Abstract
The Negative Elongation Factor (NELF) is a transcription regulatory complex that induces stalling of RNA polymerase II (Pol II) during early transcription elongation and represses expression of several genes studied to date, including Drosophila Hsp70, mammalian proto-oncogene junB, and HIV RNA. To determine the full spectrum of NELF target genes in Drosophila, we performed a microarray analysis of S2 cells depleted of NELF and discovered that NELF RNAi affects many rapidly inducible genes involved in cellular responses to stimuli. Surprisingly, only one-third of NELF target genes were, like Hsp70, up-regulated by NELF-depletion, whereas the majority of target genes showed decreased expression levels upon NELF RNAi. Our data reveal that the presence of stalled Pol II at this latter group of genes enhances gene expression by maintaining a permissive chromatin architecture around the promoter-proximal region, and that loss of Pol II stalling at these promoters is accompanied by a significant increase in nucleosome occupancy and a decrease in histone H3 Lys 4 trimethylation. These findings identify a novel, positive role for stalled Pol II in regulating gene expression and suggest that there is a dynamic interplay between stalled Pol II and chromatin structure.
Keywords: Gene expression, transcription elongation, polymerase stalling, chromatin structure
Gene expression by RNA Polymerase II (Pol II) can be regulated at multiple steps in the transcription cycle. In addition to numerous mechanisms for controlling the recruitment of Pol II and transcription factors to a gene promoter, Pol II escape from the promoter-proximal region and its transition to productive elongation are also subject to regulation (for review, see Saunders et al. 2006). Manipulating the efficiency of transcription elongation through the initially transcribed region by a process called promoter-proximal pausing or stalling governs transcription output in systems as diverse as bacteriophage λ, Drosophila, humans, and the HIV virus (Krumm et al. 1992; Strobl and Eick 1992; Laspia et al. 1993; Lis 1998; Roberts et al. 1998). However, despite reports of polymerase stalling in a wide variety of biological systems, relatively few genes were known to be regulated during early transcription elongation in vivo, leading many to view promoter-proximal stalling as a rare phenomenon.
Recently, genome-wide location analyses of Pol II have challenged this view by demonstrating that >1000 of ∼18,000 Drosophila genes exhibit hallmarks of Pol II stalling within their promoter-proximal regions (Muse et al. 2007; Zeitlinger et al. 2007). Moreover, similar ChIP–chip analyses of Pol II distribution in human cells suggested that regulation of transcription elongation represents a rate-limiting step in the expression of a significant fraction of genes (Guenther et al. 2007). Interestingly, these studies found an enrichment in highly regulated genes such as those involved in development and responses to stimuli among the genes that possessed stalled Pol II (Guenther et al. 2007; Muse et al. 2007; Zeitlinger et al. 2007). As a result, there is a growing interest in understanding the mechanisms that regulate transcription through the promoter-proximal region and the potential physiological and developmental roles for Pol II stalling.
The Drosophila heat-shock (Hsp) genes were the first genes shown to be regulated by promoter-proximal stalling of Pol II. Under uninduced conditions, Pol II is recruited to the Hsp70 promoter and begins RNA synthesis, but pauses 20–45 nucleotides (nt) downstream from the transcription start site (Gilmour and Lis 1986; Rougvie and Lis 1988; Rasmussen and Lis 1993; Lis 1998). The stalled elongation complex remains stably engaged within the promoter-proximal region; in this complex, the C-terminal domain (CTD) of the Pol II Rpb1 subunit is phosphorylated at Ser 5, and the nascent RNA is partially capped (Rougvie and Lis 1988; Rasmussen and Lis 1993; Boehm et al. 2003). Escape of Pol II from the stalled state is highly regulated and is rate-limiting for Hsp70 gene expression. Release of polymerase into the gene takes place extremely quickly after heat induction, allowing for a rapid and robust increase in RNA levels. Regulation of Pol II stalling requires the coordinated action of negative and positive elongation factors; however, the mechanisms that govern this process are poorly defined.
Several proteins associated with stalled Pol II at Hsp70 under uninduced conditions have been identified by chromatin immunoprecipitation (ChIP). DSIF, a heterodimeric complex of Spt4 and Spt5, colocalizes with Pol II at Hsp70 prior to and during heat shock (Andrulis et al. 2000; Wu et al. 2005). DSIF is associated with active Pol II throughout the genome and has been shown to elicit both negative and positive effects on transcription, depending on the assay conditions (Hartzog et al. 1998; Wada et al. 1998; Andrulis et al. 2000). One prime candidate for regulating Pol II promoter-proximal stalling is the Negative Elongation Factor, NELF. Although few targets of NELF have been defined, NELF is known to influence expression of several genes that are regulated by Pol II stalling, including Drosophila Hsp70, mammalian junB, and HIV RNA (Fujinaga et al. 1998; Wu et al. 2003; Aida et al. 2006; Zhang et al. 2007). NELF requires DSIF to interact with Pol II, and the two factors together inhibit transcription elongation in vitro by increasing the duration of intrinsic pauses (Yamaguchi et al. 1999; Renner et al. 2001; Palangat et al. 2005; Cheng and Price 2007). Importantly, NELF is present at the uninduced Hsp70 promoter, but dissociates upon gene activation, and depletion of NELF reduces Pol II stalling at Hsp70 both in vitro and in vivo (Wu et al. 2003, 2005).
Biochemical assays indicate that the kinase activity of P-TEFb relieves transcription repression by NELF (Renner et al. 2001; Peterlin and Price 2006; Cheng and Price 2007). P-TEFb phosphorylates the Pol II CTD at Ser 2, which facilitates Pol II elongation and provides a binding platform for elongation and 3′-processing factors (Peterlin and Price 2006 and references therein). In addition, P-TEFb can phosphorylate both Spt5 and NELF; this has been suggested to trigger de-repression by releasing NELF from the elongation complex and converting Spt5 from a negative to a positive elongation factor (Fujinaga et al. 1998; Ivanov et al. 2000; Kim and Sharp 2001; Lavoie et al. 2001; Yamada et al. 2006). In agreement with a role for P-TEFb in releasing stalled Pol II, artificial recruitment of P-TEFb to the Hsp70 promoter under uninduced conditions led to an increase in basal transcription levels (Lis et al. 2000).
The NELF complex is conserved in higher eukaryotes, but is absent from Caenorhabditis elegans, Saccharomyces cerevisiae, and Arabidopsis thaliana (Narita et al. 2003). It is composed of four subunits, NELF-A, NELF-B, NELF-C/D, and NELF-E. The mammalian NELF-A gene, also called WHSC2, is a potential contributor to Wolf-Hirschhorn syndrome (Wright et al. 1999). The NELF-B subunit, referred to as COfactor of BRCA1 (COBRA-1), interacts with the BRCA-1 protein in a yeast two-hybrid assay and has been reported to bind ER-α and AP-1 family members (Ye et al. 2001; Aiyar et al. 2004; Zhong et al. 2004). NELF C/D are translation variants of the same mRNA and are homologous to the mammalian protein TH1-like. NELF-E has an RNA recognition motif (RRM), which is proposed to bind nascent RNA as it emerges from Pol II (Narita et al. 2003).
With so few defined targets of NELF, its physiological function and the prevalence of NELF-mediated regulation of gene expression remained unknown. To address these questions, we performed a genome-wide search for NELF-regulated genes in Drosophila. We discovered that NELF impacts the expression of many genes that are induced in response to stimuli. Moreover, our data reveal that NELF-dependent Pol II stalling can both enhance and repress gene expression in vivo, illuminating a role for promoter-proximally stalled polymerase in transcription activation. We demonstrate that loss of Pol II stalling and diminished gene expression are coupled to increased nucleosome occupancy of promoter regions, demonstrating a novel, dynamic relationship between Pol II stalling and promoter chromatin architecture.
Results
Global identification of Drosophila genes regulated by NELF
To probe the function of the NELF complex in vivo, we compared genome-wide transcriptional profiles of Drosophila S2 cells that had been depleted of NELF using RNAi against both NELF-B and NELF-E, to those of untreated cells and cells mock-treated with dsRNA targeting β-galactosidase. Western analysis demonstrated that the levels of both NELF subunits were reduced by 90%–95% by the RNAi treatment (Supplemental Fig. S1), but that the levels of Pol II and transcription elongation factors like Spt5 remained unchanged (data not shown).
NELF depletion significantly altered the levels of 241 of ∼18,500 transcripts present on the Affymetrix Drosophila Genome 2.0 arrays (Fig. 1; Supplemental Table S1). To validate our microarray data, expression levels of more than 35 candidate NELF target genes were determined by RT–PCR using RNA isolated from independent RNAi-treated samples. For all putative NELF target genes examined, the direction and magnitude of fold change in expression levels following NELF RNAi agreed well between microarray and RT–PCR analyses (Supplemental Fig. S2).
Figure 1.
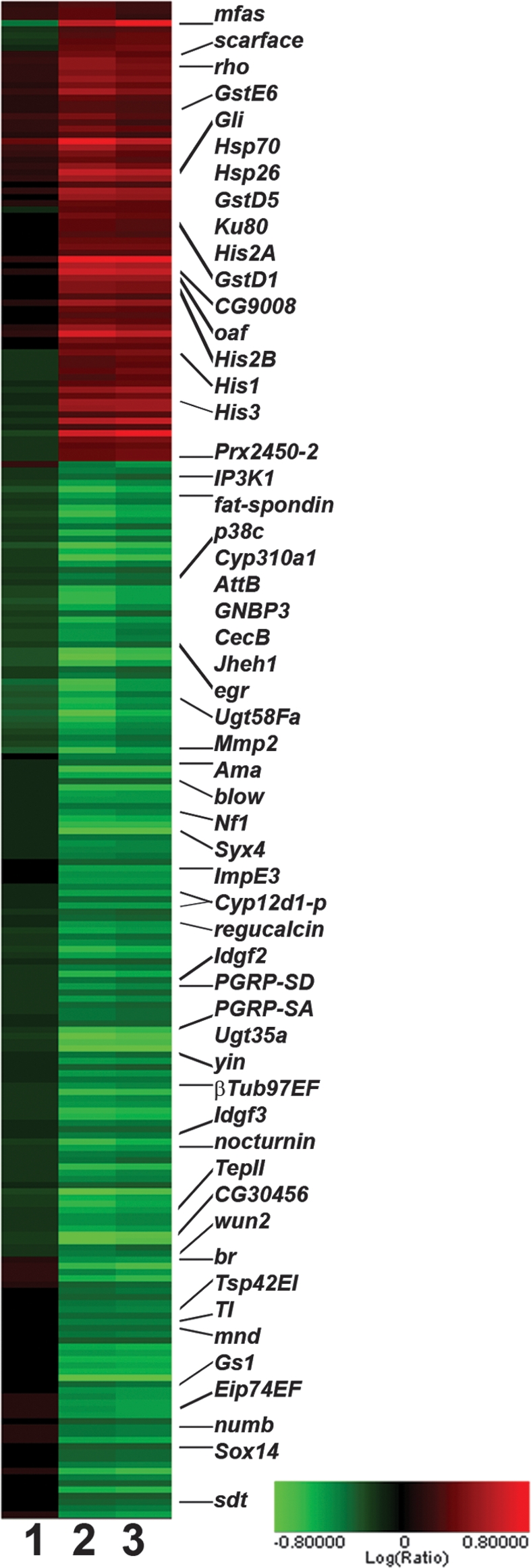
Cluster analysis showing the expression changes of transcripts significantly affected by NELF depletion. Triplicate samples for each condition were averaged, and pairwise comparisons between conditions were performed. Shown are 241 transcripts that changed >1.5-fold in NELF-depleted cells, with a P-value <0.001 as compared with both untreated and mock-treated cells: untreated versus mock-treated (lane 1); untreated versus NELF-depleted (lane 2); mock-treated versus NELF-depleted (lane 3). Several genes of interest are labeled at right. Red indicates an increase in RNA levels, while green indicates a decrease. Values are given in log base 2 units, with the color bar shown at bottom right.
Gene Ontology (GO) classification of NELF target transcripts revealed a significant enrichment in genes that respond to environmental or intracellular stimuli, including the defense/innate immune responses and responses to temperature, toxin, chemical, and protein stimulus (Fig. 2). The propensity of NELF target genes to undergo transcriptional changes in response to specific signals suggests that NELF may function at these genes to facilitate rapidly inducible gene expression. Interestingly, recent identification of genes with promoter-proximally stalled Pol II in Drosophila S2 cells also revealed an over-representation in stimulus-responsive genes, suggesting that NELF-mediated Pol II stalling plays key roles in the rapid and precise control of gene expression.
Figure 2.
NELF target genes are involved in cellular responses to stimuli. A query of the Gene Ontology Database with a list of the 168 NELF target genes that have annotated Biological Processes reveals several classes that are significantly over-represented in this gene list (P-value cutoff <0.001). The number of genes in each Gene Ontology class, the percentage of total genes in that class that are NELF-regulated, and the associated P-values are given.
Consistent with previous work showing that NELF affects promoter-proximal stalling at Hsp70 (Wu et al. 2003), expression of Hsp70 and two other heat-shock protein genes, Hsp26 and Hsp22, increased 2-5-fold upon NELF depletion (Supplemental Table S1). Because NELF inhibits transcription elongation in vitro, we expected that NELF would repress gene expression in vivo and thus the expression of most NELF target genes would increase in response to NELF depletion. Contrary to these expectations, we found that levels of more than two-thirds of NELF target transcripts decreased following NELF RNAi (170 of 241) (Supplemental Table S1), indicating that NELF could play a positive role in transcription of this group of genes. To make a distinction between these two classes of NELF target genes, we refer herein to genes like Hsp70 whose transcript levels increase upon NELF RNAi as up-regulated genes and to genes whose transcription decreases upon NELF RNAi as down-regulated genes.
NELF is associated with promoter-proximally stalled Pol II at NELF target genes
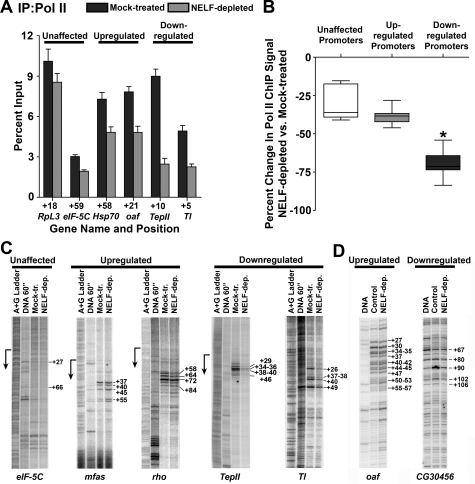
ChIP analyses of Pol II association with the newly identified NELF target genes show high levels of Pol II focused near the gene promoters and little Pol II in the downstream regions, a hallmark of polymerase stalling (Fig. 3; Lis 1998; Muse et al. 2007). In addition, NELF ChIP signal was readily detectable at all NELF target promoters where Pol II occupancy was observed (Fig. 3; Supplemental Fig. S3). The NELF ChIP signal observed was in proportion to the Pol II signal in the promoter regions, consistent with the idea that NELF interacts with the Pol II complex at these genes (Yamaguchi et al. 2002). Interestingly, significant levels of NELF were detected at promoters whose transcription was not affected by NELF RNAi under these conditions, including eIF-5C and RpL3 (Fig. 3A). This finding is in agreement with previous data showing that NELF colocalizes broadly with Pol II on polytene chromosomes (Wu et al. 2003), and suggests that NELF globally associates with Pol II preinitiation or early elongation complexes.
Figure 3.
Pol II and NELF occupy the promoter-proximal regions of NELF target genes. ChIP material was immunoprecipitated with antibodies that recognize the Rpb3 subunit of Pol II or NELF-E. The percentage of input obtained in each ChIP sample is plotted for the genes designated, with positions given representing the center of each primer pair with respect to the transcription start site for that gene. ChIP signals are shown for NELF-unaffected genes eIF-5C and RpL3 (A); up-regulated genes Hsp70, mfas, oaf, and CG9008 (B); and down-regulated genes nocturnin, Mmp2, wun2, CG11739, egr, and TepII (C).
If genes whose expression decreases in response to NELF depletion are primary targets of NELF, then their transcript levels should respond to NELF RNAi within the same time frame as up-regulated genes; however, if the observed decreases in transcript levels are secondary effects, then the down-regulated genes could respond more slowly to NELF RNAi. To test this possibility, we monitored transcript levels of several genes over a time course of NELF depletion. Cells were treated with dsRNA targeting the NELF complex, and samples were collected after 24, 40, 56, and 72 h of RNAi treatment. RT-PCR analysis revealed that while increased expression of up-regulated genes was detectable within 40–72 h of NELF RNAi (representative examples shown in Supplemental Fig. S1), expression levels of genes down-regulated upon NELF depletion diminished even more rapidly (within 24–40 h), supporting the idea that many of these genes are direct targets of NELF.
NELF RNAi has different effects on Pol II occupancy at up-regulated versus down-regulated promoters
Our recent ChIP–chip comparison of Pol II distribution in mock-treated versus NELF-depleted cells demonstrated that NELF RNAi diminished promoter-proximal Pol II signal at genes with stalled Pol II; however, the magnitude of this effect was highly variable (Muse et al. 2007). We therefore asked whether the Pol II signals at the different classes of NELF target promoters would display different sensitivities to NELF depletion. To address this issue, we performed ChIP on several target genes that were up-regulated versus down-regulated upon NELF RNAi (Fig. 4A). We observed a striking decrease in Pol II ChIP signal near down-regulated promoters that was significantly greater than that near promoters that were up-regulated or unaffected by NELF RNAi (Fig. 4B, 70% median decrease, P < 0.0001). This marked reduction in Pol II occupancy at down-regulated genes suggests that the decreased transcript levels observed following NELF RNAi result from decreased Pol II recruitment to these promoters, and indicates that NELF-dependent Pol II stalling is important for sustained transcription of these genes.
Figure 4.
Depletion of NELF significantly alters Pol II distribution at down-regulated genes. (A) Pol II ChIP signal at the promoters of NELF-unaffected genes RpL3 and eIF-5C; up-regulated genes Hsp70 and oaf; and down-regulated genes TepII and Tl in cells that were either mock-treated or depleted of NELF using RNAi. (B) Change in Pol II ChIP signal between mock-treated and NELF-depleted cells as a fraction of mock-treated Pol II ChIP signal for nine NELF-unaffected promoters, seven up-regulated promoters, and 16 down-regulated promoters. Median Pol II ChIP signal change at down-regulated promoters (−70%) is significantly different from NELF-unaffected (−36%) and up-regulated (−38%) promoters (P < 0.0001; Kruskal-Wallis test). (C) Analysis of permanganate reactivity at NELF target genes in S2 cells. Lanes depict, from left to right: the A+G ladder used to determine the position of the promoter (shown by arrow), the permanganate reactivity of naked DNA as a control, and reactivity pattern of the indicated genes in S2 cells that were either mock-treated or depleted of NELF. (D) Analysis of permanganate reactivity in salivary glands from wild-type and NELF-D RNAi flies. Lanes from left to right depict the permanganate reactivity of naked DNA and of the indicated genes in salivary glands from either wild-type (control) or NELF-D-depleted flies.
NELF RNAi also diminished Pol II levels near the promoters of up-regulated genes, but to a smaller extent (Fig. 4A [Hsp70, oaf], B [38% median value]). These data, combined with the observed increase in RNA levels, suggest that while NELF depletion reduces the duration of Pol II stalling near these promoters, the Pol II released into up-regulated genes upon NELF RNAi is replaced by newly recruited Pol II. Depletion of NELF also modestly reduced the Pol II ChIP signal near the promoters of genes with unchanged RNA levels (Fig. 4A [RpL3, eIF-5C], B [36% median value]), consistent with the presence of NELF ChIP signal at all promoters bound by Pol II (Fig. 3) and suggesting that NELF may influence early elongation at many promoters but that this step is rate-limiting for transcription of only certain genes.
The differential effects of NELF RNAi on Pol II levels observed at down-regulated versus up-regulated promoters are in full agreement with the observed effects of NELF depletion at the 31 NELF target genes present on our partial ChIP–chip arrays (Muse et al. 2007). In that study, we probed partial Drosophila genome arrays with Pol II ChIP material from mock-treated and NELF-depleted cells, and used computational methods to characterize Pol II distribution under each condition. The 18 down-regulated genes contained on these arrays show a 49% median decrease in promoter-proximal Pol II signal upon depletion of NELF (calculated as in Muse et al. [2007], from probes spanning the region between −250 and +500 with respect to the transcription start site), whereas up-regulated and NELF unaffected genes showed only 15% and 20% median decreases, respectively. Taken together, these data confirm that NELF plays a key role in maintaining Pol II levels at down-regulated promoters, but that Pol II enrichment is preserved at up-regulated promoters in cells depleted of NELF.
In light of earlier biochemical data demonstrating a functional interaction between NELF and P-TEFb (Peterlin and Price 2006), we investigated whether NELF-dependent stalling functions at some genes to ensure that Pol II released into the gene is properly phosphorylated by P-TEFb. To test this, we evaluated the levels of Ser 2 phosphorylated Pol II in mock-treated and NELF-depleted cells and found that NELF RNAi had no detectable effect on the phosphorylation status of Pol II within NELF target genes (Supplemental Fig. S4). Thus, our data suggest that P-TEFb is recruited to these genes and efficiently phosphorylates Pol II as it transitions into active elongation, even in the absence of NELF.
Down-regulated promoters lose stalled Pol II upon NELF depletion
To gain a high-resolution view of the effect of NELF depletion on Pol II distribution at NELF-regulated genes, we used permanganate footprinting. Permanganate reacts with thymine residues in regions of single-stranded DNA, revealing the presence and location of open transcription bubbles associated with a transcriptionally engaged polymerase. We observed significant permanganate reactivity in the promoter-proximal regions of many of the newly identified NELF targets analyzed, confirming that Pol II stalls during early elongation at these genes (Supplemental Fig. S3). NELF RNAi resulted in a dramatic decrease in promoter-proximal permanganate reactivity at down-regulated genes (Fig. 4C, TepII, Tl), consistent with ChIP data indicating that maintenance of stalled Pol II near these promoters requires NELF (Fig. 4B). By comparison, there was little or no discernable decrease in promoter-proximal permanganate reactivity at up-regulated genes following NELF depletion (Fig. 4C, mfas, rho), which indicates that while depletion of NELF likely decreases the duration of Pol II stalling at the up-regulated genes, stalling is not eliminated upon NELF RNAi. Moreover, we observed that NELF RNAi released Pol II to elongate into these genes, as evidenced by increased permanganate reactivity at sites further downstream from the promoters (Fig. 4C, mfas positions +45, +55; rho positions +84 and downstream).
The differential effect of NELF depletion on Pol II at up-regulated versus down-regulated genes is not confined to S2 cultured cells. NELF target genes also demonstrated clear differences in promoter-proximal permanganate reactivity in salivary glands from flies expressing a NELF-D RNAi construct compared with those from wild-type flies (Fig. 4D). At the up-regulated gene oaf, depletion of NELF-D did not change permanganate reactivity in the region from +27 to +47, but increased the reactivity downstream from +52 to +82 (Fig. 4D), demonstrating that Pol II recruitment and stalling persists at this gene. In contrast, NELF-D RNAi simply decreased permanganate reactivity at the down-regulated gene CG30456, from +90 to +102, indicating a substantial reduction of stalled Pol II at this promoter.
Loss of NELF-mediated stalling alters the chromatin architecture at down-regulated promoters
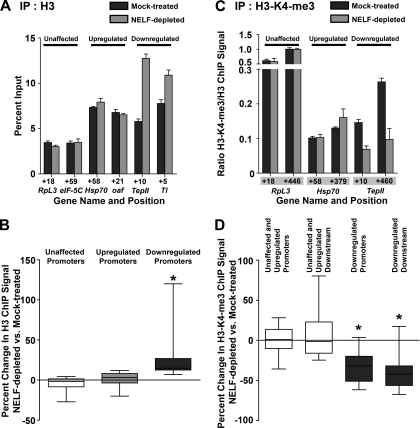
ChIP against histone H3 at genes down-regulated upon NELF depletion revealed that the decreases in transcript levels and Pol II stalling were accompanied by changes in chromatin architecture. Histone H3 ChIP signal increased at the promoters of down-regulated genes in response to NELF RNAi (Fig. 5A [TepII, Tl], B), suggesting that the stalled Pol II lost from these promoters is replaced by nucleosomes. This increase was significantly greater than that observed at the promoters of up-regulated or NELF-unaffected genes (Fig. 5A [RpL3, eIF-5C, Hsp70, oaf], B [P < 0.0001]), where Pol II occupancy is not changed markedly upon NELF depletion. Taken together, these results suggest that the presence of a stalled Pol II might protect the promoter regions of some genes against nucleosome occupancy, and that the genes that lose stalled Pol II upon NELF RNAi might be down-regulated because their promoters become obstructed by nucleosomes.
Figure 5.
Depletion of NELF alters the chromatin environment at down-regulated genes. (A) Histone H3 ChIP signal at the promoters of NELF-unaffected genes RpL3 and eIF-5C; up-regulated genes Hsp70 and oaf; and down-regulated genes TepII and Tl. (B) Change in H3 ChIP signal between mock-treated and NELF-depleted cells as a fraction of mock-treated H3 ChIP signal. Median H3 ChIP signal change at down-regulated promoters (n = 16) is significantly different from at NELF-unaffected (n = 9) and up-regulated (n = 7) promoters (P < 0.0001). (C) H3-K4-me3 ChIP signals obtained with an antibody against H3-K4-me3 and expressed as a fraction of H3 ChIP signal are shown at promoters and downstream regions of RpL3, Hsp70, and TepII. (D) Change in H3-K4-me3 ChIP signal between mock-treated and NELF-depleted cells as a fraction of mock-treated H3-K4-me3 ChIP signal for 16 NELF-unaffected and up-regulated promoters and downstream regions, and 16 down-regulated promoters and downstream regions. Median H3-K4-me3 ChIP signal change at down-regulated promoters and downstream regions is significantly different from at NELF-unaffected and up-regulated promoters and downstream regions (P < 0.0001).
We also found that the ChIP signal for histone H3 trimethylation at Lys 4 (H3-K4-me3), a hallmark of active transcription, decreased substantially at down-regulated genes in cells depleted of NELF relative to control cells (Fig. 5C [TepII], D [32% and 42% decreases in the promoter and downstream regions, respectively]), consistent with inactivation of these promoters following NELF RNAi. In contrast, up-regulated and NELF-unaffected genes showed no trend in H3-K4-me3 change upon NELF depletion (Fig. 5D).
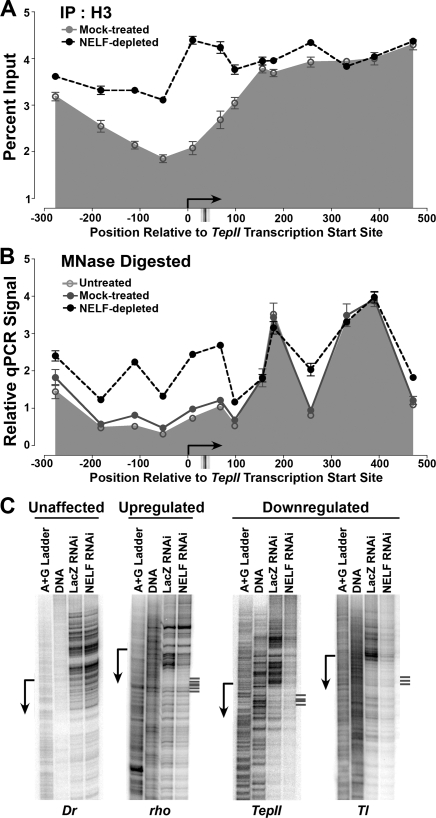
To better define the changes in chromatin structure that take place at down-regulated genes upon NELF RNAi, we analyzed histone H3 ChIP signal using tiled PCR amplicons spanning the promoter-proximal regions of the TepII and Tl genes (Fig. 6A; Supplemental Fig. S5). Many active Drosophila genes possess a canonical nucleosome organization with a nucleosome-free region near the promoter bracketed by well-positioned upstream and downstream nucleosomes, referred to as the −1 and +1 nucleosomes, respectively (Mavrich et al. 2008). In agreement with this, we found low histone H3 ChIP signal surrounding both of these promoters, with increased nucleosome occupancy beginning downstream from the position where Pol II stalling occurs (Fig. 6A; Supplemental Fig. S5). Upon NELF depletion and loss of stalled Pol II, we observed large increases in histone H3 ChIP signal promoter-proximally, accompanied by increased nucleosome density within the upstream region (Fig. 6A; Supplemental Fig. S5). These data indicate that, when Pol II stalling is lost, nucleosome occupancy increases over the DNA regions where newly recruited Pol II and transcriptional activators would be expected to bind.
Figure 6.
Obstruction of promoter regions by nucleosomes in response to NELF depletion. (A) Histone H3 ChIP signal at the down-regulated gene TepII in mock-treated and NELF-depleted S2 cells, revealed using primer pairs spanning the TepII transcription start site. For comparison, major sites of permanganate reactivity indicating the locations of stalled Pol II are shown below. (B) Nucleosome locations at TepII are altered by depletion of NELF. Chromatin from untreated, NELF-depleted, and mock-treated cells was digested with MNase and PCR performed as above. (C) DNase I hypersensitivity at promoters of NELF target genes. Lanes depict, from left to right: the A + G ladder used for position determination, the reactivity of naked DNA as a control, and the DNase I hypersensitivity of the indicated genes in nuclei isolated from S2 cells that were either mock-treated or NELF-depleted. Major sites of permanganate reactivity indicating the locations of stalled Pol II at the rho, TepII, and Tl genes are shown at right.
More detailed MNase analysis of nucleosome positioning at TepII and Tl confirmed the presence of a nucleosome-depleted region surrounding the promoters, and revealed well-positioned +1 and +2 nucleosomes downstream from the transcription start site (Fig. 6B; Supplemental Fig. S5). NELF depletion led to significant MNase protection in the TepII nucleosome-free region, suggestive of newly assembled, positioned nucleosomes centered at approximately +50 bp and −120 bp relative to the transcription start site. In addition, although the location of the peak in protection against MNase digestion by the +1 and +2 nucleosomes was unaltered by NELF RNAi, we detected increased protection in the internucleosome interval, indicative of increased nucleosome “shuffling” or “fuzziness.” Similar changes in MNase protection were detected near the Tl promoter (Supplemental Fig. S5). Thus, decreased NELF-dependent Pol II stalling leads to breakdown of canonical nucleosome organization and increased promoter occlusion by nucleosomes at these genes.
DNase I hypersensitivity assays also support a connection between a loss of Pol II stalling and changes in chromatin structure at down-regulated, but not up-regulated, genes. DNase I sensitivity was significantly decreased upstream of the transcription start sites of the down-regulated genes TepII and Tl in NELF-depleted cells relative to mock-treated cells (Fig. 6; Supplemental Fig. S6), suggesting that the surrounding chromatin adopts a more nucleosome-dense, repressive state upon NELF depletion. In contrast, genes whose expression was unaffected or up-regulated by NELF RNAi did not exhibit major changes in DNase I hypersensitivity following NELF depletion (Fig. 6; e.g., Dr, rho; Supplemental Fig. S6). Interestingly, the up-regulated gene rho showed unchanged DNase I reactivity in the upstream region but had somewhat diminished DNase I sensitivity downstream from the transcription start site following NELF RNAi (Fig. 6); this change may reflect the downstream redistribution of stalled Pol II observed at this promoter by permanganate mapping (Fig. 4C).
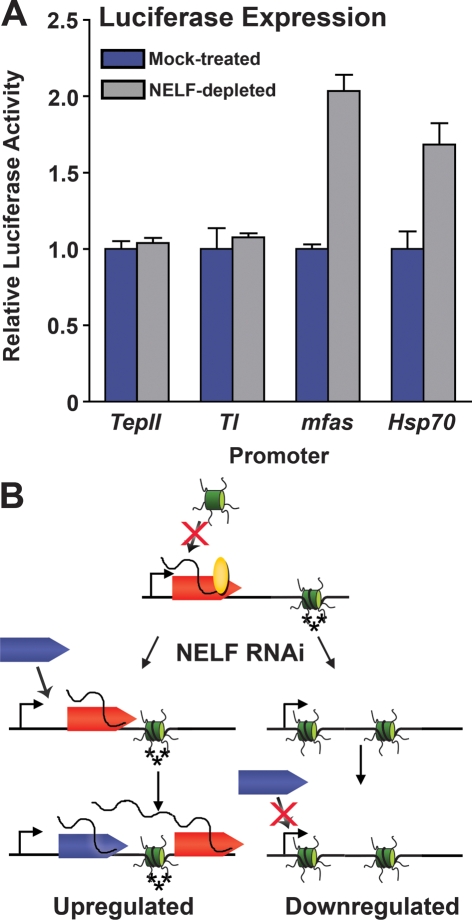
We next examined the effects of removing NELF-regulated promoters from their normal chromatin context. We made use of the fact that promoters carried on transiently transfected DNA templates are inefficiently assembled into chromatin, and would not be anticipated to exhibit a chromatin architecture similar to that of endogenous genes. Promoter regions of the down-regulated genes TepII and Tl and the up-regulated genes mfas and Hsp70 were each used to drive expression of a transiently transfected luciferase reporter. We found that NELF depletion led to an increase in expression driven by up-regulated promoters, both from the transiently transfected constructs and at the endogenous loci (Fig. 7; data not shown). In contrast, there was no effect of NELF RNAi on expression driven by plasmid-borne TepII and Tl promoters (Fig. 7), whereas control experiments confirmed that the endogenous genes were down-regulated as expected by NELF depletion (data not shown). Importantly, these data indicate that decreased transcription of down-regulated genes in response to NELF depletion is not a spurious effect stemming from incidental knockdown of a positive regulatory factor. Instead, they support the idea that chromatin context plays a role in determining how down-regulated genes, but not up-regulated genes, respond to NELF depletion and diminished Pol II stalling.
Figure 7.
Regulation of gene expression by NELF, Pol II stalling, and chromatin. (A) Differing effects of NELF-RNAi upon expression driven by transiently transfected down-regulated or up-regulated promoters. S2 cells were transfected with vectors expressing firefly luciferase driven by the promoter regions of down-regulated genes Tl and TepII or up-regulated genes mfas and Hsp70. (B) Model for the dual roles of NELF and Pol II stalling in regulating gene expression. Pol II (rocket) initiates transcription, and NELF (oval) induces Pol II to stall in the promoter-proximal region. At up-regulated genes (left panel), NELF RNAi diminishes stalling such that polymerase moves into the gene and is replaced by newly recruited Pol II, thereby increasing transcriptional output. The promoter retains H3-K4 trimethylation and low nucleosome density. At down-regulated genes, NELF RNAi leads to a significant decrease in occupancy of the promoter-proximal region by stalled Pol II, which is associated with the establishment of a more repressive chromatin structure, including increased nucleosome occupancy of the promoter region, and a reduction in histone H3-K4 trimethylation (depicted as additional nucleosome and loss of asterisks). The increased nucleosome density further inhibits recruitment of additional Pol II and decreases transcription of these genes.
Discussion
Identification of NELF target genes
Our results establish that the NELF complex regulates transcription of a significant number of genes in Drosophila S2 cells (241 transcripts) (Fig. 1; Supplemental Table S1). As exemplified by Hsp70, the only previously defined target of Drosophila NELF (Wu et al. 2003), promoter-proximally stalled Pol II is a common feature of these genes (Figs. 3 and 4; Supplemental Fig. S3). However, in contrast to Hsp70, where NELF-mediated Pol II stalling represses transcription, we found that nearly 70% of NELF target genes are down-regulated upon NELF RNAi (Fig. 1). NELF is critical for maintaining Pol II stalling at these down-regulated genes, and stalled Pol II helps preserve a nucleosome-free region around these promoters (Figs. 5, 6). Although our work is the first to investigate the mechanisms of NELF’s positive effects on gene expression, we note that previous microarray analyses of NELF-depleted human cells also detected significant numbers of transcripts that were down-regulated upon NELF depletion (Aiyar et al. 2007; Narita et al. 2007).
Our data also identify many genes in which depletion of NELF produces no change in transcriptional output but does lead to a detectable reduction in Pol II promoter occupancy (Fig. 4), in agreement with previous results (Muse et al. 2007). We suggest that at these genes, early elongation is not rate-limiting for transcription under our experimental conditions. However, we expect that NELF could influence transcriptional output of some of these genes under other conditions, in other cell types, or at different developmental stages.
Importantly, many NELF target genes are involved in responses to stimuli (Fig. 2). This finding is consistent with our recent genome-wide search for genes with promoter-proximally stalled Pol II, which also found a significant enrichment in stimulus-responsive genes among the ∼1000 Drosophila genes that were determined to possess stalled polymerase (Muse et al. 2007). We propose that inducible genes possess a poised Pol II and open chromatin architecture in order to facilitate rapid gene activation in response to environmental signals.
Effects of NELF-dependent Pol II stalling on gene expression
A key result of this study is that NELF-mediated stalling of polymerase can have both positive and negative effects on gene expression. We find that Pol II stalling can function either to attenuate the expression of genes like Hsp70 under noninducing conditions or, alternatively, to maintain basal expression levels by preventing transcription inactivation and the assembly of promoter-proximal nucleosomes. Based on our results and on the previously defined role of NELF in inhibiting transcription elongation, we propose the following model for NELF activity (Fig. 7B): NELF binds to the Pol II complex and collaborates with DSIF and/or other factors to induce promoter-proximal stalling (Renner et al. 2001; Yamaguchi et al. 2002; Wu et al. 2003). Depletion of NELF reduces the duration of Pol II stalling, allowing the polymerase to resume elongation and move away from the promoter region.
At up-regulated genes such as Hsp70 (Fig. 7, left), NELF-mediated Pol II stalling represses expression and NELF RNAi allows Pol II to be released into the gene more rapidly after initiation, resulting in an increase in transcriptional output. Significant Pol II ChIP signal remains at up-regulated promoters upon NELF RNAi (Fig. 4A,B; Muse et al. 2007), and permanganate footprinting of Pol II complexes shows similar levels of reactivity within the initially transcribed regions of these genes (Fig. 4C,D), demonstrating that the released Pol II is efficiently replaced by newly recruited polymerase molecules. Furthermore, we observe permanganate reactivity at sites slightly further downstream from the promoter of the up-regulated genes in both NELF-depleted S2 cells and flies (Fig. 4C,D), reflecting Pol II elongation into these genes. Interestingly, these results suggest that the depletion of NELF reduces, but does not completely abolish, Pol II stalling near these promoters and that inhibition of early elongation by NELF is not the only mechanism that affects promoter-proximal transcription efficiency at up-regulated genes.
Consistent with the continued presence of promoter-proximal Pol II at up-regulated genes following NELF RNAi, we do not detect an increase in nucleosome occupancy of these promoters or a change in local histone H3-K4-me3 levels (Figs. 5, 6). In addition, we do not find any evidence that the accessibility of the upstream region of the up-regulated genes is altered by NELF RNAi (Fig. 6) and conclude that the chromatin architecture at these genes is not substantially affected by a reduction in Pol II stalling duration.
In contrast, at down-regulated genes, the Pol II released upon NELF RNAi is not replaced by recruitment of additional Pol II (Fig. 7, right). We observe a striking reduction in promoter-proximal Pol II occupancy and stalling at these promoters (Fig. 4). Moreover, this decrease in polymerase occupancy is accompanied by an increase in nucleosome density and nuclease protection near the down-regulated promoters and a loss of the active chromatin mark H3-K4-me3 (Figs. 5, 6). Taken together, these data suggest that the presence of a promoter-proximally stalled Pol II complex might positively influence transcription of these genes by preventing nucleosome assembly near their promoters. It remains to be determined if the stalled polymerase achieves this task by sterically blocking the promoter region, thereby protecting it from encroaching nucleosomes, or if the stalled Pol II complex recruits or stabilizes other protein complexes near these promoters. In either case, we envision that the resulting permissive chromatin structure would enhance subsequent transcription by facilitating recruitment of the general transcription machinery and additional Pol II molecules.
Interactions between stalled Pol II and chromatin structure at down-regulated genes
Previous work has shown that mutations within the Hsp70 promoter sequence that reduced Pol II stalling also decreased the accessibility of the upstream DNA region to the heat-shock activator HSF, and led to a slower, less efficient heat-shock response (Lee et al. 1992; Shopland et al. 1995). These results indicated that the stalled Pol II plays a critical role in establishing the normal, open promoter architecture at Hsp70 that is required for both activator binding and Pol II recruitment during heat induction. We propose that preventing promoter-proximal nucleosome assembly is a general role for stalled Pol II. In this way, we envision that Pol II stalling can play a positive role in gene expression by “poising” the promoter for further or future activation.
The idea that there is interplay between the stalled polymerase and the +1 nucleosome has recently received support from a whole-genome analysis of nucleosome positioning in Drosophila (Mavrich et al. 2008). These data revealed the presence of a highly positioned +1 nucleosome located in proximity to the promoter, in a position to influence Pol II stalling. The +1 nucleosome was shifted slightly downstream at genes with stalled Pol II, suggesting that Pol II engages this nucleosome and influences its location. Our data are consistent with this possibility and, furthermore, suggest that stalled Pol II can affect the occupancy as well as the positioning of promoter-proximal nucleosomes.
We note that although we detect an increased signal for histone transcripts in our microarray analysis, this arises from aberrant polyadenylated histone transcripts produced in the absence of NELF rather than an actual increase in transcription of histone genes (Supplemental Fig. S7). These results are consistent with the recently demonstrated role of human NELF in stimulating histone mRNA processing and indicate that this function is conserved in metazoans (Narita et al. 2007). Importantly, this change in histone mRNA processing does not lead to observable changes in histone protein levels (Supplemental Fig. S7).
Differences between genes that are up-regulated versus down-regulated by NELF RNAi
While it is currently unknown what dictates whether a given gene would be repressed, stimulated, or unaffected by NELF-mediated Pol II stalling, this is likely to involve interactions among the stalled polymerase, transcription activators like P-TEFb, chromatin-modifying complexes, and general transcription factors. We envision that differences in the inherent rate of Pol II recruitment and initiation at individual genes would affect for how long the promoter was left unoccupied following release of stalled Pol II. Under conditions in which the Pol II recruitment rate is fast, the gene might maintain high Pol II occupancy and a nucleosome-free structure even in the absence of stalling. In contrast, a gene with a slow rate of recruitment might be more susceptible to encroaching nucleosomes when stalled Pol II is released artificially, such as through depletion of NELF. Likewise, the duration of Pol II stalling at a given gene, and how this compared with the rate of Pol II recruitment, would play an important role in determining the transcription outcome in response to NELF depletion.
Both this work and our previous ChIP–chip analyses demonstrated that promoters with stalled Pol II exhibit variable responses to NELF RNAi. Whereas the majority of promoters with stalled Pol II on our partial genome arrays exhibited dramatic decreases in Pol II levels upon NELF depletion, approximately one-third of these genes retained promoter-proximal enrichment of Pol II in the absence of NELF (Muse et al. 2007). These data suggested that while most genes require NELF to establish stalled Pol II, a subset of genes possess mechanisms for recruiting and retaining Pol II within the promoter region that are independent of NELF. Many up-regulated genes, including Hsp70, fall into this latter class. Based on the wealth of data describing promoter-proximal Pol II stalling at Hsp70, we suggest that aspects of the initially transcribed sequence and the binding of GAGA factor upstream would help to establish stalling at these genes (Lis 1998).
To address the possibility that inherent sequence properties of the promoter may play a role in determining the role of NELF-mediated stalling, we analyzed promoter-proximal sequences of up-regulated and down-regulated genes (from −250 to +50 bp). We found that up-regulated promoters are significantly more likely to contain TATA elements than are down-regulated genes or Drosophila promoters genome-wide (Supplemental Table S2). This result is intriguing, since TATA-containing promoters as a class have been shown to exhibit a less canonical nucleosome organization and lack a well-positioned +1 nucleosome centered near +150. Instead, these genes appear to establish gene-specific chromatin structures with the help of chromatin remodeling complexes (Albert et al. 2007; Mavrich et al. 2008). Thus, the TATA-containing, up-regulated genes might possess redundant mechanisms to maintain the appropriate chromatin structure. This is clearly the case at heat-shock genes, where GAGA factor, TFIID and the stalled polymerase have all been shown to contribute to maintaining a nucleosome-depleted upstream region and positioned +1 nucleosome located well downstream of +200 (Wu 1980; Costlow and Lis 1984; Lee et al. 1992; Shopland et al. 1995; Leibovitch et al. 2002). Although other up-regulated genes have not been studied as extensively, MNase footprinting performed on the promoter regions of mfas, oaf, and rho showed no evidence of a well-positioned +1 nucleosome positioned upstream of +200 (data not shown), suggesting that they too lack a canonical nucleosome distribution. We propose that these up-regulated genes, like Hsp70, might use chromatin remodeling complexes to help maintain their gene-specific nucleosome structure, making them less reliant on NELF-mediated Pol II stalling for maintenance of promoter accessibility.
In contrast, the down-regulated promoters investigated have tightly positioned nucleosomes that are located close to the transcription start site and a canonical nucleosome pattern, typical of TATA-less genes. We find a positioned +1 nucleosome centered at approximately +150 bp downstream from the TepII, Tl, and nocturnin promoters, as well as defined nucleosome-free promoter and upstream regions (Fig. 6; Supplemental Fig. S5; data not shown). We suggest that, in the absence of specific chromatin remodeling activities, the down-regulated genes depend on promoter-proximally stalled Pol II as a place-holder in order to maintain a nucleosome free promoter region and potentiate gene expression.
Conclusions
In summary, we find that NELF-mediated Pol II stalling is not a strictly repressive process, but, unexpectedly, can also function to enhance transcription. The fact that many NELF target genes are involved in responses to stimuli suggests that Pol II stalling plays a role in dynamic, signal-dependent activation of transcription. Consistent with this idea, we find that the stalled Pol II maintains a chromatin environment around NELF target genes that could facilitate rapid recruitment of additional Pol II to these genes. It will be interesting in future studies to determine in detail how the stalled Pol II impacts local nucleosome architecture and promoter accessibility.
Materials and methods
RNAi and microarray analysis
Drosophila S2 cells in triplicate were untreated, treated with dsRNA against β-galactosidase (mock-treated), or against NELF subunits as described in Adelman et al. (2006). Total RNA was isolated using the RNeasy kit (Qiagen). Gene expression analysis was conducted using Drosophila Genome 2.0 Genechip arrays (Affymetrix). Data were imported into the Rosetta Resolver system (version 5.1). Fold changes and P-values, based on ratios built in the system, were exported for further analysis. Ratios were built as follows: mock-treated:Untreated, NELF-depleted:Untreated, and NELF-depleted:mock-treated. Based on these ratios, genes with a P-value <0.001 and an absolute fold change >1.5 in both comparisons were clustered hierarchically using average linkage clustering and the cosine correlation. Microarray data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession number GSE6141.
Validation of microarray results and RNAi time-course analysis were performed using three independent biological replicates for each treatment group (Supplemental Fig. S1). Unless noted otherwise, cDNA was generated using oligo(dT) for reverse priming, and RNA levels were determined using quantitative PCR (qPCR) and gene-specific primer pairs (primer sequences available upon request). All values were normalized to actin5c transcript levels. Expression levels were plotted relative to the mean of three untreated samples. Gene Ontology classification was performed using NetAffx (Affymetrix) and confirmed using Genespring software (Agilent Technologies).
ChIP and tandem RNAi-ChIP
Untreated cross-linked and RNAi-ChIP samples were prepared as described in Adelman et al. (2006) with the following modifications: formaldehyde cross-linking proceeded for 10 min and 7.5 × 106 cells were used per IP. Both NELF-B and NELF-E were targeted with dsRNA for preparation of NELF-RNAi ChIP samples. The Pol II (Rbp3 subunit) and NELF antibodies were described previously (Wu et al. 2003; Adelman et al. 2005) and were used at 12 μL and 14 μL per IP, respectively. Anti-Histone H3 antibody (ab1791; Abcam) was used at 20 μL per IP, while the H3-K4-me3 (#07-473) antibody (Upstate Biotechnologies) was used at 30 μL per IP. qPCR was performed as described above. The values given are the average of three biological replicates, and the error represents the SEM. The Kruskal-Wallis test was used for statistical analysis of the differences between median ChIP signals from the different classes of genes. Changes deemed significant had P-values <0.0001.
Permanganate footprinting
Permanganate footprinting in dissected salivary glands was performed as previously described (Wu et al. 2003). Permanganate footprinting in S2 cells was done similarly to salivary glands but with the following modifications. Approximately 1 × 107 cells were spun down to remove the culture media. The cell pellet was resuspended in 1 mL of ice-cold 10 mM KMnO4 solution in 1× PBS and incubated on ice for 60 sec. The reaction was stopped by the addition of 0.5 mL of stop solution (20 mM Tris-HCl at pH 8.0; 40 mM EDTA; 1% SDS; 0.5 M β-mercaptoethanol). DNA was purified as has been described for salivary glands (Wu et al. 2003). Oligonucleotide sequences used in ligation-mediated PCR reactions are available upon request.
NELF RNAi transgenic fly lines
RNAi-encoding transgenes were expressed under control of the Gal4 UAS sequence. Expression was induced in salivary glands by mating flies containing the RNAi transgene with Blooming stock #1824 as described previously (Wu et al. 2003). The NELF-D RNAi transgene was constructed in the plasmid pUAST similarly to the NELF-E RNAi transgene with the following modifications: A 573-nt region of NELF-D was amplified with the primers ggcgaattcAAAAATGGAAGTGGAATACGA and ggctagatctGAGAAAACCTCAATCTGCTG, and subcloned as an EcoRI/BglII cut fragment into EcoRI/BglII cut pUAST. (Uppercase letters designate NELF sequences, and lowercase letters sequence designate sequence added for cloning.) A 703-nt region of NELF D was amplified with the primers ccgtctaga AAAAATGGAAGTGGAATACGA and ggctagatctGATGAGC ACCTGGGAGTAT, and subcloned as a BglII/XbaI fragment into the pUAST derivative just described. The resulting plasmid contains two regions of the NELF-D gene in opposing orientation that generate the NELF-D RNAi.
DNase I hypersensitivity analysis
DNase I hypersensitivity analysis in S2 cells was performed as described in Wu et al. (1979) with the following modifications. Nuclei were treated with 5 U/mL DNase I for 10 min, unless noted otherwise. After termination of the DNase I cleavage by the addition of EDTA, nuclei were treated with RNAse Cocktail (Ambion) for 10 min at room temperature, and DNA was purified using the DNeasy Blood and Tissue Kit (Qiagen). DNase I hypersensitive sites were visualized with a modified ligation-mediated PCR protocol. Following gene-specific primer extension to generate blunt ends at cleavage sites, DNA was ligated to a linker and amplified using 20 cycles of PCR with gene-specific and linker-specific primers. Amplified sequences were labeled and run on an 8% acrylamide 8 M urea gel. Oligonucleotide sequences used in ligation-mediated PCR reactions are available upon request.
MNase protection analysis
S2 cells were cross-linked for 45 sec with 1% formaldehyde, quenched, and washed in TBS. Cells were resuspended in 10 mM Tris (pH 8), 10 mM KCl, 3 mM CaCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, Protease Inhibitors (Roche), and Triton X-100 was added to 0.1%. After incubation on ice, cells were lysed with a dounce homogenizer and washed once with Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM EDTA, Protease Inhibitors). Cells were resuspended in Buffer B and incubated on ice for 15 min. Chromatin was isolated by centrifugation and resuspended in Digest Buffer (15 mM Tris at pH 8, 60 mM KCl, 15 mM NaCl, 1 mM DTT, 0.25 M sucrose, 1 mM CaCl2). Twenty units of MNase were used to digest 200 μL of chromatin for 2 h at 25°C. Stop Solution (1% SDS, 100 mM sodium bicarbonate, 25 mM EDTA) was added, and material was incubated for 90 min at 65°C. After overnight digestion with Proteinase K, DNA phenol:chloroform-extracted and precipitated. Pellets were resuspended in water, treated with RNAse Cocktail, and separated on a 1.2% agarose gel. This procedure yielded ∼80% DNA from mononucleosomes (∼150 bp) and ∼20% DNA from di- and trinucleosomes (∼300 and ∼450 bp, respectively). The mono-nucleosome DNA was excised from the gel, gel-purified, and used as a qPCR template. DNA sheared by sonication was used as a reference standard.
Luciferase reporter assays
S2 cells were transfected with modified pGL3 vectors (Promega) where firefly luciferase expression was driven by promoter regions of down-regulated genes TepII (−334 to +188 relative to the transcription start site) and Tl (−422 to +139) or up-regulated genes mfas (−411 to +209) and hsp70 (−336 to +235). Each promoter drove luciferase expression to a level >20-fold that of a promoterless control construct. The vector pRL-polIII Renilla expressing Renilla luciferase driven by the promoter of Drosophila RpIII128 (−155 to +11) was co-transfected to control for transfection efficiency. Forty hours after transfection, cells were depleted of NELF or mock-treated as described above. After 72 h of RNAi treatment, cells were harvested, and lysates were assayed for firefly and Renilla luciferase activity using the Dual Luciferase Reporter Assay System (Promega). Results were plotted as the ratio of firefly to Renilla luminescence and were normalized to the signal from mock-treated cells.
Acknowledgments
We thank John Lis, Paul Wade, and Mike Resnick for critical reading of the manuscript, and Mike Marr for generously providing antibodies. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to K.A. (Z01 ES101987) and NIH grant GM47477 to D.S.G.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1643208.
Note added in proof
The distribution of NELF across the Drosophila genome has been described recently (Lee et al. 2008). One-hundred-fifty-four of 226 of our NELF-regulated genes were found to have significant NELF binding within 200 bp of the transcription start.
References
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Adelman K., Wei W., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T. Drosophila paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar S.E., Sun J.L., Blair A.L., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., et al. Attenuation of estrogen receptor α-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes & Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar S.E., Blair A.L., Hopkinson D.A., Bekiranov S., Li R. Regulation of clustered gene expression by cofactor of BRCA1 (COBRA1) in breast cancer cells. Oncogene. 2007;26:2543–2553. doi: 10.1038/sj.onc.1210047. [DOI] [PubMed] [Google Scholar]
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Guzman E., Doring P., Werner J., Lis J.T. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A.K., Saunders A., Werner J., Lis J.T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Price D.H. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Costlow N., Lis J.T. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:1853–1863. doi: 10.1128/mcb.4.9.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K., Cujec T.P., Peng J., Garriga J., Price D.H., Grana X., Peterlin B.M. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D.S., Lis J.T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 1986;6:3984– 3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog G.A., Wada T., Handa H., Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Kwak Y.T., Guo J., Gaynor R.B. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B., Sharp P.A. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 2001;276:12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- Krumm A., Meulia T., Brunvand M., Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes & Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Laspia M.F., Wendel P., Mathews M.B. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J. Mol. Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- Lavoie S.B., Albert A.L., Handa H., Vincent M., Bensaude O. The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J. Mol. Biol. 2001;312:675–685. doi: 10.1006/jmbi.2001.4991. [DOI] [PubMed] [Google Scholar]
- Lee H., Kraus K.W., Wolfner M.F., Lis J.T. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes & Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Lee C., Li X., Hechmer A., Eisen M., Biggin M.D., Venters B.J., Jiang C., Li J., Pugh B.F., Gilmour D.S. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitch B.A., Lu Q., Benjamin L.R., Liu Y., Gilmour D.S., Elgin S.C. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol. 2002;22:6148–6157. doi: 10.1128/MCB.22.17.6148-6157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lis J.T., Mason P., Peng J., Price D.H., Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Mavrich T.N., Jiang C., Ioshikhes I.P., Li X., Venters B.J., Zanton S.J., Tomsho L.P., Qi J., Glaser R.L., Schuster S.C., et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D.K., Hasegawa J., Omori M., Inukai N., et al. Human transcription elongation factor NELF: Identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Palangat M., Renner D.B., Price D.H., Landick R. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl. Acad. Sci. 2005;102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rasmussen E.B., Lis J.T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner D.B., Yamaguchi Y., Wada T., Handa H., Price D.H. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- Roberts J.W., Yarnell W., Bartlett E., Guo J., Marr M., Ko D.C., Sun H., Roberts C.W. Antitermination by bacteriophage λ Q protein. Cold Spring Harb. Symp. Quant. Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Rougvie A.E., Lis J.T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Saunders A., Core L.J., Lis J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Shopland L.S., Hirayoshi K., Fernandes M., Lis J.T. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Strobl L.J., Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Sugimoto S., Yano K., Hartzog G.A., Winston F., et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright T.J., Costa J.L., Naranjo C., Francis-West P., Altherr M.R. Comparative analysis of a novel gene from the Wolf-Hirschhorn/Pitt-Rogers-Danks syndrome critical region. Genomics. 1999;59:203–212. doi: 10.1006/geno.1999.5871. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P.M., Livak K.J., Holmgren R., Elgin S.C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979;16:797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Yamaguchi Y., Benjamin L.R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Lee C., Fan R., Smith M.J., Yamaguchi Y., Handa H., Gilmour D.S. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–1279. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Wada T., Watanabe D., Takagi T., Hasegawa J., Handa H. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Inukai N., Narita T., Wada T., Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Hu Y.F., Zhong H., Nye A.C., Belmont A.S., Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 2001;155:911–921. doi: 10.1083/jcb.200108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Klatt A., Gilmour D.S., Henderson A.J. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 2007;282:16981–16988. doi: 10.1074/jbc.M610688200. [DOI] [PubMed] [Google Scholar]
- Zhong H., Zhu J., Zhang H., Ding L., Sun Y., Huang C., Ye Q. COBRA1 inhibits AP-1 transcriptional activity in transfected cells. Biochem. Biophys. Res. Commun. 2004;325:568–573. doi: 10.1016/j.bbrc.2004.10.079. [DOI] [PubMed] [Google Scholar]