (sense and antisense probes) were prepared by PCR, cloned, and transcribed using T7 RNA polymerase. Primer sequences are specified in Supplemental Table S1. To label the RNA probes, digoxigenin-11-UTP (Roche) was incorporated using the AmpliScribe T7 kit (Epicentre Technologies).
m-GPx4 overexpression and functional analyses
Grsf1 siRNA-treated embryos were transfected with pcDNA3.1 m-GPx4 overexpression vector before cultivation at the E7.5 stage. Extracted plasmid vector in 50 ng/μL concentrations was injected into the yolk sac of the embryos. Untreated controls (sham operated) and negative controls by microinjecting vector plasmid without m-GPx4 sequence were included in the in vitro embryo culture. The developing embryos were removed at the E9.5 stage for morphological examination. m-GPx4 overexpression was confirmed by qRT–PCR and whole mount in situ hybridization as described above. Protein expression was then assessed by immunohistochemistry analysis. Immunohistochemical staining of m-GPx4 was performed using a monoclonal anti-human m-GPx4 antibody raised against the pure recombinant Sec46Cys-mutant and cross-reactive with murine m-GPx4 (Borchert et al. 2006). After in situ hybridization, embryos were fixed in 4% p-formaldehyde, washed at 4°C in PBS, dehydrated, embedded in paraffin wax, and cut into 5-μm sections. Apoptotic cells were stained by standard TUNEL technique. Tissue sections were incubated with TdT enzyme and conjugated with anti-digoxigenin peroxidase (Chemicon) for color development. Oxidative stress in embryos was quantified by 8-isoprostane ELIZA methods as described (Chan et al. 2002).
Acknowledgments
Special thanks to Ms. Dawn Lui and Nina Chu for their technical support in immunohistochemistry analysis and the TUNEL assay. This work was supported in part by research grants of the European Commission (FP6,LSHM-CT-2004-0050333) to H.K. and of Deutsche Forschungsgemeinschaft to B.J.T. (Th 459/5-1) as well as by two research grants (Special Equipment Grant [Medicine/ASTP] and Li Ka Shing Institute of Health Sciences) to C.C.W., and to C.U. and E.E.B. a grant from The Leverhulme Trust.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.466308.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D.S., Buter N., Stumpf C., Wickens M. Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. doi: 10.1016/S1046-2023(02)00015-4. [DOI] [PubMed] [Google Scholar]
- Birney E., Kumar S., Krainer A.R. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert A., Savaskan N.E., Kuhn H. Regulation of expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Tissue-specific expression pattern and identification of functional cis- and trans-regulatory elements. J. Biol. Chem. 2003;278:2571–2580. doi: 10.1074/jbc.M209064200. [DOI] [PubMed] [Google Scholar]
- Borchert A., Wang C.C., Ufer C., Schiebel H., Savaskan N.E., Kuhn H. The role of phospholipid hydroperoxide glutathione peroxidase isoforms in murine embryogenesis. J. Biol. Chem. 2006;281:19655–19664. doi: 10.1074/jbc.M601195200. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic. Biol. Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R., Maurer S., Lotzer K., Bol G., Kallionpaa H., Lehtolainen P., Viita H., Yla-Herttuala S. Overexpression of PHGPx inhibits hydroperoxide-induced oxidation, NFκB activation and apoptosis and affects oxLDL-mediated proliferation of rabbit aortic smooth muscle cells. Atherosclerosis. 2000;152:307–316. doi: 10.1016/s0021-9150(99)00486-4. [DOI] [PubMed] [Google Scholar]
- Chan L.Y., Chiu P.Y., Siu N.S., Wang C.C., Lau T.K. Diclofenac-induced embryotoxicity is associated with increased embryonic 8-isoprostaglandin F2α level in rat whole embryo culture. Reprod. Toxicol. 2002;16:841–844. doi: 10.1016/s0890-6238(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahling M., Mrowka R., Steege A., Nebrich G., Perlewitz A., Persson P.B., Thiele B.J. Translational control of collagen prolyl 4-hydroxylase-α(i) gene expression under hypoxia. J. Biol. Chem. 2006;281:26089–26101. doi: 10.1074/jbc.M604939200. [DOI] [PubMed] [Google Scholar]
- Gilbert C., Kristjuhan A., Winkler G.S., Svejstrup J.Q. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Hall-Pogar T., Liang S., Hague L.K., Lutz C.S. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H., Silver P.A. A systems view of mRNP biology. Genes & Dev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- Imai H., Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- Imai H., Hirao F., Sakamoto T., Sekine K., Mizukura Y., Saito M., Kitamoto T., Hayasaka M., Hanaoka K., Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem. Biophys. Res. Commun. 2003;305:278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- Jiao X., Trifillis P., Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- Kash J.C., Cunningham D.M., Smit M.W., Park Y., Fritz D., Wilusz J., Katze M.G. Selective translation of eukaryotic mRNAs: Functional molecular analysis of GRSF-1, a positive regulator of influenza virus protein synthesis. J. Virol. 2002;76:10417–10426. doi: 10.1128/JVI.76.20.10417-10426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp E.A., Arndt T.L., Eng K.L., Caldwell M., LeBoeuf R.C., Deeb S.S., O’Brien K.D. Murine phospholipid hydroperoxide glutathione peroxidase: cDNA sequence, tissue expression, and mapping. Mamm. Genome. 1999;10:601–605. doi: 10.1007/s003359901053. [DOI] [PubMed] [Google Scholar]
- Kuhn H., Borchert A. Regulation of enzymatic lipid peroxidation: The interplay of peroxidizing and peroxide reducing enzymes. Free Radic. Biol. Med. 2002;33:154–172. doi: 10.1016/s0891-5849(02)00855-9. [DOI] [PubMed] [Google Scholar]
- Lickert H., Cox B., Wehrle C., Taketo M.M., Kemler R., Rossant J. Dissecting Wnt/β-catenin signaling during gastrulation using RNA interference in mouse embryos. Development. 2005;132:2599–2609. doi: 10.1242/dev.01842. [DOI] [PubMed] [Google Scholar]
- Maele-Fabry G.V., Delhaise F., Picard J.J. Morphogenesis and quantification of the development of post-implantation mouse embryo. Toxicol. In Vitro. 1990;4:149–156. doi: 10.1016/0887-2333(90)90037-t. [DOI] [PubMed] [Google Scholar]
- Maiorino M., Scapin M., Ursini F., Biasolo M., Bosello V., Flohe L. Distinct promoters determine alternative transcription of gpx-4 into phospholipid-hydroperoxide glutathione peroxidase variants. J. Biol. Chem. 2003;278:34286–34290. doi: 10.1074/jbc.M305327200. [DOI] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- McKee A.E., Minet E., Stern C., Riahi S., Stiles C.D., Silver P.A. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C., Wingler K., Brigelius-Flohe R. 3′UTRs of glutathione peroxidases differentially affect selenium-dependent mRNA stability and selenocysteine incorporation efficiency. Biol. Chem. 2003;384:11–18. doi: 10.1515/BC.2003.002. [DOI] [PubMed] [Google Scholar]
- Nakamura A., Sato K., Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nam S., Nakamuta N., Kurohmaru M., Hayashi Y. Cloning and sequencing of the mouse cDNA encoding a phospholipid hydroperoxide glutathione peroxidase. Gene. 1997;198:245–249. doi: 10.1016/s0378-1119(97)00321-1. [DOI] [PubMed] [Google Scholar]
- Niessing D., Blanke S., Jackle H. Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes & Dev. 2002;16:2576–2582. doi: 10.1101/gad.240002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.W., Wilusz J., Katze M.G. Regulation of eukaryotic protein synthesis: Selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. 1999;96:6694–6699. doi: 10.1073/pnas.96.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone-Bizzozero N., Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 2002;68:121–126. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- Pfeifer H., Conrad M., Roethlein D., Kyriakopoulos A., Brielmeier M., Bornkamm G.W., Behne D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15:1236–1238. [PubMed] [Google Scholar]
- Puglisi R., Tramer F., Panfili E., Micali F., Sandri G., Boitani C. Differential splicing of the phospholipid hydroperoxide glutathione peroxidase gene in diploid and haploid male germ cells in the rat. Biol. Reprod. 2003;68:405–411. doi: 10.1095/biolreprod.102.006544. [DOI] [PubMed] [Google Scholar]
- Qian Z., Wilusz J. GRSF-1: A poly(A)+ mRNA binding protein which interacts with a conserved G-rich element. Nucleic Acids Res. 1994;22:2334–2343. doi: 10.1093/nar/22.12.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Q., Gu M., Van Remmen H., Strong R., Roberts J.L., Richardson A. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J. Neurosci. Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- Reimann I., Huth A., Thiele H., Thiele B.J. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J. Mol. Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- Roveri A., Flohe L., Maiorino M., Ursini F. Phospholipid-hydroperoxide glutathione peroxidase in sperm. Methods Enzymol. 2002;347:208–212. doi: 10.1016/s0076-6879(02)47020-6. [DOI] [PubMed] [Google Scholar]
- Savaskan N.E., Borchert A., Brauer A.U., Kuhn H. Role for glutathione peroxidase-4 in brain development and neuronal apoptosis: Specific induction of enzyme expression in reactive astrocytes following brain injury. Free Radic. Biol. Med. 2007;43:191–201. doi: 10.1016/j.freeradbiomed.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Schaub M.C., Lopez S.R., Caputi M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J. Biol. Chem. 2007;282:13617–13626. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]
- Schnabel D., Salas-Vidal E., Narvaez V., Sanchez-Carbente Mdel R., Hernandez-Garcia D., Cuervo R., Covarrubias L. Expression and regulation of antioxidant enzymes in the developing limb support a function of ROS in interdigital cell death. Dev. Biol. 2006;291:291–299. doi: 10.1016/j.ydbio.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Schnurr K., Belkner J., Ursini F., Schewe T., Kuhn H. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase controls the activity of the 15-lipoxygenase with complex substrates and preserves the specificity of the oxygenation products. J. Biol. Chem. 1996;271:4653–4658. doi: 10.1074/jbc.271.9.4653. [DOI] [PubMed] [Google Scholar]
- Sneddon A.A., Wu H.C., Farquharson A., Grant I., Arthur J.R., Rotondo D., Choe S.N., Wahle K.W. Regulation of selenoprotein GPx4 expression and activity in human endothelial cells by fatty acids, cytokines and antioxidants. Atherosclerosis. 2003;171:57–65. doi: 10.1016/j.atherosclerosis.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Sun X., Li X., Moriarty P.M., Henics T., LaDuca J.P., Maquat L.E. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol. Biol. Cell. 2001;12:1009–1017. doi: 10.1091/mbc.12.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.P., Geiger P.G., Maiorino M., Ursini F., Girotti A.W. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochim. Biophys. Acta. 1990;1045:252–260. doi: 10.1016/0005-2760(90)90128-k. [DOI] [PubMed] [Google Scholar]
- Tramer F., Micali F., Sandri G., Bertoni A., Lenzi A., Gandini L., Panfili E. Enzymatic and immunochemical evaluation of phospholipid hydroperoxide glutathione peroxidase (PHGPx) in testes and epididymal spermatozoa of rats of different ages. Int. J. Androl. 2002;25:72–83. doi: 10.1046/j.1365-2605.2002.00327.x. [DOI] [PubMed] [Google Scholar]
- Tramer F., Vetere A., Martinelli M., Paroni F., Marsich E., Boitani C., Sandri G., Panfili E. cAMP-response element modulator-tau activates a distinct promoter element for the expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Biochem. J. 2004;383:179–185. doi: 10.1042/BJ20040974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufer C., Borchert A., Kuhn H. Functional characterization of cis- and trans-regulatory elements involved in expression of phospholipid hydroperoxide glutathione peroxidase. Nucleic Acids Res. 2003;31:4293–4303. doi: 10.1093/nar/gkg650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F., Bindoli A. The role of selenium peroxidases in the protection against oxidative damage of membranes. Chem. Phys. Lipids. 1987;44:255–276. doi: 10.1016/0009-3084(87)90053-3. [DOI] [PubMed] [Google Scholar]
- Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohe L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- Wang C.C., Chu K.O., Chong W.S., Li W.Y., Pang C.P., Shum A.S., Lau T.K., Rogers M.S. Tea epigallocatechin-3-gallate increases 8-isoprostane level and induces caudal regression in developing rat embryos. Free Radic. Biol. Med. 2007;43:519–527. doi: 10.1016/j.freeradbiomed.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Webster P.J., Liang L., Berg C.A., Lasko P., Macdonald P.M. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes & Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L.J., Ran Q., Rao L., Van Remmen H., Shibatani T., Belter J.G., Motta L., Richardson A., Prolla T.A. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- Yeo W., Gautier J. Early neural cell death: Dying to become neurons. Dev. Biol. 2004;274:233–244. doi: 10.1016/j.ydbio.2004.07.026. [DOI] [PubMed] [Google Scholar]