Abstract
Aims
Cyclin-dependent kinase inhibitors (CDKIs) play a critical role in negatively regulating the proliferation of cardiomyocytes, although their role in cardiac differentiation remains largely undetermined. We have shown that the most prominent CDKI in Xenopus, p27Xic1(Xic1), plays a role in neuronal and myotome differentiation beyond its ability to arrest the cell cycle. Thus, we investigated whether it plays a similar role in cardiomyocyte differentiation.
Methods and results
Xenopus laevis embryos were sectioned, and whole-mount antibody staining and immunofluorescence studies were carried out to determine the total number and percentage of differentiated cardiomyocytes in mitosis. Capped RNA and/or translation-blocking Xic1 morpholino antisense oligonucleotides (Xic1Mo) were microinjected into embryos, and their role on cardiac differentiation was assessed by in situ hybridization and/or PCR. We show that cell-cycling post-gastrulation is not essential for cardiac differentiation in Xenopus embryos, and conversely that some cells can express markers of cardiac differentiation even when still in cycle. A targeted knock-down of Xic1 protein by Xic1Mo microinjection decreases the expression of markers of cardiac differentiation, which can be partially rescued by co-injection of full-length Xic1 RNA, demonstrating that Xic1 is essential for heart formation. Furthermore, using deleted and mutant forms of Xic1, we show that neither its abilities to inhibit the cell cycle nor the great majority of CDK kinase activity are essential for Xic1’s function in cardiomyocyte differentiation, an activity that resides in the N-terminus of the molecule.
Conclusion
Altogether, our results demonstrate that the CDKI Xic1 is required in Xenopus cardiac differentiation, and that this function is localized at its N-terminus, but it is distinct from its ability to arrest the cell cycle and inhibit overall CDK kinase activity. Hence, these results suggest that CDKIs play an important direct role in driving cardiomyocyte differentiation in addition to cell-cycle regulation.
Keywords: p27Xic1, CDK inhibitor, Cardiomyocyte, Differentiation, Cell cycle
1. Introduction
The adult mammalian heart is generally considered a terminally differentiated organ due to the absence of a significant regenerative ability, following incidences such as ischaemic attack,1,2 the major cause of morbidity and mortality in the Western world. It has been argued that the acquired specialization of the differentiated adult cardiomyocytes renders the majority of them incapable of proliferation, whereas a number of studies have demonstrated that the embryonic cardiomyocytes have considerable ability to divide.2 In the mammalian heart, this gradual developmental loss of cardiomyocyte proliferative capacity, leading to a transition from hyperplastic to hypertrophic growth, has been attributed to a block in the cell cycle with the majority of adult cardiomyocytes blocked in the G0/G1 phase of the cell cycle.3 It has been demonstrated that the expression of negative regulators of cell cycle, and in particular the cyclin-dependent kinase inhibitors (CDKIs) p21Cip1 and p27Kip1 significantly increases during rat myocyte development.4,5 Recently, it has become clear that CDKIs, while playing a crucial role in cell-cycle exit in multiple tissues of the developing organism, may also play addition roles in specification and differentiation, over and above their ability to regulate cell division.6 In particular, CDKIs have been shown to play roles beyond the cell cycle during neuronal differentiation.7,8 A cell-cycle-independent role has been confirmed by studies of mammalian p27Kip1 in the developing mouse cerebral cortex. p27Kip1 promotes both differentiation and radial migration of cortical projection neurons and both these functions are independent of its ability to regulate the cell cycle.9 Although it is generally accepted that there is an inverse relationship between proliferation and differentiation, and molecules that induce cell-cycle exit promote differentiation,10 the processes of cardiac differentiation and cell-cycle withdrawal have generally been investigated in isolation, and the links between these processes have not been fully elucidated. Thus, the identification of molecules that might play a pivotal role in the decision to proliferate or differentiate is crucial for a clearer understanding of cardiac development.
Here, we investigate the role of cell-cycle exit and CDKI function in the development of Xenopus embryonic heart, showing that by stage-29/30, when the heart can be distinguished morphologically, the vast majority of the cardiomyocytes have exited the cell cycle. However, a small proportion (∼1.5%) remains cycling even up to stage-41. By blocking cell division from the gastrula stage, we show that cell proliferation is not essential for differentiation of cardiomyocytes, and the resulting heart remains essentially normal in size, as determined by the expression of markers of cardiac differentiation. The CDKI Xic1 is expressed in the developing heart and while overexpression of Xic1 has little effect, depletion of Xic1 protein using antisense-morpholino- oligonucleotides (Mo) leads to a substantial reduction in the expression of markers of cardiac differentiation. This is due to decrease in the number of differentiated cardiomyocytes, resulting in a reduction in heart size, an effect that can be rescued by restoring Xic1 protein. We further demonstrate that Xic1 is required at later stages of cardiac differentiation. Significantly, the activity of Xic1 required for cardiac differentiation is localized within its N-terminus and is distinct from Xic1’s CDK inhibitory activity.
2. Materials and methods
2.1. Xenopus embryos
Xenopus laevis embryos were injected by standard methods using either β-galactosidase mRNA (500 pg injected/embryo), or Biotin-Dextran (70 kDa, 10 ng injected/embryo) developed with ExtrAvidin-alkaline phosphatase (Sigma) and BCIP, as lineage tracers. Only embryos expressing the lineage tracers in the appropriately targeted regions were analysed further. The investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85–23, revised 1996).
2.2. mRNA injection and morpholinos
Xic1 morpholino (Xic1Mo) previously showed to block Xic1 translation8 and a new control morpholino (CTRMo) with the five-nucleotide substitutions were used (Supplementary material online, Figure S1A). Capped RNAs were synthesized in vitro using the SP6 Message Machine kit (Ambion) from nuc-β-gal (500 pg),11 full-length WT-Xic1, 30 pg; CT-Xic1, 30 pg; and Xic1(35–96), 30 pg; NT-Xic1, 15 pg12 (Supplementary material online, Figure S2). Xic1 mRNA not targeted by Xic1Mo (FL-Xic1, NT-Xic1, and Xic1CK-) used for rescuing experiments had eight translationally silent nucleotide substitutions (underlined), 5′-AUGGCUGCAUUUCAUAUAGCGUUAC-3′, (Supplementary material online, Figures S1B and S2).
2.3. Reverse transcriptase–PCR
Total RNA was isolated from three whole embryos, per stage and condition, using the RNeasy kit (Qiagen) and treated with DNaseI before reverse transcription using 100 ng of RNA/reaction (SuperscriptIII-RT, Invitrogen). Two microlitre of the synthesized cDNA was then used as a template in a 20 µL PCR reaction incorporating 32P-dATP.13 The optimum cycle number for each primer pair was determined as the cycle number at the linear range of a standard curve, constructed using a range of cDNA dilutions at different cycle numbers. xODC, 55°C, 20-cycles14 also used for qPCR; p27Xic1, 55°C, 22-cycles;8 cardiac Troponin I (TIc)-F: 5′-TCGGTCCTATGCCACAGAACCAC-3′; TIc-R: 5′-TTTTGAACTTGCCACGGAGG-3′; 63°C/28-cycles. xMHCα, 66°C/28-cycles and xNkx2.5, 59°C/30-cycles.15 xMLC1v-F: 5′-CCATGCTCCAACACATTTCCA-3′; xMLC1v-R: 5′-CAGCTCCCATTACCGTACCATT-3′; xMLC1v-Taqman™ probe: 5′-FAM-ACATACGAAGACTTTGTTGAAGGGCTGC-3′-BHQ1, extension/acquisition: 60°C/1 min.
2.4. Whole-mount in situ hybridization and antibody staining
Linearized plasmids from xTIc (HindIII/T7), xNkx2.5 (EcoR1/T7), xNkx2.10 (Not1/T7), and p27Xic1 (BamH1/T7) were used to generate digoxigenin-11-UTP-labelled (Boehringer Mannheim) antisense RNA probes. Whole-mount in situ hybridization (ISH) was carried out as described previously16 with BM-purple substrate. Whole-mount antibody staining for α-phospho-histone H3 (phH3) (1:500, TCS Biologicals) was performed as described previously.16
2.5. Immunofluorescence
Embryos were fixed in 4% paraformaldehyde in PBS for 45 min and embedded in O.C.T. (Sakura Finetek, Tissue-Tek). Consecutive 10 µm cryostat sections were immunostained using standard methods with mouse α-Tropomyosin (CH1, 1:500; Developmental Studies Hybridoma Bank), or α-phH3. Secondary antibodies were Alexa Fluor 546-coupled α-mouse or Alexa Fluor 488-coupled α-rabbit.
All statistical analyses were performed using one-way analysis of variance and Bonferonni t-test.
3. Results
3.1. Terminally differentiated embryonic Xenopus cardiomyocytes maintain proliferative capacity
To investigate proliferative capacity of Xenopus hearts during embryonic development, we determined the total number and percentage of cardiomyocytes that were proliferating by immunostaining embryo sections at various stages expressing Tropomyosin, a marker of heart differentiation along with a marker for mitotic cells, phosphorylated histone H3 (phH3) (Figure 1A). The total number of cardiomyocytes in the heart gradually increased from stages-29/30 to -39 but no further significant change occurred from stages-39 to -41 (Figure 1B). Statistical analyses confirmed a significant increase in total number of myocytes when comparing stages-37/8 to -33/4 and all earlier stages (Figure 1B, P < 0.025) with a doubling in myocyte number between stage-32 (509 ± 45) and stage-37/8 (1164 ± 184). We observed a significant increase in myocyte number from stage-39, when compared to all earlier stages (Figure 1B, P < 0.05).
Figure 1.
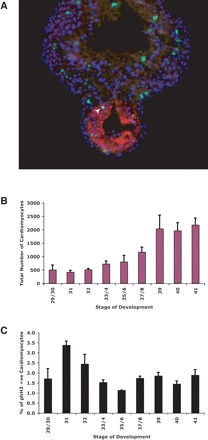
Division of embryonic cardiomyocytes. (A) Stage-33/34 embryo, Tropomyosin (red) in the cardiac tube, phH3 (green), DNA in nuclei (Hoechst, blue). Arrowhead: dividing cardiomyocyte (phH3 and Tropomyosin positive). (B) Total number of Tropomyosin-positive cardiomyocytes at increasing embryonic-stages (n ≥ 3 embryos/stage). (C) Percentage of dividing cardiomyocytes (both phH3 and Tropomyosin positive) at increasing stages.
Some Tropomyosin-expressing cells also expressed phH3 (e.g. Figure 1A), indicating that the structural genes are expressed in Xenopus cardiomyocytes that have not stably exited the cell cycle. Indeed, at all stages of heart development studied, we observed a small percentage of cells co-expressing both phH3 and Tropomyosin (Figure 1C), with this increasing significantly at stages-31 (P < 0.04, Figure 1C). Thus, while the vast majority of differentiated cardiomyocytes are not proliferating, a small percentage remains in cycle.
3.2. Xenopus cardiac differentiation can occur normally when the cell cycle is inhibited
Here we investigated the contribution of cell proliferation in determining the size of the linear heart-tube formed. In addition, we investigated the extent to which cells are able to respond to cardiac differentiation cues if cell cycling is prevented in post-gastrulating Xenopus embryos. It has previously been reported that blocking cell cycling in Xenopus embryos just post-gastrulation, using the DNA replication inhibitors, Hydroxyurea and Aphidicolin (HUA), results in no gross phenotypic abnormalities, although their cells are considerably larger.17 We incubated stage-13 embryos in HUA, and investigated the effects on formation and differentiation of the heart at both the cardiac plate and linear heart-tube stages (stages-29/30 and 33/4, respectively). phH3 staining was performed at stage-29/30 to confirm that HUA treatment was inhibiting cell-cycle progression (Figure 2A and B) and this resulted in a very substantial decrease (>50-fold, n = 5) in the number of mitotic cells (Figure 2A–C). Moreover, phH3-expressing cells were substantially decreased even in internal embryonic tissues demonstrating penetration of the HUA (Figure 2D vs. E). phH3 cells in the linear heart-tube at stage-33/34, which were already at a low level (Figure 1C), were barely detectable in sections of HUA-treated embryos at stage-33/34 (Figure 2D′ vs. E′).
Figure 2.
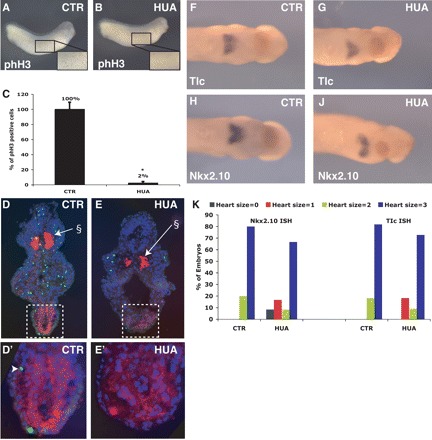
Cell division is not required for cardiomyocyte differentiation. Embryos stained for phH3 expression after incubation without (A) or with HUA (B) from stage-13. (C) Percentage of phH3-positive cells in the epidermis are decreased after HUA treatment vs. untreated control (matched areas, n ≥ 3 separate embryos ± SD; *P < 0.001). Transverse sections of stage-33/34 embryos after incubation of embryos in normal media (D and D′) or media containing HUA (E and E′), Tropomyosin expression (red), phH3 (green), and DNA in nuclei (Hoechst, blue); §Myotome is also positively labelled by tropomyosin. Note: Stage-29/30 embryos after control (F and H) or HUA (G and J) incubation showing TIc (F and G) or Nkx2.10 (H and J) expressions. (K) Heart size in control or HUA-incubated embryos as measured by TIc and Nkx2.10 expression area (see Figure 4A–H for examples).
ISH for markers of cardiac differentiation, TIc and Nkx2.10, were performed on cardiac plate-stage embryos (Figure 2F–J) and these were scored according to area of expression, i.e. size of the heart field. Heart-field size can vary somewhat both between batches of embryos and between embryos from the same batch of eggs so quantification of absolute heart size by area measurement does not allow statistically meaningful comparison between experiments. Hence, we adopted a blind-scoring method of 0 to 3, where 0 denotes no detectable heart and 3 denotes a heart of maximal size for that stage of development (Figure 4A–D and Supplementary material online, Figure S1D).
Figure 4.
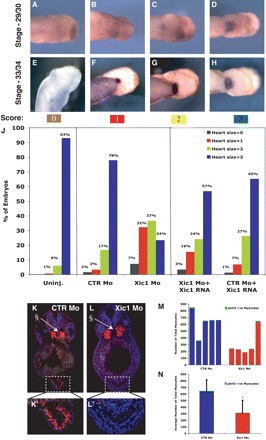
Xic1 is required for heart formation. TIc expression in Xic1Mo injected embryos at stage-29/30 (A–D) and stage-33/34 (E–H), demonstrating the range of cardiac phenotypes from normal expression of TIc (D and H, score = 3) to no expression of TIc (A and E, score = 0). (J) Graph summarizing the percentage of stage-29/30 embryos with hearts of sizes 0 to 3, as assayed by TIc expression, following injection with 20 ng of CTRMo or Xic1Mo and rescued by co-injection with Xic1mRNA (30 pg). n = 58–130 embryos per injection. Transverse sections of stage-33/34 embryos, after injection with CTRMo (K and K′) or Xic1Mo (L and L′), Tropomyosin (red), phH3 (green), DNA in nuclei (Hoechst, blue). §Myotome is also positively labelled by tropomyosin. (M) Total number of cardiomyocytes (blue or red) and the proportion of phH3 cardiomyocytes (green) in five separate stage-33/34 embryos, injected with either CTRMo or Xic1Mo. (N) Average total number of cardiomyocytes (blue or red), and the number of dividing cardiomyocytes (green) from embryos shown in graph M (*P = 0.022, n = 5).
Although a small number of HUA-treated embryos had smaller than average hearts, in the large majority of embryos, the area of the heart field was unchanged (Figure 2K), indicating that, following gastrulation, cell proliferation is not absolutely essential for differentiation of a cardiac plate of normal size. Moreover, antibody staining of the linear heart tube at stage-33/34 showed that the expression of Tropomyosin was also generally minimally affected (Figure 2D′ vs. E′), indicating that later events up to linear heart-tube formation were not substantially perturbed. Therefore, in post gastrulation Xenopus embryos, cardiac differentiation can occur in the absence of cell-cycle progression.
3.3. Xic1 is expressed in the Xenopus embryonic heart
We first investigated whether the CDKI Xic1 is expressed at the right time and in the right place to be playing a role in cardiac development. To this end, we performed ISH on embryos of increasing stage.
Starting at stage-27, a ventral view of the embryo reveals a region of Xic1 expression posterior to the anterior placodes and cement gland, in a position consistent with a low level of expression in the developing heart (Figure 3A, arrow), and this staining remains visible throughout tailbud stages (data not shown). By stage-31, these swimming tadpoles are expressing Xic1 more intensely in the head, eye, and myotome region, and additionally in the ventral heart region just posterior to the cement gland7,8 (data not shown). Expression in this region is maintained at least until stage-41 (Figure 3E, arrow).
Figure 3.
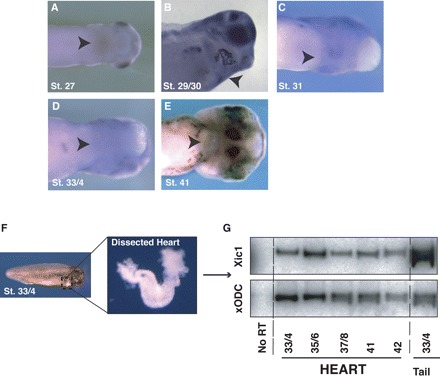
Xic1 is expressed in the differentiating heart. In situ hybridization to detect Xic1 in ventral or lateral view (A–E) of stage 27–41 embryos. Xic1 is weakly expressed in the hearts of embryos from stage-27 onwards (arrows). Picture B was taken following clearing embryo with Benzyl-benzoate:Benzyl-alcohol. (G) RT–PCR confirming expression of Xic1 transcript in the dissected hearts (F) of stages-33/4 to -42.
To confirm Xic1 is expressed in the heart itself and not in overlying ectoderm, we took an RT–PCR approach. Linear heart-tube formation in Xenopus is completed by stage-3218 and the tube can be dissected out as a discrete entity from stage-33/34 onwards. We dissected hearts from stage-33/4 to stage-42 embryos (Figure 3F) along with tailbud tissue, where Xic1 is strongly expressed8 as a positive control, and RT–PCR was performed to detect Xic1 expression. Xic1 message is clearly detectable in the heart and levels do not change dramatically from stages-33/4 to stage-42 (Figure 3G). Therefore, Xic1 is expressed at the right time and in the right place to play a role in cell-cycle exit and differentiation of the Xenopus heart.
3.4. Xic1 depletion results in loss of differentiated heart
To investigate the effect of Xic1 overexpression, we injected mRNA encoding either full-length Xic1 or deletion-constructs encompassing the N- or C-terminal regions, targeted to the developing heart region. Interestingly, injection of these constructs had no significant effect on heart formation (Supplementary material online, Figure S3).
As Xic1 is expressed in the developing heart, we also investigated the effect of Xic1 depletion on heart formation in Xenopus. Antisense morpholino-oligonucleotide (Mo) injection blocks translation of Xic1 mRNA in Xenopus embryos,8 resulting in undetectable levels of the protein (Supplementary material online, Figure S1C). Xic1Mo, or a specific CTRMo, was targeted to the heart region by injection into two dorsal-vegetal blastomeres at the 8-cell stage. Embryos were allowed to develop to stage-29/30, when the extent of cardiac differentiation in embryos was visualized as two symmetrical flat plates expressing TIc (Figure 4A–D) and/or Nkx2.10 (not shown), either side of the ventral midline. The area of the cardiac plate was scored blindly on a scale of 0 to 3, as described earlier. Embryos were also observed at stage-33/34 (Figure 4E–H) when the linear heart-tube has formed but before morphological looping has commenced.
Targeted injection of Xic1Mo resulted in a significant decrease in the size of the differentiating heart, as indicated by expression of TIc, when compared to sibling-uninjected embryos (e.g. Figure 1A–C and E–G, and Supplementary material online, Figure S1D). Although differentiated heart tissue was clearly diminished in Xic1 depleted embryos, nevertheless myocardial wall closure commenced normally, since the linear heart-tube was clearly visible at stage-33/4 (Supplementary material online, Figure S1D). Following injection of Xic1Mo, 39% of embryos exhibited a substantially reduced heart (0 or 1 based on TIc expression) whereas only 5% of CTRMo-injected embryos exhibited a similar phenotype (Figure 4J). Interestingly, we observed no difference in the heart rate of stage-37/8 Xic1 depleted embryos (49.6 ± 4.8, n = 16), where hearts were visible, compared to stage matched uninjected (50.1 ± 3.9, n = 15) or CTRMo-injected (50.3 ± 4.0, n = 19) embryos. Thus, upon depletion of Xic1, late-stage embryos still generally retain a broadly functional, albeit smaller, hearts. However, we occasionally observed alive embryos without a detectable heart beat, presumably corresponding to embryos which exhibited no expression of cardiac differentiation markers in the ISH studies (scored as ‘0’).
To demonstrate specificity for loss of Xic1 protein, we co-injected Xic1Mo with mRNA coding for Xic1, which could not be targeted by the morpholino (Supplementary material online, Figure S1A and B). Co-injection of CTRMo with this Xic1 RNA had only a very small effect on heart size similar to Xic1 mRNA on its own (Supplementary material online, Figure S3B), where hearts remained predominantly size 3 (Figure 4J). However, the reduction in heart size seen after Xic1Mo injection was significantly rescued by co-injection with Xic1 mRNA; the percentage of embryos with a severely reduced heart (score of 0 or 1) dropped by half, from 39 to 19%, while the number of size 3 hearts rose from 24 to 57% (Figure 4J). Therefore, Xic1 is required for heart formation in Xenopus.
CTRMo (Figure 4K) or Xic1Mo (Figure 4L) injected embryos were sectioned, and stained for Tropomyosin and phH3. As we saw by ISH for TIc, we observed a range of phenotypes (Supplementary material online, Figure S4). The majority of Xic1Mo injected embryos showed a marked reduction in the total number of differentiated cardiomyocytes when compared to CTRMo (Figure 4M). Indeed in some embryos, few if any, differentiated cardiomyocytes could be detected in any of the sections (Figure 4L′). On average, we observed a 50% reduction in total number of cardiomyocytes in sections from Xic1Mo injected embryos (310 ± 175) when compared to sections from CTRMo injected embryos (639 ± 192) (n = 5, P = 0.022) (Figure 4N). Depletion Xic1 did not result in a detectable induction of apoptosis, as no increase TUNEL positive nuclei following injection of Xic1Mo (16 ± 22, n = 5) was seen, compared to uninjected (30 ± 15, n = 5) or CTRMo injected (46 ± 13, n = 5) embryos.
Upon injection of Xic1Mo, we did not observe a significantly change in the number of phH3-expressing cells in the remaining Tropomyosin-positive tissue (1.4 ± 0.6%, percentage of phH3 positive cardiomyocytes, n = 5), when compared to sections from CTRMo injected embryos (1.9 ± 0.9%) (Figure 4N), or in the surrounding area (Figure 4L′ and data not shown). These results suggest that Xic1 is probably not the central regulator of cell-cycle exit in this tissue at this time, but it may be present to fulfil an alternative function in differentiation.
3.5. The cardiac differentiation function of Xic1 is located N-terminally but is distinct from its CDK kinase inhibitory activity
It has previously been reported that Xic1 is required for differentiation of primary neurons and myotomal muscle, independent of its ability to arrest the cell cycle, and this function is localized to the N-terminus of Xic1.7,8 Hence, we wished to determine whether it is the cell-cycle inhibitory function of Xic1 that is required for cardiac differentiation, or whether an additional function of Xic1 beyond cell-cycle regulation is required. To this end, we attempted rescue of the reduced cardiac phenotype seen after injection of Xic1Mo using Xic1 mutant constructs.
Embryos were injected with CTRMo or Xic1Mo, either with or without mRNA for non-targeted Xic1-FL (Supplementary material online, Figure S1A and B) or truncation mutants of Xic1 (Supplementary material online, Figure S2). Following targeted injections, embryos were allowed to develop to the cardiac plate stage, stage-29/30, and Nkx2.10 and TIc expression was determined. As expected, the majority of embryos injected with Xic1Mo (Figure 5C, J, and P) demonstrated a significant decrease in the expression of these markers of cardiac differentiation compared to uninjected (Figure 5A and H) and CTRMo (Figure 5B and I) injected controls (Figure 5T). This Xic1Mo-induced cardiac phenotype could be effectively rescued by co-injection with either FL-Xic1 (Figure 5D, K, and Q) or NT-Xic1 (Figure 5E and L), where the percentage of embryos with size 3 hearts rose from 14 to 54% and 41%, respectively (Figure 5T). Interestingly, Xic1(35–96), which can inhibit overall CDK-kinase activity, has only a small rescuing ability (Figure 5G, N, and T) while the C-terminus of Xic1 shows no rescuing ability (Figure 5F, M, and T). These results indicate that requirement for Xic1 in heart formation goes beyond its ability to control cell-cycle progression since both the N-terminus and C-terminus are able to inhibit the cell cycle, yet only the N-terminus can rescue cardiac differentiation.
Figure 5.
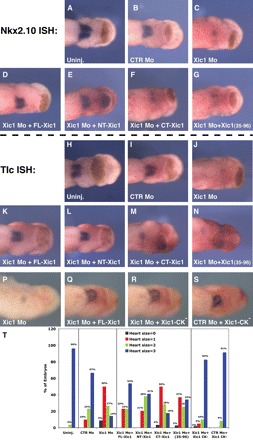
The requirement for Xic1 in cardiomyocyte differentiation is distinct from its ability to arrest the cell cycle. Stage-29/30 embryos either uninjected (A and H) or injected with CTRMo (B and I), Xic1Mo (C, J, and P), or Xic1Mo plus FL-Xic1 (30 pg) (D, K, and Q), NT-Xic1 (15 pg) (E and L), CT-Xic1 (30 pg) (F and M), Xic1(35–96) (30 pg) (G and N), Xic1CK- (R), or CTRMo plus Xic1CK- (S) detecting either Nkx2.10 (A–G) or TIc (H–S). (T) Percentage of stage-29/30 embryos with heart sizes 1–3 (cf. Figure 4) following injection with Mo and mRNA combinations as described. n = 26 – 53 embryos/treatment group.
Since our results indicted that the N-terminal segment of Xic1, which encompasses the Cyclin:CDK binding domain (Supplementary material online, Figure S2), is important for its role in cardiac differentiation, we investigated whether binding to cyclins and CDKs is required for Xic1 to promote cardiac differentiation. The residues within Xic1 which are critical for cyclin and CDK bindings have been previously identified by mutagenesis and a Xic1 construct with mutations within both the cyclin-binding domain (R33A and L35A) and CDK-binding domains (F65A and F67A) has been described, which is unable to bind to CDK2 and Cyclin E,19 the predominant CDK/cyclin pair at this stage in development. Hence, we constructed a Xic1 mutant, Xic1CK-, engineered to prevent targeting by Xic1Mo, and performed rescue experiments by co-injection with Xic1Mo (Supplementary material online, Figure S2). Interestingly, this mutant still had the ability to rescue the loss of cardiac differentiation observed following injection of Xic1 Mo (Figure 5R and T).
3.6. Xic1 depletion inhibits later stages of cardiomyocyte differentiation
Our results indicate that Xic1 is required for cardiomyocyte differentiation and this effect is detectable by stage-29/30. However, we wished to determine whether Xic1 is required early in specification of cardiomyocytes or later, during cardiomyocyte differentiation. Thus, first we investigated the expression of Xic1 transcripts in the presumptive cardiac field during early stages of development and compared its expression to that of Nkx2.5.20 In our hands, Nkx2.5 is first detected at stage-15 (Figure 6A) and in agreement with previous reports, its expression, although somewhat diffuse, is localized to the ventral region of Xenopus embryos.20 In contrast, at the same stages of development, the expression of Xic1 transcript is predominantly localized dorsally although a low level of anterior-ventral expression can be seen (Figure 6B, stage-16/7 and stage-18).
Figure 6.
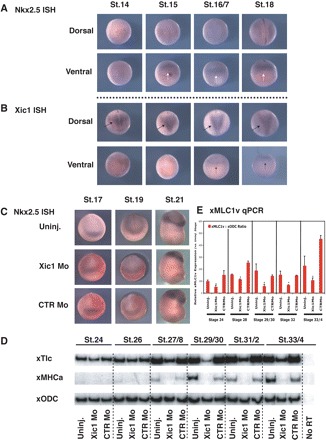
Xic1 is required for late stages of cardiomyocyte differentiation. Expression of Nkx2.5 (A) or Xic1 (B) by in situ hybridization at described stages. Nkx2.5 staining is largely localized to the ventral side of embryos in the cardiac crescent (A, white arrows). In contrast, Xic1 expression is predominantly dorsal (B, Dorsal view, black arrows) and only weakly detected at the ventral side (B, ventral view, black star-arrowhead). (C) Xic1Mo does not affect the cardiac field as measured by Nkx2.5 expression at stages-17, -19 and -21 of development. (D) RT–PCR from uninjected embryos or embryos injected with either CTRMo or Xic1Mo to detect expression of TIc and MHCα with ODC as loading control, stages as described. (E) Quantitative RT–PCR demonstrating the relative expression of xMLC1v in embryos described in (D), following normalization against xODC.
Nkx2.5 expression can be seen as a ventral crescent spanning the midline by stage-17, the presumptive cardiac field, with levels increasing through to stage-21 (Figure 6A and C). After targeted injection of Xic1Mo or CTRMo, Nkx2.5 expression was not significantly altered, indicating that Xic1 is not required for formation of the cardiac field at early stages of development (Figure 6).
Loss of Xic1 has a marked effect at later stages of development, resulting in defective cardiac differentiation (Figure 4). To determine where in the cardiomyocyte differentiation program Xic1 is required, Xic1Mo or CTRMo-injected embryos were harvested at increasing stages of development. cDNA was prepared and assayed by semi-quantitative RT–PCR for expression of TIc and MHCα, late markers of cardiomyocyte differentiation. While at stages-24 and -26, the effect of Xic1Mo on TIc was unclear, by stage-27/28 the expression of TIc was reduced in the Xic1-depleted embryos and this persisted up to stage-33/34 (Figure 6D). MHCα was undetectable at earlier stages but by stage-27/28 when levels were increasing, MHCα expression was absent in embryos injected with Xic1Mo, through to stage-33/34 (Figure 6D). These results confirm that Xic1 is required at these later stages when cardiomyocyte differentiation occurs. xTIc and xMHCa are both expressed in the primary heart-tube, throughout the embryonic myocardium at later stages of development. In contrast, MLC1v is found in the primary heart-tube and later, only specifically in the ventricles but not in the atria.21 The quantitative-PCR analysis shows that MLC1v is also downregulated after Xic1 depletion (Figure 6E), demonstrating that at least the primary heart field is affected by the loss of Xic1.
4. Discussion
We aimed to determine the extent to which cell-cycle progression is required for either the specification of the presumptive cardiac field or for differentiation of Xenopus cardiomyocytes. We found mitotic phH3-expressing cells co-expressing the cardiac differentiation marker Tropomyosin (Figure 1A), suggesting that at least up to stage-41 of Xenopus embryonic development, differentiating cardiomyocytes can still undergo active cell division. These observations are in agreement with early reports of observed cytokinesis in dividing embryonic rat and mouse cardiomyocytes which are gradually diminished at early postnatal stage.2 We observed an increase in the total number of cardiomyocytes in the developing Xenopus heart resulting in a four-fold increase from stage-29/30 to stage 41 (Figure 1B). The total number of cardiomyocytes doubled from stage-32 to stage-37/8, consistent with a previous report in the same species by Goetz et al.15 and with the morphological changes that occur as the linear heart-tube undergoes right-hand looping. Indeed, a small percentage of cells remain in cycle even as late as stage-41 (Figure 1C). Interestingly, at stages-31 and -32 we observed a statistically significant increase in the percentage cardiomyocytes that were mitotic (Figure 1C). These findings are consistent with the reported increase in the length of linear heart-tube in Xenopus embryo, where proliferation may be a prerequisite for the looping to commence.18
It has been proposed that cardiomyocytes have an intrinsic program of cell division that dictates the number of cell cycles before differentiation can occur.22 However, it is not clear whether cell division per se is required for cardiac specification and differentiation, as is the case in other cell types such as cortical neurons.23 Neither is it clear whether, in Xenopus, cell division of cardiac precursors is actually required for formation of a cardiac plate of normal size. Therefore, we addressed these questions by looking at specification and differentiation of the cardiac plate after the cell cycle had been substantially inhibited by incubation of embryos in HUA. Since cell-fate determination and specification of cardiogenic precursor cells of the primary heart field is thought to occur during gastrulation,24 cell-cycle arrest in our embryos by incubation in HUA from stage-13 should not interfere with this process. Tropomyosin (Figure 2D and E), TIc (Figure 2F and G), and Nkx2.10 (Figure 2H and J) expression was minimally affected by HUA treatment, indicating that the final cardiac plate area is not substantially determined by proliferation of a small number of committed cardiac progenitors post-gastrulation, and indeed cell proliferation beyond gastrulation is not absolutely required for specification or differentiation of cardiomyocytes in the cardiac plate (Figure 2). Our results are consistent with observations in chick embryos, where Soufan et al.25 demonstrated that a four-fold increase in the length of the primary heart-tube had occurred when the tube hardly displayed any proliferative activity, as quantified by a very low phH3-labelling index. These authors demonstrated that the majority of the growth in the linear heart-tube was attributed to the ‘recruitment’ of precursor cells to the linear heart-tube.25
Although Xenopus has three CDKIs,12,26 Xic1 is the predominant CDKI expressed at early developmental stages, sharing homology with all members of the mammalian CIP/KIP family.27 We see that Xic1 is expressed in developing heart (Figure 3). The timing of expression of Xic1 in cardiomyocytes at late tail-bud stages of development fits well with a role in controlling cardiomyocyte differentiation rather than a role in earlier specification. We explored this further. We investigated whether Xic1 is required for formation of the Xenopus heart, and if so, whether it acted at an early stage of specification or later stages in cardiomyocyte differentiation. Previous reports demonstrate that overexpression of Xic1 results in both an increase in the number of primary neurons and an expanded myotome.7,8 However, we did not observe any substantial alteration in Xenopus heart size following overexpression of Xic1 or Xic1 mutants thereof (Supplementary material online, Figure S3B).
Why does overexpression of Xic1 expand the myotome but not the heart? Myotomal muscle can be induced by a single myogenic factor, MyoD. We have previously demonstrated that both MyoD and Xic1 must be above threshold levels to promote myotomal muscle differentiation.8 However, unlike the myotome, cardiomyocyte differentiation involves numerous cardiogenic transcription factors acting in concert. Therefore, even though Xic1 may be required for cardiomyocyte differentiation, elevation of Xic1 levels alone may be insufficient to promote further cardiogenesis, leading to a bigger heart.
To determine whether Xic1 is required for heart formation, we inhibited Xic1 using Xic1Mo. Loss of Xic1 resulted in a substantial decrease in the area of the cardiac plate expressing Nkx2.10 (Figure 5C), TIc (Figure 5J and P), and also cardiac-actin and MHCα (not shown) at stage-29/30, with a corresponding decrease in the size of the heart- tube at stage-33/34 (Figure 4K and L, and Supplementary material online, Figure S1D). Moreover, the depletion of Xic1 resulted in a statistically significant decrease in the number of differentiated cardiomyocytes expressing Tropomyosin at stage-33/34 (Figure 4M and N). The loss of differentiated cardiomyocytes was specifically due to loss of Xic1, as this phenotype could be rescued to a large extent by co-injection of Xic1 mRNA (Figure 4J, 5D, K, and T).
At neurula stages, Xic1 is not strongly expressed in the cardiac field (Figure 6B), suggesting that Xic1 is unlikely to play a role in the early stages of cardiac development. However, its later expression in the anterio-ventral region of tailbud stage embryos where cardiac differentiation occurs (Figure 3A–E) indicates that Xic1 may function in later stages of cardiomyocyte differentiation. Loss of Xic1 had no effect on expression of Nkx2.5 at early stages, indicating that Xic1 levels do not influence the specification of the presumptive cardiac field (Figure 6C). However, expression of later markers of cardiac differentiation were reduced by Xic1Mo (Figures 4–6). As Xic1 depletion also resulted in diminishing expression of xMLc1v, this suggests that Xic1 is needed for the differentiation of the primary heart-tube. It would be interesting to know if Xic1 also affects the differentiation of the artrioventricular canal, the atrium and/or the secondary heart field and septation of atrium which occurs at stages-44 and beyond in Xenopus. However, this may not be possible in Xenopus embryos using the same strategy, as these events occur at stages beyond those at which morpholinos are likely to be effective.
To determine whether Xic1’s role in differentiation is solely to promote cell-cycle exit in cardiomyocytes, we rescued Xic1Mo-induced loss of cardiac differentiation with different Xic1 deletion constructs, which all have the ability to inhibit cell-cycle progression (Supplementary material online, Figure S3A) but by different mechanisms. FL-Xic1 (Figure 5D, K, Q, and T) and NT-Xic1 (Figure 5E, L, and T) both inhibit CDK-kinase activity and can rescue cardiomyocyte differentiation. In contrast CT-Xic1, which inhibits the cell cycle by binding PCNA, cannot rescue the loss of differentiated heart tissue seen with Xic1Mo (Figure 5F, M, and T). Xic1(35–96) can block the cell cycle and inhibit the majority of CDK kinase activity but it cannot promote neurogenesis or myogenesis in the early tadpole.7,8 However, Xic1(35–96) can partially rescue the loss of cardiomyocyte differentiation, albeit more weakly that FL-Xic1 and NT-Xic1 constructs (Figure 5T).
Our results suggest that Xic1 is required for late stages of cardiomyocyte differentiation. Is this because it must drive cells to exit the cell cycle to terminally differentiate? Our evidence argues against this possibility: first, Xenopus cardiomyocytes can still divide even when expressing markers of terminal differentiation (Figure 1 and Goetz et al.15), indicating that cell-cycle exit is not an absolute prerequisite for this process. Second, the C-terminus of Xic1 is able to inhibit the cell cycle (Supplementary material online, Figure S3A) yet it cannot rescue the loss of heart seen when endogenous Xic1 is lost (Figure 6F, M, and T). Third, we see no significant difference in the total number of phH3 expressing mitotic cardiomyocytes following injection of Xic1Mo (1.4 ± 0.6%), when compared to sections from CTRMo injected embryos (1.9 ± 0.9%) (Figure 4N), indicating that Xic1 does not regulate cell-cycle exit in this tissue.
However, our results did not rule out the possibility that the ability of Xic1 to promote cardiac differentiation might be due to the ability of Xic1 to bind to and inhibit Cyclin/CDK2 activity. FL-Xic1, NT-Xic1 and Xic1(35–96) all share the ability to inhibit overall CDK2 kinase activity and all are able to restore heart differentiation to a greater or lesser extent when co-injected with Xic1Mo. Indeed, Xic1(35–96) produces a less efficient rescue that FL-Xic1 and NT-Xic1 (Figure 5T), and this might be because it is fully able to inhibit Cyclin E/CDK2, the predominant CDK2 kinase at these stages, but it has a reduced ability to inhibit Cyclin A2/CDK2.28
Thus, to address this possibility, we performed rescue experiments using a Xic1 mutant (Xic1CK-) harbouring mutations within the cyclin (R33A, L35A) and CDK (F65A, F67A) binding domains (Supplementary material online, Figure S2). It had previously been reported that these mutations abolish the binding ability of Xic1 to CDK2 and cyclin E.19 Xic1CK- is effective at rescuing the loss of cardiac differentiation seen with Xic1Mo injection (Figure 5R, T). This indicates that inhibition of cyclin E/CDK2, the predominant cyclin/CDK pair at this stage, is not essential for Xic1’s role in cardiac differentiation. Nevertheless, we cannot rule out the possibility that the Xic1CK- mutant may retain some ability to bind to and inhibit the activity of cyclin A2/CDK2 or another cyclin/CDK pair, which regulates cardiomyocyte differentiation. However, we would note that cyclin A2 expression is essentially undetectable in the Xenopus heart up to stage-33/4 of development.29
Loss of the mammalian CDKI, p27Kip1, in mice leads to bigger organs, including the heart, demonstrated to be due to an increase in the total cell number.30 However, this may only occur in species where cell proliferation of precursors contributes substantially to growth of the heart, and this appears not to be the case in Xenopus (Figure 2D–K), and redundancy may allow other CDKIs to perform essential differentiation functions in mammals following knockout of a single CDKI.
So how is Xic1 playing a role in cardiac differentiation, if not via its cell-cycle regulatory function? Interestingly, the mammalian CDKI p57Kip2 has been shown to physically interact with the myogenic factor MyoD, further stabilizing and increasing its half-life.31 It is possible that Xic1 might be binding to one of the many transcription factors that drive cardiac differentiation, leading to its stabilization and thus further promoting cardiogenesis, although, we have been unable to demonstrate a physical interaction between in vitro translated Xic1 and either Nkx2.5, Tbx5, GATA4, or Myocardin proteins by immunoprecipitation (data not shown).
In summary, here we provide evidence that a CDKI plays an important role in cardiac differentiation beyond its ability to arrest the cell cycle. Currently, much is being learned about the multi-functionality of CDK inhibitors, and if we are to understand the versatility of these molecules, it will be important to investigate CDKI function in systems where effects on the cell cycling can be distinguished from other less obvious roles.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by British Heart Foundation project grant PG/03/068.
Supplementary Material
Acknowledgements
We are indebted to Ruth Cosgrove who provided the data in Supplementary material online, Figure S1C, Tim Mohun, Branko Latinik, and Paul Krieg for reagents, Ian Horan for impeccable technical assistance and Tim Mohun, Peter Weissberg, Christelle Fiore-Heriche, Shinichi Ohnuma, Elizabeth Jones, and Toshiyaki Mochizuki for helpful discussions. We thank J-C Lin and the DSHB, University of Iowa for antibodies.
Conflict of interest: none declared.
References
- 1.Mummery CL. Cardiology: solace for the broken-hearted? Nature. 2005;433:585–587. doi: 10.1038/433585a. [DOI] [PubMed] [Google Scholar]
- 2.Rumyantsev PP. Interrelations of the proliferation and differentiation processes during cardiact myogenesis and regeneration. Int Rev Cytol. 1977;51:186–273. [PubMed] [Google Scholar]
- 3.Poolman RA, Gilchrist R, Brooks G. Cell cycle profiles and expressions of p21CIP1 AND P27KIP1 during myocyte development. Int J Cardiol. 1998;67:133–142. doi: 10.1016/s0167-5273(98)00320-9. [DOI] [PubMed] [Google Scholar]
- 4.Poolman RA, Brooks G. Expression and activities of cell cycle regulatory molecules during the transition from myocyte hyperplasia to hypertrophy. J mol Cell Cardiol. 1998;30:2121–2135. doi: 10.1006/jmcc.1998.0808. [DOI] [PubMed] [Google Scholar]
- 5.Koh K, Kang M, Frith-Terhune A, Park S, Kim I, Lee C, et al. Persistent and heterogenous expression of the cyclin-dependent kinase inhibitor, p27KIP1, in rat hearts during development. J Mol Cell Cardiol. 1998;30:463–474. doi: 10.1006/jmcc.1997.0611. [DOI] [PubMed] [Google Scholar]
- 6.Cremisi F, Philpott A, Ohnuma S. Cell cycle and cell fate interactions in neural development. Curr Opin Neurobiol. 2003;13:26–33. doi: 10.1016/s0959-4388(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 7.Vernon AE, Devine C, Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130:85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- 8.Vernon AE, Philpott A. A single cdk inhibitor, p27Xic1, functions beyond cell cycle regulation to promote muscle differentiation in Xenopus. Development. 2003;130:71–83. doi: 10.1242/dev.00180. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, et al. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Studzinski GP, Harrison LE. Differentiation-related changes in the cell cycle traverse. Int Rev Cytol. 1999;189:1–58. doi: 10.1016/s0074-7696(08)61384-4. [DOI] [PubMed] [Google Scholar]
- 11.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 12.Ohnuma S, Philpott A, Wang K, Holt CE, Harris WA. p27Xic1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell. 1999;99:499–510. doi: 10.1016/s0092-8674(00)81538-x. [DOI] [PubMed] [Google Scholar]
- 13.Philpott A, Porro EB, Kirschner MW, Tsai LH. The role of cyclin-dependent kinase 5 and a novel regulatory subunit in regulating muscle differentiation and patterning. Genes Dev. 1997;11:1409–1421. doi: 10.1101/gad.11.11.1409. [DOI] [PubMed] [Google Scholar]
- 14.Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- 15.Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sive H, Grainger R, Harland R. Early Development of Xenopus Laevis: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 17.Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- 18.Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The morphology of heart development in Xenopus laevis. Dev Biol. 2000;218:74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- 19.Chuang LC, Yew PR. Regulation of nuclear transport and degradation of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1. J Biol Chem. 2001;276:1610–1617. doi: 10.1074/jbc.M008896200. [DOI] [PubMed] [Google Scholar]
- 20.Tonissen KF, Drysdale TA, Lints TJ, Harvey RP, Krieg PA. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- 21.Smith SJ, Ataliotis P, Kotecha S, Towers N, Sparrow DB, Mohun TJ. The MLC1v gene provides a transgenic marker of myocardium formation within developing chambers of the Xenopus heart. Dev Dyn. 2005;232:1003–1012. doi: 10.1002/dvdy.20274. [DOI] [PubMed] [Google Scholar]
- 22.Burton PB, Yacoub MH, Barton PJ. Cyclin-dependent kinase inhibitor expression in human heart failure. A comparison with fetal development. Eur Heart J. 1999;20:604–611. doi: 10.1053/euhj.1998.1231. [DOI] [PubMed] [Google Scholar]
- 23.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 24.Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- 25.Soufan AT, van den Berg G, Ruijter JM, de Boer PA, van den Hoff MJ, Moorman AF. Regionalized sequence of myocardial cell growth and proliferation characterizes early chamber formation. Circ Res. 2006;99:545–552. doi: 10.1161/01.RES.0000239407.45137.97. [DOI] [PubMed] [Google Scholar]
- 26.Daniels M, Dhokia V, Richard-Parpaillon L, Ohnuma S. Identification of Xenopus cyclin-dependent kinase inhibitors, p16Xic2 and p17Xic3. Gene. 2004;342:41–47. doi: 10.1016/j.gene.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 27.Chuang LC, Zhu XN, Herrera CR, Tseng HM, Pfleger CM, Block K, et al. The C-terminal domain of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1, is both necessary and sufficient for phosphorylation-independent proteolysis. J Biol Chem. 2005;280:35290–35298. doi: 10.1074/jbc.M506430200. [DOI] [PubMed] [Google Scholar]
- 28.Hartley RS, Sible JC, Lewellyn AL, Maller JL. A role for cyclin E/Cdk2 in the timing of the midblastula transition in Xenopus embryos. Dev Biol. 1997;188:312–321. doi: 10.1006/dbio.1997.8647. [DOI] [PubMed] [Google Scholar]
- 29.Vernon AE, Philpott A. The developmental expression of cell cycle regulators in Xenopus laevis. Gene Expr Patterns. 2003;3:179–192. doi: 10.1016/s1567-133x(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 30.Poolman RA, Li J-M, Durand B, Brooks G. Altered expression of cell cycle proteins and prolonged duration of cardiac myocyte hyperplasia in p27KIP1 knockout mice. Circ Res. 1999;85:117–127. doi: 10.1161/01.res.85.2.117. [DOI] [PubMed] [Google Scholar]
- 31.Reynaud EG, Leibovitch MP, Tintignac LA, Pelpel K, Guillier M, Leibovitch SA. Stabilization of MyoD by direct binding to p57(Kip2) J Biol Chem. 2000;275:18767–18776. doi: 10.1074/jbc.M907412199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


