Abstract
The recent discovery of H3K27me3 demethylases suggests that H3K27me3 may dynamically regulate gene expression, but this potential role in mammalian tissue homeostasis remains uncharacterized. In the epidermis, a tissue that balances stem cell self-renewal with differentiation, H3K27me3, occupies the promoters of many differentiation genes. During calcium-induced differentiation, H3K27me3 was erased at these promoters in concert with loss of PcG protein occupancy and increased binding by the H3K27me3 demethylase, JMJD3. Within epidermal tissue, JMJD3 depletion blocked differentiation, while active JMJD3 dominantly induced it. These results indicate that epigenetic derepression by JMJD3 controls mammalian epidermal differentiation.
Keywords: Chromatin, differentiation, stem cell, epigenetics, gene regulation, histones
Post-translational modifications of core histone octamer proteins, in the form of phosphorylation, acetylation, ubiquitination, and methylation, have been increasingly appreciated as powerful regulators of gene expression. In Drosophila and mammals, the histone 3 Lys 27 trimethylation mark (H3K27me3) is associated with repression of gene transcription (Lund and van Lohuizen 2004;Barski et al. 2007). Polycomb group proteins (PcG) and their antagonists, the histone demethylases, are epigenetic regulators that control gene expression by modifying the methylation status of H3K27 (Grimaud et al. 2006; Swigut and Wysocka 2007). The PcG protein, Enhancer of Zest Homolog 2 (EZH2), is a histone methyltransferase that can trimethylate H3K27. Two other PcG proteins, Embryonic Ectoderm Development (EED) and Suppressor of Zeste 12 homolog (SUZ12), are necessary for EZH2 to function properly (Pasini et al. 2004). Genome-wide mapping of H3K27me3 marks and PcG target sites in embryonic stem cells has recently suggested a major role for PcG proteins in maintaining H3K27me3 to allow for repression of the differentiated state and promotion of embryonic stem cell self-renewal, with significant enrichment of these proteins on genes encoding homeodomain proteins and other putative differentiation factors (Boyer et al. 2006; Lee et al. 2006). The mechanism reversing such potential repressive influences on differentiation gene expression by H3K27me3 and other histone modifications in somatic tissue is not fully characterized.
Among potential mediators of differentiation gene derepression are newly characterized Jumanji C (JmjC) domain-containing proteins, UTX and JMJD3, which are enzymes capable of demethylating promoters marked by H3K27me3. Recently, the H3K27 demethylase activity of UTX and JMJD3 has been shown to act at HOX gene promoters to derepress HOX gene transcription (Agger et al. 2007; Lan et al. 2007; Lee et al. 2007). These histone demethylases were demonstrated in mammalian cells and in live zebrafish to antagonize PcG gene silencing by modifying chromatin to permit gene transcription. During inflammation, bacterial products and cytokines induce JMJD3, which then removes H3K27me3 marks to derepress genes involved in macrophage transdifferentiation (De Santa et al. 2007). In neural stem cells, overexpression of JMJD3 was shown to activate a subset of neural differentiation genes (Jepsen et al. 2007). These data raise the possibility that H3K27 demethylases may play a role in the normal induction of differentiation in adult somatic tissues.
Mammalian epidermis comprises a self-renewing stratified epithelial tissue barrier that prevents dehydration while providing a first line of defense against microbes and toxic substances on the skin. Human epidermis has a turnover rate of ∼4 wk, undergoing continual replenishment from stem cells that reside in the innermost epidermal basal layer. As progeny of these stem cells differentiate, they withdraw from the cell cycle, detach from the basement membrane, and migrate outwards to form the spinous layer (Blanpain and Fuchs 2006). In the spinous layer, cells express the key cytoskeletal proteins keratin 1 (K1) and 10 (K10) to assemble a network of intermediate filaments that provide the mechanical strength to withstand physical stress. As differentiation progresses, the cells generate the granular layer, which is characterized by the expression of filaggrin and loricrin along with other cornified envelope proteins that are assembled on the inner face of the plasma membrane (Segre 2006). During the final stages of differentiation, transglutaminases crosslink cornified envelope proteins, while lipids are extruded to form an intercellular sealant (Eckert et al. 2005). Terminally differentiated cornified epithelial cells embedded in these lipids form the water-impermeable outermost epidermal layer known as the stratum corneum. Although expression patterns of structural genes in the epidermis have been well characterized, much less is known about the mechanism by which differentiation genes are suppressed in undifferentiated cells and induced during outward migration.
We examined whether histone demethylases dynamically regulate differentiation in epidermis. We found that H3K27me3 marks were enriched in a subset of epidermal differentiation gene promoters in undifferentiated cells. These marks were erased during calcium-induced differentiation in concert with JMJD3 binding and loss of SUZ12 occupancy at differentiation gene promoters. Expression of active JMJD3 induced premature epidermal differentiation, as did depletion of SUZ12 or EZH2. JMJD3 depletion, in contrast, prevented normal induction of epidermal differentiation. Unexpectedly, this differentiation gene regulation was also seen at some promoters lacking H3K27me3 marks. Our findings indicate that the JMJD3 histone demethylase acts outside HOX targets to induce differentiation in mammalian epidermis.
Results and Discussion
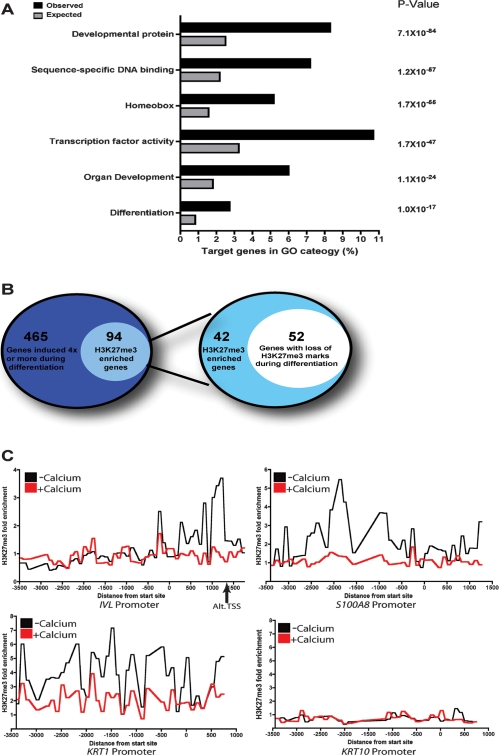
To study the role of H3K27me3 demethylases in regulating epidermal differentiation, we globally mapped sites of H3K27me3 enrichment in human keratinocytes. The epidermis displays increasing concentrations of calcium in its more differentiated outer layers, and calcium is a well-characterized stimulus of the epidermal differentiation program in cultured epidermal keratinocytes (Hennings et al. 1980; Boyce and Ham 1983; Punnonen et al. 1993). Anti-H3K27me3 antibody was used to immunoprecipitate chromatin from keratinocytes cultured in growth (−Ca2+) or differentiation medium (+Ca2+) conditions for 72 h. Genomic DNA enriched for H3K27me3 marks was hybridized to tiling microarrays covering genomic regions from 3500 bp upstream to 750 bp downstream from transcription start sites of 24,659 human genes. A significant enrichment for H3K27me3 was found on 2335 gene promoters in growth conditions (9.5% of total promoters assayed), as listed in Supplemental Table 1. To determine the biological role of H3K27me3-regulated genes in keratinocytes, we used Gene Ontology (GO) to determine which biological functions were overrepresented. H3K27me3 was highly enriched at genes involved in regulating transcription, development, and differentiation (Fig. 1A). These included multiple non-HOX genes along with >100 genes encoding homeobox-containing proteins that belong to gene families such as PAX, LHX, and DLX (Supplemental Table 2).
Figure 1.
Global analysis of H3K27me3 enrichment at genomic promoters during epidermal differentiation. (A) Gene Ontology (GO) analysis of promoters enriched for H3K27me3 marks. Black bars represent the observed percentage of H3K27me3 enriched sites in a certain GO category; gray bars represent the expected percentage. P-values were obtained using hypergeometric distribution. (B) Classification of epidermal differentiation genes. The left diagram depicts the number of H3K27me3 enriched epidermal differentiation genes. The right diagram shows the number of H3K27me3 enriched differentiation genes that lose methylation during differentiation. (C) H3K27me3 enrichment ratios on genomic tiling arrays comparing individual differentiation gene promoters for all probes in cells cultured in growth medium (−calcium) or differentiation medium (+calcium). The arrow below the IVL graph denotes an alternate transcription start site.
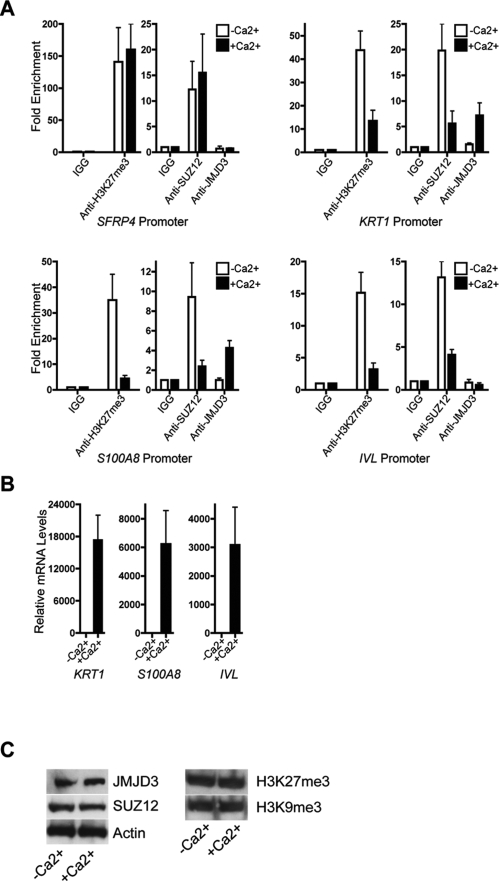
To determine whether H3K27 methylation status plays a role in epidermal differentiation, we identified genes up-regulated during calcium-induced keratinocyte differentiation (Supplemental Table 3) that also had enrichment for H3K27me3 in undifferentiated cells. As expected, many established epidermal differentiation genes such as KRT1, KRT10, IVL, and the SPRR family were strongly up-regulated during differentiation (Supplemental Fig. 1; Supplemental Table 3). Of the 559 genes induced by fourfold or more during differentiation, 94 had enrichment for H3K27me3 marks. Out of the 94 genes with H3K27me3 enrichment, the majority, 52 genes (55%), had twofold or more loss of H3K27me3 marks during differentiation (Fig. 1B; Supplemental Table 4). Loss of H3K27me3 is also specific to epidermal differentiation genes as addition of calcium only caused the loss of H3K27me3 on 134 of the 2241 (5.98%) nonepidermal differentiation gene promoters (Supplemental Table 5). Key epidermal differentiation genes such as IVL, S100A8, and KRT1 showed substantial loss of H3K27me3 during calcium-induced differentiation, raising the possibility that H3K27me3 may help regulate a subset of epidermal differentiation genes (Fig. 1C). Reduction of H3K27me3 marks at a subset of epidermal differentiation genes during differentiation suggested potential for reciprocal action by PcG proteins and histone demethylases in this process. To explore this, we performed chromatin immunoprecipitation (ChIP) with antibodies against H3K27me3, SUZ12, and JMJD3. Consistent with our ChIP–chip results, H3K27me3 was considerably reduced on the promoters of well-characterized epidermal differentiation genes KRT1, S100A8, and IVL with the onset of calcium-induced differentiation (Fig. 2A). SFRP4, a known H3K27me3 marked gene that is not regulated by differentiation (Schlesinger et al. 2007), showed no decrease in H3K27me3. H3K27me3 loss was mirrored by a marked reduction in SUZ12 occupancy at IVL, S100A8, and KRT1 promoters, with a corresponding rise in JMJD3 binding at S100A8 and KRT1 promoters. However, there was no significant enrichment for JMJD3 binding to the IVL promoter in −Ca2+ or +Ca2+ conditions, suggesting that the decrease in H3K27me3 at the IVL promoter could be due to passive loss of methylation resulting from reduction of SUZ12 binding. The loss of H3K27me3 and SUZ12 as well as the increase in JMJD3 on the promoters of differentiation genes correlated with the induction on the mRNA level of those genes (Fig. 2B). JMJD3 and SUZ12 protein levels remained constant during calcium-induced differentiation, implying that selective targeting, rather than relative expression levels, of these proteins is responsible for changes in H3K27me3 on differentiation promoters. Furthermore, differentiation did not induce wholesale erasure of H3K27me3 or another repressive mark H3K9me3 (Fig. 2C). Reciprocal binding of PcG components and the JMJD3 demethylase on structural epidermal differentiation genes suggests a mechanism of direct regulation of these non-HOX target genes.
Figure 2.
JMJD3 binds to the promoters of epidermal differentiation genes. (A) ChIP using H3K27me3, SUZ12, or JMJD3 antibody on the promoters of three differentiation genes, KRT1, S100A8, and IVL, in the absence or presence of calcium. SFRP4 was used as a positive control for H3K27me3 binding. (B) QPCR analysis on the mRNA levels of differentiation genes in the absence or presence of calcium. (C) Western blot depicting levels of JMJD3, SUZ12, H3K27me3, and H3K9me3 in keratinocytes grown in the absence or presence of calcium. Actin was used as a loading control.
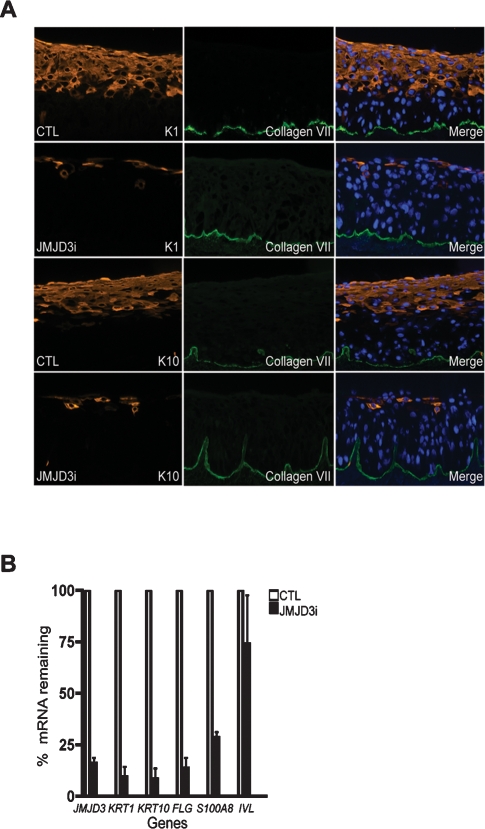
Direct JMJD3 binding to differentiation gene promoters suggested JMJD3 may be required for normal induction of differentiation genes in epidermis. To test this, we depleted JMJD3 expression in organotypic human skin. This human tissue approach recapitulates spatially accurate epidermal stratification and differentiation in the architecturally faithful context of normal human epidermal keratinocytes and intact extracellular matrix in the form of human dermis (Truong et al. 2006). JMJD3 depletion blocked induction of KRT1 and S100A8 (Fig. 3A,B); similar results were seen using another siRNA targeting JMJD3 (Supplemental Fig. 2). JMJD3 is therefore necessary for the induction of H3K27me3-marked differentiation genes such as KRT1 and S100A8. No effect is seen with the absence of JMJD3 on expression of IVL, whose promoter JMJD3 does not physically bind (Figs. 2A, 3B). This suggests that IVL may be regulated via passive loss of methylation due to reduced SUZ12 binding or by the action of an unidentified demethylase. Next we wanted to characterize the extent of the differentiation defects seen in JMJD3-depleted tissue. We assayed for the expression of critical structural differentiation genes such as KRT10 and FLG. The importance of these epidermal differentiation genes has been demonstrated in human clinical disease, as seen with KRT1 or KRT10 mutations, resulting in epidermolytic hyperkeratosis, and FLG mutations, causing ichthyosis vulgaris (Fuchs and Cleveland 1998; Gruber et al. 2007). Epidermal tissue treated with siRNAs against JMJD3 demonstrated a near complete absence of K10 and filaggrin (Fig. 3). Thus, a subset of differentiation genes, notably, KRT10 and FLG, also depend on the presence of JMJD3 for its induction despite having no H3K27me3 marks in studied promoter regions (Figs. 1C, 3A,B; Supplemental Table 1).
Figure 3.
JMJD3 is required for epidermal differentiation. (A) Epidermal cells receiving siRNA against JMJD3 (JMJD3i) or nonfunctional control siRNA (CTL) were used to regenerate epidermal tissue on human dermis in organotypic culture. Tissue sections were stained for the presence of keratin 1 (K1) and keratin 10 (K10). Type VII collagen (green) was used as a costain and marks the boundary between the epidermis and dermis. Note the loss of differentiation-specific keratins with JMJD3 depletion. (B) Quantitation of tissue mRNA levels by QPCR analysis, normalized to GAPDH.
To further demonstrate that JMJD3 is necessary for the induction of differentiation genes, we performed rescue experiments by overexpressing JMJD3 in JMJD3-depleted cells. Keratinocytes were transduced with control or JMJD3 expression constructs, and 4 d post-transduction, cells were depleted of JMJD3 using siRNAs and placed on organotypic cultures. Again, JMJD3 depletion blocked epidermal differentiation, whereas overexpression of JMJD3 rescued the differentiation defect even in the presence of siRNAs against JMJD3 (Supplemental Fig. 3). To establish that JMJD3 has a role in vivo in the regulation of epidermal differentiation, we depleted JMJD3 from keratinocytes using siRNAs and grafted the JMJD3-depleted cells on immune-deficient mice. As shown in Supplemental Figure 4, JMJD3-depleted epidermises were defective in differentiation although the effect was more dramatic in organotypic cultures which is likely due to the higher expression of JMJD3 in the grafts. Moreover, the differentiation defects observed in JMJD3-depleted cells were specific as no effects on differentiation were seen when another recently identified H3K27me3 demethylase, UTX, was knocked down in organotypic tissue (Supplemental Fig. 5). JMJD3 thus plays a nonredundant role in epidermal differentiation.
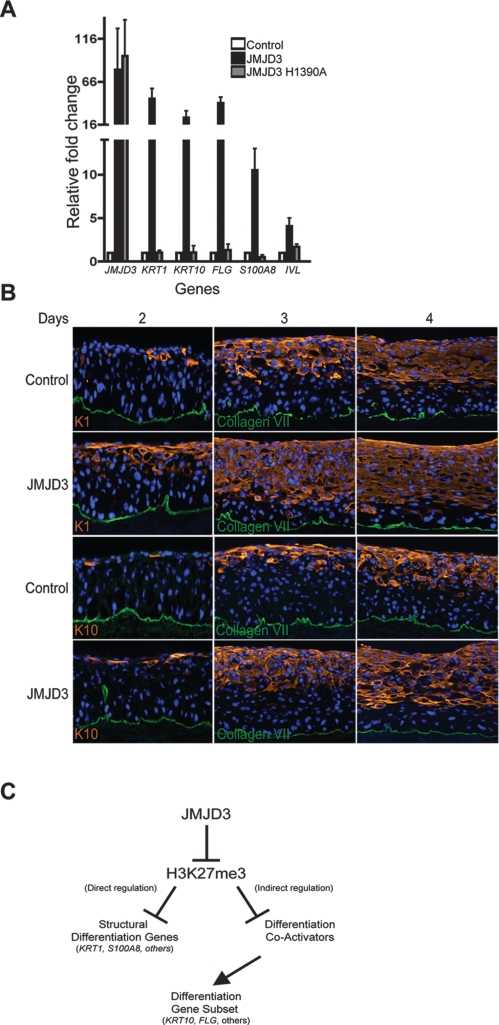
The JMJD3 requirement for induction of epidermal differentiation suggests that active JMJD3 should also trigger premature differentiation. Enforced expression of JMJD3 in epidermal cells grown in low calcium media led to ectopic induction of differentiation genes, including KRT1, KRT10, S100A8, FLG, and IVL (Fig. 4A). To examine if the premature differentiation induced by JMJD3 was due to its demethylase activity, we generated a demethylase defective JMJD3 mutant by mutating histidine 1390 to alanine. This disrupts the iron chelating function of the JmjC domain necessary for demethylase activity (Xiang et al. 2007). As Figure 4A shows, expression of mutant JMJD3 completely abolished the differentiation-inducing effects of wild-type JMJD3. Since active JMJD3 induced the expression of differentiation genes, we hypothesized that PcG depletion should also result in premature differentiation. To determine if PcG proteins are necessary for repression of differentiation genes in undifferentiated cells, we produced shRNA retroviral constructs to deplete SUZ12 and EZH2 expression. Keratinocytes were transduced with shRNA constructs targeting SUZ12 and EZH2 along with GFP shRNA control (Supplemental Fig. 6A). EZH2 or SUZ12 knockdown resulted in derepression of epidermal differentiation genes in the absence of calcium (Supplemental Fig. 6B,C). Similar results were seen with all functional shRNAs targeting either SUZ12 or EZH2. Next, we expressed active JMJD3 in epidermal tissue and examined differentiation gene expression over a timecourse during epidermal regeneration in organotypic culture. Cytoskeletal proteins, K1 and K10, which are hallmarks of epidermal differentiation, are expressed earlier in JMJD3 overexpressing cells (Fig. 4B). Thus, JMJD3 can act dominantly to induce epidermal differentiation.
Figure 4.
JMJD3 can induce premature epidermal differentiation. (A) Keratinocytes were transduced with retroviral expression constructs for empty vector control, JMJD3, and the JMJD3 H1390A demethylase-defective point mutant and grown in low calcium conditions. RNA was isolated from the samples 5 d post-transduction and QPCR was used to determine levels of derepression of epidermal differentiation genes. (B) JMJD3-expressing and control regenerated organotypic tissue was harvested at days 2, 3, and 4 to determine the impact of JMJD3 expression on differentiation. Tissue sections were stained for keratin 1 and keratin 10. Note earlier expression of differentiation proteins and their localization nearer to the epidermal basement membrane in tissue engineered to express JMJD3. (C) Model of epigenetic control of epidermal differentiation.
Here we mapped H3K27me3 occupancy at promoter sites in epidermal keratinocytes, identifying a role for these repressive marks in maintaining progenitor cell identity by repressing transcription of differentiation genes. H3K27me3 marks disappear on a subset of epidermal gene promoters upon differentiation. SUZ12 was found on H3K27me3 marked differentiation promoters in the absence of differentiation signals, but was replaced by JMJD3 during differentiation. The binding of JMJD3 to promoters suggests a direct mode of action on a subset of epidermal structural differentiation genes. Indeed, siRNA-mediated depletion of JMJD3 blocked the normal activation of JMJD3-bound differentiation genes, while overexpression of JMJD3 ectopically induced them. However, gain of JMJD3 function and loss of SUZ12/EZH2 function also resulted in premature induction of several differentiation genes that did not show H3K27me3 occupancy at baseline, while loss-of-function JMJD3 prevented their induction. These non-H3K27me3 marked differentiation genes may be regulated by H3K27me3 at distal regulatory regions not covered by tiling array regions studied; alternatively, H3K27me3 may indirectly regulate the expression of these genes via direct regulation of other effectors. Non-H3K27me3 marked differentiation genes such as KRT10 and FLG may be regulated by H3K27me3 marked “master regulators.” Potential H3K27me3 marked regulators include PITX1, POU2F3, FOXC1, NDRG4, and FOXQ1 (Supplemental Table 4). POU2F3, also known as SKN-1a, and FOXQ1 are both transcription factors involved in keratinocyte differentiation and hair shaft development, respectively (Hildesheim et al. 2001; Hong et al. 2001; Cabral et al. 2003; Potter et al. 2006). HOX and forkhead transcription factors such as PITX1 and FOXC1 have potential roles as regulators of differentiation (Gage et al. 1999; Lehmann et al. 2003). Our results suggest a model in which PcG proteins methylate H3K27 in undifferentiated epithelial cells, resulting in repression of differentiation genes (Fig. 4C). Upon induction of differentiation, promoter-bound PcG proteins are replaced by JMJD3 on a subset of key epidermal differentiation genes, and possibly on other indirect effector genes, enabling activation of a differentiation-specific transcriptional cascade. In summary, these data indicate a functional role for JMJD3 in the induction of epidermal differentiation.
Materials and methods
Tissue culture and gene transfer
Primary human keratinocytes were derived from freshly discarded surgical skin specimens. Cells were grown in KSF-M (GIBCO-BRL) supplemented with epidermal growth factor (EGF) and bovine pituitary extract (BPE). Additional details are available in the Supplemental Material.
siRNAs and retroviral constructs
All siRNAs used were generated from Dharmacon. Detailed information about siRNAs, shRNAs, and retroviral constructs are described in the Supplemental Material.
Protein expression
Forty micrograms of the cell lysates were used for immunblotting and resolved on 10% SDS-PAGE and transferred to PVDF membranes. Membranes were incubated in primary and secondary antibodies for 1 h each. Details on antibodies can be found in the Supplemental Material.
mRNA expression analysis
Total RNA from cells cultured in growth and differentiation (3 d) medium was extracted using the RNeasy mini kit (Qiagen) and quantified by Nanodrop. Detailed information on quantitative PCR and Microarray analysis is described in the Supplemental Material.
ChIP
ChIP-on-chip was performed as described previously (Rinn et al. 2007). Additional information on ChIP analysis is described in the Supplemental Material.
Acknowledgments
We thank Y. Shi for reagents, helpful advice, and presubmission review, as well as M. Fuller, A. Oro, J. Crabtree, and M. Scott for presubmission review, and J. Reuter and T. Ridky for helpful advice, reagents, and technical help. This work was supported by the U.S. Veterans Affairs Office of Research and Development and by NIH/NIAMS grant AR45192 to P.A.K and NIH NRSA 5T32 CA09302 to G.L.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1673508.
References
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce S.T., Ham R.G. Calcium-regulated differentiation of normal human epidermal keratinocytes in chemically defined clonal culture and serum-free serial culture. J. Invest. Dermatol. 1983;81:33s–40s. doi: 10.1111/1523-1747.ep12540422. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cabral A., Fischer D.F., Vermeij W.P., Backendorf C. Distinct functional interactions of human Skn-1 isoforms with Ese-1 during keratinocyte terminal differentiation. J. Biol. Chem. 2003;278:17792–17799. doi: 10.1074/jbc.M300508200. [DOI] [PubMed] [Google Scholar]
- De Santa F.,, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Eckert R.L., Sturniolo M.T., Broome A.M., Ruse M., Rorke E.A. Transglutaminase function in epidermis. J. Invest. Dermatol. 2005;124:481–492. doi: 10.1111/j.0022-202X.2005.23627.x. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Cleveland D.W. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Gage P.J., Suh H., Camper S.A. The bicoid-related Pitx gene family in development. Mamm. Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- Grimaud C., Negre N., Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Gruber R., Janecke A.R., Fauth C., Utermann G., Fritsch P.O., Schmuth M. Filaggrin mutations p.R501X and c.2282del4 in ichthyosis vulgaris. Eur. J. Hum. Genet. 2007;15:179–184. doi: 10.1038/sj.ejhg.5201742. [DOI] [PubMed] [Google Scholar]
- Hennings H., Michael D., Cheng C., Steinert P., Holbrook K., Yuspa S.H. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980;19:245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- Hildesheim J., Kuhn U., Yee C.L., Foster R.A., Yancey K.B., Vogel J.C. The hSkn-1a POU transcription factor enhances epidermal stratification by promoting keratinocyte proliferation. J. Cell Sci. 2001;114:1913–1923. doi: 10.1242/jcs.114.10.1913. [DOI] [PubMed] [Google Scholar]
- Hong H.K., Noveroske J.K., Headon D.J., Liu T., Sy M.S., Justice M.J., Chakravarti A. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29:163–171. doi: 10.1002/gene.1020. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Solum D., Zhou T., McEvilly R.J., Kim H.J., Glass C.K., Hermanson O., Rosenfeld M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Di Croce L., Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lehmann O.J., Sowden J.C., Carlsson P., Jordan T., Bhattacharya S.S. Fox’s in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- Lund A.H., van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C.S., Peterson R.L., Barth J.L., Pruett N.D., Jacobs D.F., Kern M.J., Argraves W.S., Sundberg J.P., Awgulewitsch A. Evidence that the satin hair mutant gene Foxq1 is among multiple and functionally diverse regulatory targets for Hoxc13 during hair follicle differentiation. J. Biol. Chem. 2006;281:29245–29255. doi: 10.1074/jbc.M603646200. [DOI] [PubMed] [Google Scholar]
- Punnonen K., Denning M., Lee E., Li L., Rhee S.G., Yuspa S.H. Keratinocyte differentiation is associated with changes in the expression and regulation of phospholipase C isoenzymes. J. Invest. Dermatol. 1993;101:719–726. doi: 10.1111/1523-1747.ep12371682. [DOI] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E., et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Segre J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T., Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Truong A.B., Kretz M., Ridky T.W., Kimmel R., Khavari P.A. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes & Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Lin H., Xu L., Chen C.D. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]